Significance
We combined molecule counting and superresolution microscopy to observe the size, shape, and biomolecular composition of vesicles in the endocytic pathway. From a large number of steady-state snapshots, we follow the maturation of vesicles to endosomes. First, phosphatidylinositol 3-phosphate (PI3P) is generated on newly formed vesicles, principally after they shed their clathrin coat. Next, PI3P sites increase (∼100-fold) on small vesicles whose size is tightly regulated. Finally, the GTPase Vps21 (Rab5 homolog) is associated with small endosomal structures during the initial phase of PI3P production, whereas Ypt7 (Rab7 homolog) localizes to larger endosomal structures, which exhibit saturating levels of PI3P.This new technique opens up the possibility of accurately measuring molecular stoichiometries across several orders of magnitude while resolving subdiffraction-limit organelles.
Keywords: phosphoinositides, GTPases, endocytosis, PALM, single-molecule
Abstract
Cells tightly regulate trafficking of intracellular organelles, but a deeper understanding of this process is technically limited by our inability to track the molecular composition of individual organelles below the diffraction limit in size. Here we develop a technique for intracellularly calibrated superresolution microscopy that can measure the size of individual organelles as well as accurately count absolute numbers of molecules, by correcting for undercounting owing to immature fluorescent proteins and overcounting owing to fluorophore blinking. Using this technique, we characterized the size of individual vesicles in the yeast endocytic pathway and the number of accessible phosphatidylinositol 3-phosphate binding sites they contain. This analysis reveals a characteristic vesicle maturation trajectory of composition and size with both stochastic and regulated components. The trajectory displays some cell-to-cell variability, with smaller variation between organelles within the same cell. This approach also reveals mechanistic information on the order of events in this trajectory: Colocalization analysis with known markers of different vesicle maturation stages shows that phosphatidylinositol 3-phosphate production precedes fusion into larger endosomes. This single-organelle analysis can potentially be applied to a range of small organelles to shed light on their precise composition/structure relationships, the dynamics of their regulation, and the noise in these processes.
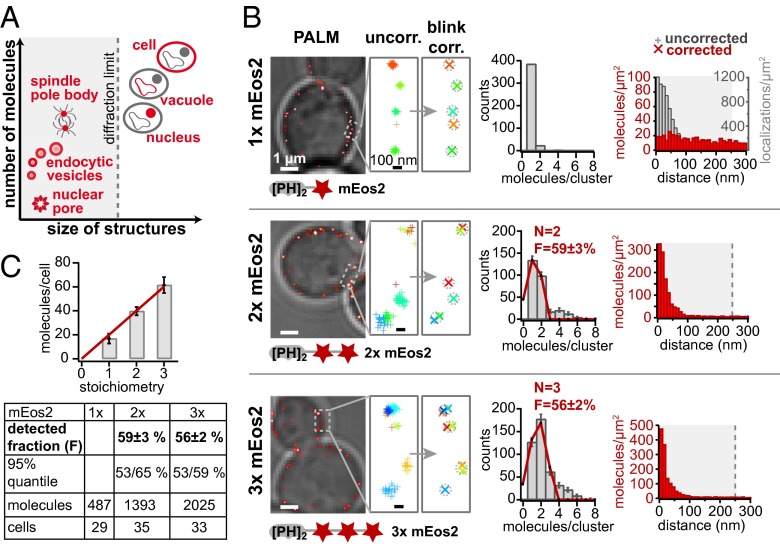
Single-cell analysis of protein abundance has revealed cell-to-cell variation in the form of phenotypic (extrinsic) heterogeneity and intrinsic variation (1). Dynamic processes such as the cell cycle, endocytosis, and meiosis are regulated but also influenced by stochastic events involving small numbers of molecules (e.g., DNA transcription) (2, 3). Similarly, the size and molecular composition of small subcellular organelles are dynamically regulated but still subject to stochastic noise. Studying the biomolecular composition and size of organelles will illuminate the regulation and noise in these dynamically stable systems. A major challenge for such exploration, however, is the development of a combined approach for both resolving small structures below the optical diffraction limit and simultaneously counting biomolecules over several orders of magnitude (Fig. 1A).
Fig. 1.
Intracellular calibration of superresolution microscopy allows counting of biomolecules in subdiffraction-limit structures. (A) Intracellular structure size and number of biomolecules span several orders of magnitude. (B) Resolution and counting accuracy are determined using constructs with different mEos2 stoichiometry. (Left) Superresolution images of calibration constructs (red) are superimposed on transmitted light images of yeast cells (gray). Single (1×, Top), double (2×, Middle) and triple (3×, Bottom) repeats of mEos2 were constitutively expressed as fusions with the membrane-localized PH domain of Plcδ. (Center) The magnified fields depict uncorrected (+) and corrected (X) single molecule positions. Each molecule and localization is color-coded by frame number. The number of molecules per cluster is fitted to a binomial distribution B(N,F) (red line). (Right) Pair-correlation functions of corrected images reflect the average distance between molecules, which is constant for the single mEos2 repeat and peaked for the 2× and 3× repeat owing to colocalization. The peak width of corrected 2× and 3× data are narrower than for uncorrected data (only shown for 1×) and reflects the increase in resolution by combining photons from all fluorescent bursts to the molecule's position. (C) Summary of calibration results. The ensemble values of corrected molecules per cell exhibit a linear relation with an expected two- and threefold higher number of mEos2 molecules of the 2× and 3× repeat compared with the single repeat. The table summarizes fraction of observable mEos2 molecules F from the single molecule counting data.
In the endocytic pathway, both vesicle morphology and biomolecular composition are dynamically regulated along a maturation path. Formation of vesicles, tethering, fusion, and maturation to endosomes are controlled by over 60 proteins in concert with conversion of phosphoinositides (PIs) (4, 5). Phosphatases and PI-kinases produce phosphatidylinositol 3-phosphate (PI3P) from plasma membrane phospholipids (6). PI3P is required for endocytosis (7) and membrane transport to early and late endosomes (8, 9). PI3P regulates the recruitment of proteins (10), such as Rab GTPases, which coordinate many aspects of vesicle identity (11, 12) and maturation to endosomes, including tethering and fusion (13–16). These master regulators also modulate PI3P production; thus, there is mutual regulation between PI3P and the Rabs (17).
Fluorescence microscopy has documented the localization and dynamic exchange of proteins and PIs at different stages in the endocytic pathway (18–20). In addition, structural studies using electron microscopy complemented fluorescence studies by revealing the shape and size of vesicles below the optical diffraction limit together with qualitative protein localization (21, 22). However, owing to limitations of high-resolution counting techniques, little is known about the relationship between the size and the exact biomolecular composition of endocytic and endosomal vesicles because these properties cannot be simultaneously measured by the same technique. Because size and biomolecular composition are major determinants of vesicle and endosome identity, such knowledge could reveal mechanistic insights into how protein and lipid conversion and vesicle fusion are regulated along the maturation path. A precise measurement of these parameters would allow us to study noise caused by stochastic fluctuations of involved components in the endosomal maturation pathway.
Here, we develop an approach to simultaneously measure vesicle size and composition using superresolution microscopy. Single-molecule superresolution microscopy methods, more commonly known as stochastic optical reconstruction microscopy (23) or photoactivated localization microscopy (24), are based on localizing individual photoactivatable fluorophores through fitting their point-spread functions. These methods offer potential information about the local number of molecules, especially when using photoactivatable fluorescent proteins (PAFPs) as the probe. However, they have associated over- and undercounting errors that must be corrected. For example, blinking of PAFPs such as mEos2 has been shown to cause overcounting artifacts (25). Although previous studies devoted effort to calibrating these effects (25–28), variation of photophysical properties with experimental conditions for compatible PAFPs complicates quantitative image reconstruction. In addition, undetected fluorescent proteins owing to problems such as misfolding lead to undercounting. Here, we develop an intracellular calibration approach for single-molecule superresolution microscopy by measuring the relevant photophysical parameters in cells under imaging conditions to correct for blinking. By counting the number of molecules in protein complexes of defined stoichiometry, we then determine the fraction of undetected PAFPs, allowing us to calibrate and count absolute numbers of molecules.
Applying this method to the yeast endocytic pathway allows us to simultaneously measure the size of vesicles and endosomes and the number of associated biomolecules, revealing a regulated maturation trajectory of lipid conversion and fusion. Established markers for different stages of vesicle-to-endosome maturation (such as GTPases) colocalize with distinct regimes on this maturation trajectory and reveal that the number of accessible PI3P binding sites on vesicles increases about 100-fold before they start to fuse. By analyzing vesicle-to-vesicle variability within and between cells, we identify noise sources in this maturation trajectory and confirm our results by colocalization. Our approach sheds light on the endocytic pathway and paves the way for fundamental studies on the relationship between the number of biomolecules and the size of small intracellular structures.
Results
Calibration of Single-Molecule Superresolution Microscopy Eliminates Over- and Undercounting.
The first challenge for accurately counting fluorescently labeled molecules is overcounting owing to blinking. As shown in previous studies, blinking of PAFPs can be corrected by combining fluorescent bursts from a single molecule based on a cutoff time determined from the measured characteristic dark-time distribution (25, 27). In addition, it has been previously reported that the detectable fraction of PAFPs can be substantially smaller than 100% (29–31). Both the blink characteristics and the detectable fraction are sensitive to the experimental conditions. To solve this problem, we measured both effects in cells under the exact same conditions as our actual experiments. Specifically, we created three yeast strains for calibration, each constitutively expressing low levels of either a single, double, or triple repeat of mEos2 (32). The mEos2 repeats were fused to the plasma membrane-localized Pleckstrin homology (PH) domain of Plcδ, resulting in a ring of fluorophores around the periphery of the cell (Fig. 1B, Left).
Before correction, blinking single mEos2 molecules falsely appear closely clustered in time and space (Fig. 1B, Top). By combining closely separated photoactivation events, we obtain the photon-weighted average position of each molecule (Fig. 1B, Top and Fig. S1). The counting histogram was created by counting corrected molecule positions closer than 50 nm and by subtracting background counts from cells lacking mEos2 (Fig. S2). The small fraction of observed double counts is caused by randomly overlapping molecules.
To evaluate our blink correction algorithm, we analyzed superresolution images with the pair-correlation function (33) (Fig. 1B, Right), which represents the distribution of distances between any pair of two localization points. Uncorrected data from 1×-mEos2 cells produces a pair-correlation function with a pronounced peak (Fig. 1B, Top Right) reflecting the clustering of localizations caused by blinking. Blink correction results in a flat pair-correlation function, verifying the random distribution of fluorophores and proper blink correction (Fig. 1B, Top Right).
In the blink-corrected data of 3×-mEos2 constructs, up to three fluorophores appear in close proximity (<50 nm separation). A portion of mEos2 molecules is undetected, resulting in observation of fewer than three molecules at same location. As a result, the counting histogram follows a binomial distribution B(N, F) (Fig. 1B, Bottom) and again exhibits a shoulder owing to randomly overlapping 3×-mEos2 molecules. When multiple fused mEos2 repeats are expressed, two or three mEos2 molecules colocalize, causing the pair-correlation functions of the 2×- and 3×-mEos2 constructs to display a peak at short distances. The width of the peak (full width at half maximum 20nm) represents the experimental localization uncertainty (Fig. 1B, Middle and Bottom Right) because the mEos2 repeats should be at the same position. This improved localization precision compared with uncorrected data (see peak width of uncorrected data, Fig. 1B, Top) results from combining all detected photons to determine each molecule’s position rather than distributing them over multiple blinks.
By fitting the distribution of the observed number of PAFPs to the known numbers in our strains (1×, 2×, and 3×-mEos2), we determined the fraction of undetected mEos2 molecules to be about 40%, allowing us to calibrate the observed number of PAFPs (Fig. 1C). Using this calibration, we can count the absolute number of PAFP-labeled molecules present in subcellular organelles and protein complexes of unknown stoichiometry with a resolution of 20 nm. As shown in SI Materials and Methods and Fig. S1, this approach holds for up to hundreds of molecules per organelle without undercounting artifacts owing to fluorescent overlap in time.
Measuring Maturing Vesicle Size and Counting Accessible PI3P Binding Sites with Superresolution Microscopy.
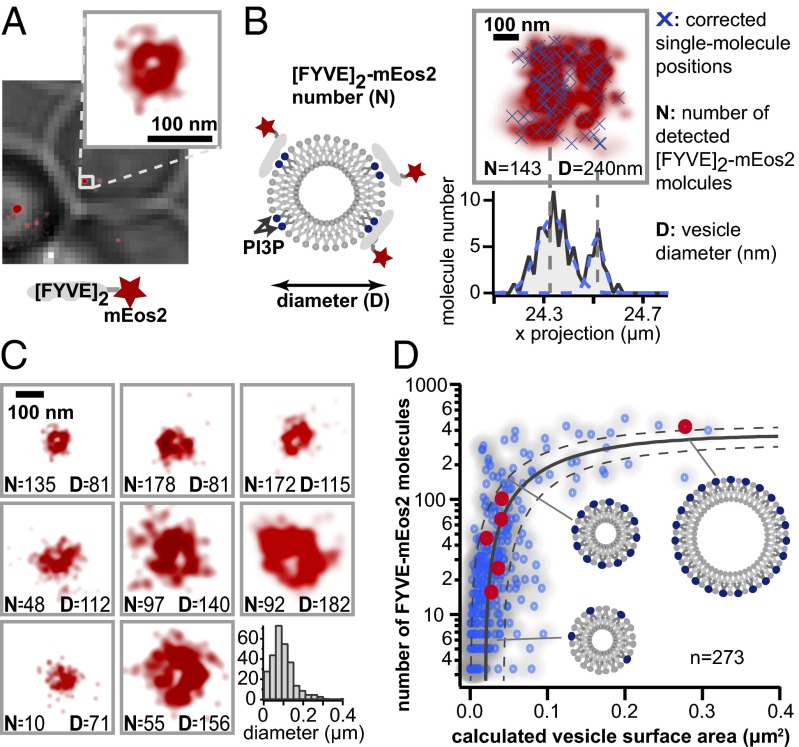
PIs define the identity of endocytic vesicles and endosomal compartments by providing specific binding sites for structural and regulatory proteins (9, 13, 17, 34, 35). To spatially resolve vesicles and simultaneously measure the number of accessible PI3P binding sites on their surfaces, we expressed mEos2 fused to a PI3P-binding domain [a double FYVE domain of EEA1 (19, 34)]. Although this reporter cannot detect PI3P molecules shielded by other proteins, it gives a stable measure (see also Fig. 3) of the exposed pool also seen by regulatory proteins containing FYVE fingers, such as Vac1p, Fab1p, and Vps27p. Constitutive FYVE–mEos2 expression is driven by the weak pCyc promoter to minimize possible interference with the endocytic pathway. In blink-corrected images, vesicles are detected with high contrast and resolved as circular structures (Fig. 2A). The high resolution not only enables us to resolve the size of vesicles but also to separate vesicles that are close together. By applying our calibrated superresolution microscopy approach, the absolute number N of molecules in each vesicle as well as its diameter D are determined (Fig. 2B). Our detection range in the vertical direction is limited by diffraction to about 500–700 nm. Because the vesicles in this study are typically smaller (mean diameter 80 nm), all molecules in a vesicle can be detected. We note, however, that the application of our technique to structures with size similar to or larger than the detection range would require imaging with different focal planes (z-stacks) to detect all molecules.
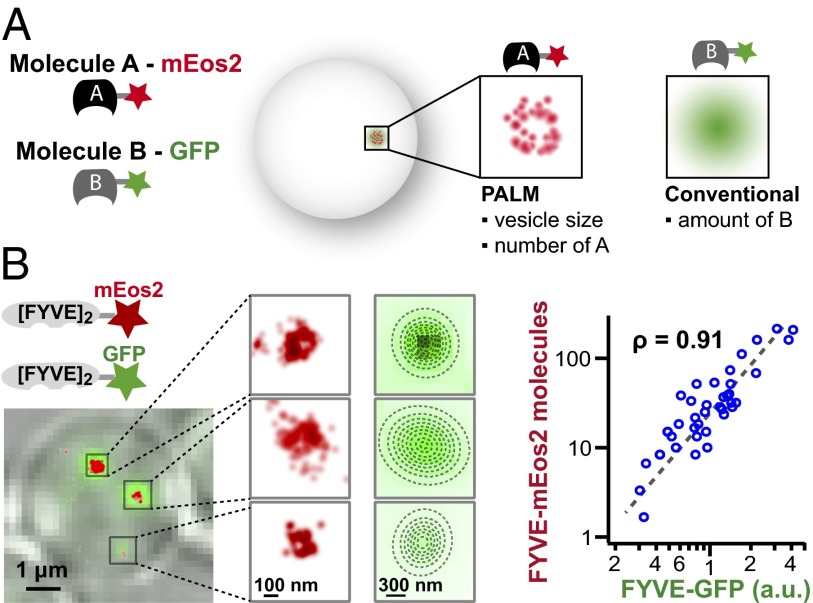
Fig. 3.
Counting of PI3P-binding sites is validated with conventional fluorescence. (A) Schematic for validation. One PI3P probe (A) is tagged with mEos2, allowing for high-resolution measurements of structure and quantification. The other (B) is fused to GFP, which can be quantified by calculating the total fluorescence intensity. (B) Quantified conventional fluorescence validates superresolution counting. FYVE–mEos2 and FYVE–GFP constructs were expressed in the same cells. A colocalization image shows transmitted light image (gray), mEos2 (red), and GFP (green). The number of mEos2 molecules determined in the red superresolution channel and the amount of GFP quantified by total fluorescence intensity on each vesicle is tightly correlated (Pearson’s coefficient, ρ= 0.91).
Fig. 2.
Vesicles in the endocytic pathway show characteristic relationship between vesicle size and PI3P content. (A) A tandem repeat of the FYVE domain of EEA1 fused to mEos2 serves as a specific probe to detect accessible PI3P binding sites on endocytic/endosomal vesicles. mEos2 (red) localizes to distinct, circular vesicles within a yeast cell (gray). (B) By applying our calibrated superresolution microscopy approach, the number of mEos2 molecules (N) bound to PI3P on each vesicle is determined (x’s). The molecular distribution exhibits a circular shape (two peaks in a 1D projection). The diameter of vesicles (D) is determined by measuring the spatial spread of molecules. (C) Vesicles display variability in PI3P content and size. Representative vesicles are shown displaying the expected circular shape. The histogram of vesicle diameters exhibits a fitted maximum at 82 nm. (D) PI3P content and vesicle surface area fall on a characteristic curve. Vesicles with fewer than 100 molecules display relatively tight size regulation, whereas structures above this threshold are significantly larger. Vesicles from each part of this characteristic distribution can be found within one individual cell (red). This path is also displayed as an exponential function (black line) fitted to box-smoothed data with 95% confidence envelope.
Number of Exposed PI3P Sites and Vesicle Surface Area Display a Maturation Path.
To study the distribution in PI3P content and size of maturing vesicles, we imaged over 50 cells and analyzed 273 vesicles. Both vesicle diameters and number of accessible PI3P binding sites display a broad range of values (Fig. 2C). The diameter of vesicles with very few PI3P binding sites may be slightly underestimated owing to an uneven distribution of detected molecules. However, the measured mean vesicle diameter (82 nm) lies within the range obtained from electron microscopy studies on clathrin-mediated endocytosis (22).
We next examined the degree to which the observed variability in size and PI3P content is random or subject to tight regulation. By plotting the number of accessible PI3P binding sites against the calculated surface area of 273 individual vesicles, we obtained a scatter plot populated in a clearly defined region (Fig. 2D). Vesicles with low to intermediate numbers of PI3P binding sites are relatively tightly distributed in size, but vesicles with several hundred binding sites show a significant increase in size and size variation. Both small and large vesicles are found within the same cell (Fig. 2D, red), suggesting that our aggregate distribution represents a superresolution snapshot of dynamic processes occurring in each cell. Because PI3P is produced on incoming vesicles, which fuse during maturation, or on early endosomes, we hypothesized that this population of vesicles represents a maturation trajectory of PI3P production and vesicle fusion.
To validate our counting of PI3P binding sites in individual vesicles, we combined superresolution with conventional fluorescence microscopy (Fig. 3A and Fig. S3). In this way, three independent parameters were obtained from each vesicle: their subdiffraction limit size, the number of one type of molecule (superresolution with mEos2), and the amount of a second molecular species (conventional fluorescence with GFP). We coexpressed our calibrated FYVE–mEos2 construct with FYVE–GFP. Each vesicle now appears in both superresolution (red) and conventional GFP (green) channels. The mEos2 counting and the GFP intensity show a high correlation (Pearson’s correlation coefficient, ρ = 0.91; Fig. 3B). This correlation validates our use of FYVE–mEos2 as a PI3P reporter.
Characteristic PI3P Sites and Vesicle Size Are Conserved Between Cells; Variation Is Due to Intrinsic and Extrinsic Noise.
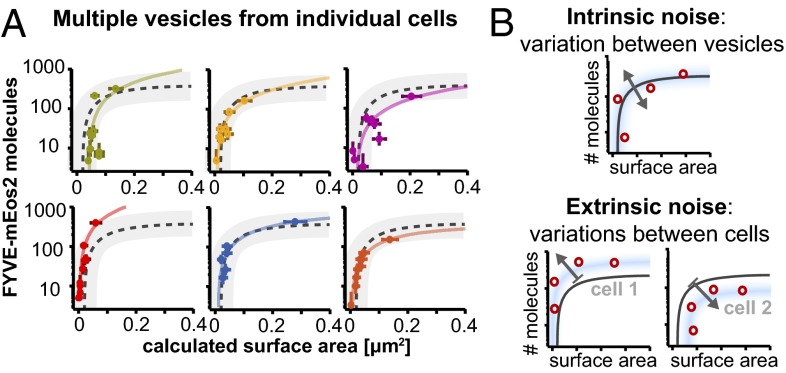
Stochastic gene expression leads to variability within a cell (intrinsic noise) as well as to phenotypic heterogeneity between cells (extrinsic noise) (1). Logically, these sources of noise might also manifest themselves on the level of small intracellular organelles. We examined whether our observed vesicle population (Fig. 2D) is caused by cell-to-cell variability or represents a trajectory of PI3P production and vesicle fusion present in each cell. Because we have verified that our fluorescent PI3P reporter is not significantly subject to intrinsic noise (Fig. 3B), our experimental setup provides the required accuracy. By analyzing multiple vesicles from individual cells (Fig. 4A, only cells with six or more vesicles are displayed), we observed that individual cells contain vesicles from all regions of the population. The majority of the spread in vesicle size and lipid content therefore seems to reflect a regulated maturation trajectory of PI3P production and vesicle fusion in the endocytic pathway. The scattering of vesicles from individual cells around the trajectory, however, displays two sources of noise: systematic deviations above or below the mean path (e.g., red, blue and orange in Fig. 4A) and intrinsic scattering within a cell around the path (e.g., green, yellow, and pink in Fig. 4A). Because both variations are larger than the experimental error, we conclude that intrinsic noise owing to variation of accessible PI3P binding sites between individual vesicles within a cell and extrinsic noise owing to variation between cells is present in the endocytic pathway (Fig. 4B).
Fig. 4.
Variation in the relationship between PI3P-binding site number and vesicle size is due to both intrinsic and extrinsic noise in the endocytic pathway. (A) Measurements made on several vesicles within one individual cell display variation around the characteristic curve. Vesicles from all regions of the PI3P number-size distribution can be found within the same cell. Vesicles within the same cell may be located above (red) or below (pink) the characteristic curve (dotted line, 95% confidence interval as gray shading). (B) Schematics showing two components of noise in PI3P content and size. (Upper) Intrinsic noise is due to a variation between vesicles within a cell and (Lower) extrinsic noise is due to systematic variations between cells.
Landmark Proteins Associate with Distinct States of the Maturation Trajectory.
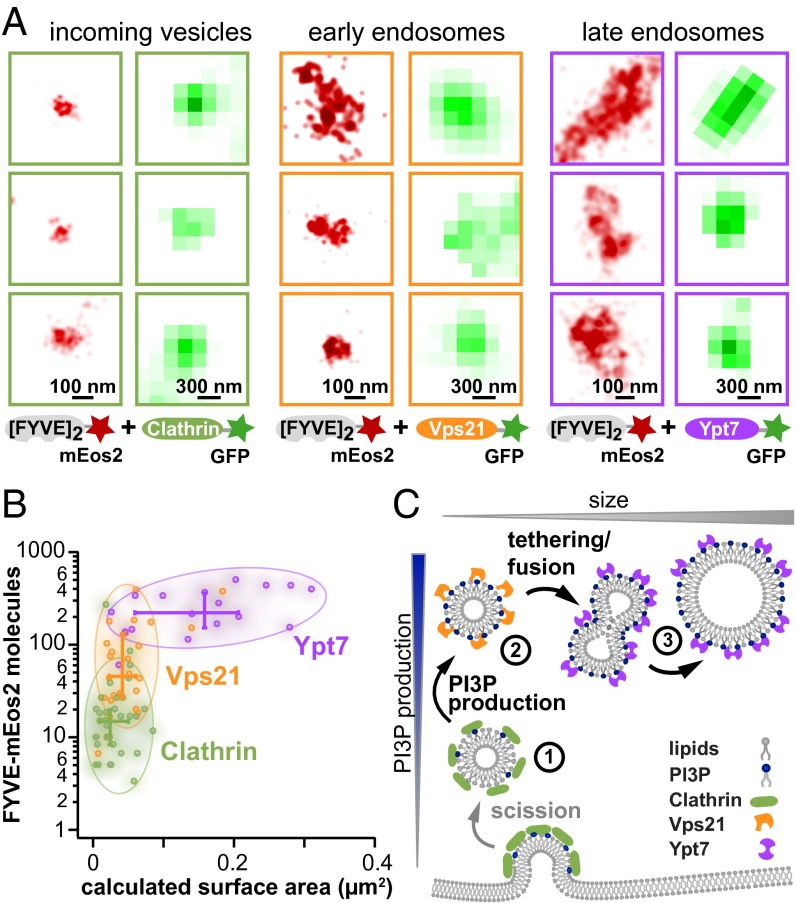
To correlate our observed PI3P-size trajectory with the timeline of vesicle maturation, we performed two-color colocalization experiments between our PI3P reporter (in the superresolution channel) and landmark endocytic pathway proteins labeled with GFP (in the conventional fluorescence channel) (Fig. 5A). We tagged three endocytic pathway proteins with GFP: (i) clathrin, present from the initial stages of membrane invagination and endocytic vesicle formation (18); (ii) the small GTPase Vps21 (yeast homolog of Rab5), an interaction partner of the PI3-kinase that associates with incoming vesicles and early endosomes; and (iii) the GTPaseYpt7 (yeast homolog of Rab7), found in late endosomes (for reviews on Rabs see refs. 16 and 36). Both GTPases are involved in tethering and fusion (15, 37) and participate in different stages of endosomal maturation (11).
Fig. 5.
Benchmarking with endocytic/endosomal landmark proteins reveals distinct phases of PI3P production and vesicle fusion. (A) Colocalization of vesicles with landmark proteins of the endocytic pathway. Representative superresolution images (red, left column of each pair) and GFP images (green, right column of each pair) for each class of protein are shown. (B) Colocalization with endocytic pathway proteins reveals distinct subclasses of vesicles. FYVE–mEos2 colocalizes with GFP-tagged clathrin (green) and GTPases Vps21p (orange) and Ypt7p (purple). For each dataset, the median, upper, and lower quartiles are displayed. (C) Model of PI3P production on vesicles in endocytic pathway followed by fusion. Phases of endocytic/endosomal protein association, enzymatic production of PI3P, and fusion to early and late endosomes (1–3) are described in B.
In each of the three sets of experiments, we counted PI3P binding sites and measured the size of individual vesicles as well as the amount of each GFP-tagged landmark protein. By plotting those vesicles with significant GFP signal (Fig. 5B and Fig. S4), we observed the same characteristic distribution of size and PI3P content. These three landmark proteins denote three distinct regimes in our observed distribution. Clathrin associates with very low PI3P-content vesicles, whose size and morphology indicate that these structures are early endocytic vesicles. In contrast, Vps21 colocalizes with vesicles containing hundreds of PI3P molecules, but both clathrin- and Vps21-associated regimes display tight regulation of vesicle size. Finally, Ypt7 associates with vesicles of significantly larger size and containing the highest numbers of PI3P binding sites. This trend is also clear in the medians of each class and hand-drawn ellipses, which include more that 80% of data points of each class (Fig. 5B, crosses and shaded ellipses, respectively). In general, more small individual vesicles are observed than large, Ypt7-associated endosomes. Given that many individual vesicles fuse to form a larger endosome, this observation is consistent with the conservation of lipid membrane along the maturation path.
Discussion
Quantifying the abundance and spatial distribution of biomolecules in cells provides fundamental insights into biological processes such as compartmentalization, polarization, and signaling. Here we take this approach one step further to study the size and biomolecular composition of subcellular compartments using calibrated superresolution microscopy. This technique opens up new and exciting opportunities to quantitatively study processes in intracellular organelles and complexes. The identity of organelles is primarily defined by their morphology and biomolecular composition, and our method provides fundamental characterization of these properties. From a large number of such measurements, insights can be obtained into how this identity is dynamically regulated along a path.
Challenges for Counting Single Molecules with Superresolution Microscopy.
Counting absolute molecule numbers remains challenging because of overcounting owing to blinking and undercounting owing to a limited PAFP maturation efficiency. Several recent studies made progress in quantifying single-molecule superresolution microscopy data by correcting for dye blinking (25–28). Other approaches calculated averaged spatial distributions of an ensemble of signaling complexes (33, 38) or determined high mEos2 molecule numbers using an average number of blinking events (39). One persistent problem is the strong environmental dependence of the photophysical parameters required for blink correction and the fraction of unobserved fluorophores. To count absolute numbers of molecules over several orders of magnitude in individual protein complexes, these two effects must be quantified, as we have done here.
Experimental conditions such as environmental oxygen and redox potential or the intensity of excitation and photo-switching affect the fluorophore’s photophysical parameters (40). These conditions may vary between cellular compartments and cell types. The fraction of undetected PAFPs also depends on environmental factors such as temperature, pH, and fixation conditions. Systematic measurement of the sum of these effects by counting fluorophores with known stoichiometry is critical for obtaining absolute molecule numbers and associated uncertainties. For future optimizations of PAFPs compatible with single-molecule superresolution microscopy, it is important to enhance not only their brightness and maturation rate but also photo-switching capability, low transition rates to dark states, and short dark times.
A Superresolution Snapshot of the Vesicle Maturation Trajectory: PI3P Production and Vesicle Fusion Occur at Different Stages.
The application of calibrated superresolution microscopy to the yeast endocytic pathway allowed us to count the number of accessible PI3P binding sites, quantify their colocalization with endocytic or endosomal proteins, and correlate these numbers with vesicle size. The observed maturation trajectory of vesicles suggests an initial phase of PI3P production on vesicles whose size is tightly regulated, followed by a later phase in which fusion of these vesicles occurs. Previously it has been shown that Vps21 mediates vesicle tethering (37), which precedes fusion. We are able to resolve tethered vesicles (e.g., in Fig. 5A, middle row and column); however, consistent with the notion that fusion is fast, such events are very rarely observed. The PI3P content is highest in large, fused structures, which we interpret to be early and late endosomes based on their morphology and Vps21/Ypt7 association. Owing to the increase in surface area in these larger structures, our findings indicate that most PI3P production occurs before vesicles start to fuse. As seen in Fig. 5A, most of the large Ypt7-associated endosomes exhibit complex, irregular morphology, consistent with EM studies in yeast that resolved the structure of endosomes and multivesicular bodies (41). Owing to this complex morphology and 2D projection of the images, the calculated surface area of those large structures may be slightly underestimated. However, this underestimation does not affect our conclusion that most of the PI3P is produced on individual vesicles before they fuse.
Because the endocytic pathway is conserved between yeast and mammalian cells, our results can be viewed in a broader context. It is well established that PI3P production is required for vesicle fusion and maturation (7). In mammalian cells, the key proteins in this process are EEA1 (homolog of yeast Fab1), which binds to PI3P (34, 42) and is thought to recruit and to effect Rab5 (homolog of yeast Vps21) (10). Rab5 in turn tethers vesicles (16, 37) and interacts with the fusion complex CORVET (43) and through EEA1 with SNARE (44). However, owing to the lack of a direct measurement, it is still debated whether PI3P is produced on incoming vesicles, fused vesicles, or endosomes. Further experiments will help to dissect maturation events following Rab5 mediated fusion in more detail and study the role of other proteins involved in these processes.
Dissecting Regulation and Noise with Multiple Parameters.
Quantifying multiple parameters of a dynamic process with many static snapshots can illuminate both regulation and variability within the pathway. For intracellular organelles, the most important parameters are morphology/size and molecular composition. Our method allows us to track these parameters and to gain mechanistic insight into the overall pathway by dissecting different regimes in the measured parameter space. In addition, we also obtain insights into intrinsic and extrinsic sources of noise by comparing parameter distributions within and between cells.
In the case of our observed vesicle maturation trajectory, we found that vesicles from individual cells cover the whole range of the distribution, indicating a regulated process in each cell. The scattering of vesicles from individual cells around the trajectory is significantly above our experimental error and displays two sources of noise: intrinsic noise owing to variation of accessible PI3P binding sites between individual vesicles within a cell and extrinsic noise owing to variation between cells. Certain endocytic proteins involved in lipid modification and protein sorting are expressed at very low levels [e.g., Fab1 (149 copies per cell) or Vps27 (172 copies per cell) (45)], creating potentially large stochastic variations between vesicles and endosomes (Fig. S5). In contrast, the higher expression level of our PI3P reporter results in a lower coefficient of variation (see also high correlation Fig. 3B) and thus less intrinsic noise than some other endocytic proteins. By synthetically introducing noise in different nodes, future experiments on modified endocytic pathways could reveal which parts are sensitive to, suppress, or amplify this variability.
Quantitative superresolution microscopy also presents the opportunity to study clustering or polarity in small intracellular compartments (46). Endosomal proteins are likely to have a complex spatial organization on vesicles that may be important for various processes in membrane trafficking and sorting. We believe that our calibrated superresolution microscopy approach together with its ability for multiparameter quantification will be useful for future mechanistic insight of dynamic processes below the diffraction limit. In addition to the endocytic pathway, a multitude of other small intracellular organelles such as processing bodies, peroxisomes, or centrosomes (47) present an exciting opportunity for exploration with this approach.
Materials and Methods
Sample Preparation.
Yeast strains (W303) (see Tables S1–S3) were grown at 30 °C overnight in synthetic media with glucose (SD), diluted 1:50 in SD, and grown for 3–4 h to log phase in the dark. After immobilization on a Con A-coated coverslip, cells were fixed for 30 min with 4% (vol/vol) formaldehyde (SI Materials and Methods gives details).
Superresolution Microscopy.
Superresolution measurements were performed on a custom-built microscope based on a Nikon Ti-E inverted microscope with the Perfect Focus System. Photoactivation (405 nm laser, 1–160 µW corresponding to a power density of roughly 0.06–10 W/cm2) and simultaneous bright-field imaging in one frame was followed by nine frames with excitation (561 nm, 17 mW corresponding to a power density of roughly 1 kW/cm2); this cycle was repeated until all mEos2 molecules were imaged and bleached (SI Materials and Methods and Fig. S3). Fluorescent intensity profiles of single mEos2 molecules were fitted with Gaussians and drift corrected (SI Materials and Methods and Fig. S1).
Calibration for Molecule Counting.
To accurately calibrate our measurements in cells under imaging conditions, we measured the dark-time histogram of well-separated [PH]2-mEos2 molecules at the plasma membrane. Using this histogram, we determined that 99% of the dark states of a single fluorophore last less than 2.66 s (SI Materials and Methods and Fig. S1). The pair-correlation function reveals that these bursts are spatially separated by less than 150 nm. Therefore, all fluorescent bursts separated by less than 150 nm and 2.66 s were combined (SI Materials and Methods and Fig. S1). For the labeling densities in this study, combining fluorescent bursts in this way does not lead to undercounting artifacts owing to overlap in time (Fig. S1B). To determine the fraction of undetectable mEos2 molecules, counting histograms of the double and triple mEos2 repeats were noise-subtracted and fitted by a binomial distribution (SI Materials and Methods and Fig. S2).
Two-Color Colocalization with Superresolution Structures.
GFP was images after mEos2 to avoid potential crosstalk (SI Materials and Methods and Figs. S3 and S4). Images were background-subtracted and corrected for uneven illumination, and the total GFP intensity per vesicle was calculated (SI Materials and Methods). Only vesicles with more than two FYVE–mEos2 molecules were analyzed (SI Materials and Methods). Vesicles with a significant GFP signal (above the 90% threshold, Fig. S4) are plotted in Fig. 5B.
Supplementary Material
Acknowledgments
We thank Eva Schmid, Mark von Zastrow, Mark Marsh, Voytek Okreglak, Vito Mennella, Marcus Taylor, and the W.A.L. and B.H. laboratories for helpful discussions. This work was supported by the Howard Hughes Medical Institute (W.A.L.) and National Institutes of Health (NIH) Grants GM55040, GM62583, and P50GM081879 (to W.A.L.) and 1DP2OD008479 (to B.H.) and NIH National Research Service Award Fellowship 5F32GM093475 (to J.M.W.) and German Research Foundation Fellowships DFG PU499/1-1 (to E.M.P.) and KA3453/1-1 (to R.K.). B.H receives the Searle Scholarship and the Packard Fellowship for Science and Engineering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309676110/-/DCSupplemental.
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 3.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440(7082):358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22(1):1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol. 2012;14(1):2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771(3):353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, et al. Evidence for phosphatidylinositol 3-kinase as a regulator of endocytosis via activation of Rab5. Proc Natl Acad Sci USA. 1995;92(22):10207–10211. doi: 10.1073/pnas.92.22.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15(5):259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84(3):699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 10.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394(6692):494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 11.Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141(3):497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122(5):735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 13.Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25(5):229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- 14.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64(5):915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 16.Lachmann J, Ungermann C, Engelbrecht-Vandré S. Rab GTPases and tethering in the yeast endocytic pathway. Small GTPases. 2011;2(3):182–186. doi: 10.4161/sgtp.2.3.16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 18.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123(2):305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 19.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19(17):4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonescu CN, Aguet F, Danuser G, Schmid SL. Phosphatidylinositol-(4,5)-bisphosphate regulates clathrin-coated pit initiation, stabilization, and size. Mol Biol Cell. 2011;22(14):2588–2600. doi: 10.1091/mbc.E11-04-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150(3):508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 22.Idrissi FZ, Blasco A, Espinal A, Geli MI. Ultrastructural dynamics of proteins involved in endocytic budding. Proc Natl Acad Sci USA. 2012;109(39):E2587–E2594. doi: 10.1073/pnas.1202789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 25.Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: Unraveling the effects of photoblinking. PLoS ONE. 2011;6(7):e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunzenhäuser J, Olivier N, Pengo T, Manley S. Quantitative super-resolution imaging reveals protein stoichiometry and nanoscale morphology of assembling HIV-Gag virions. Nano Lett. 2012;12(9):4705–4710. doi: 10.1021/nl3021076. [DOI] [PubMed] [Google Scholar]
- 27.Lee SH, Shin JY, Lee A, Bustamante C. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM) Proc Natl Acad Sci USA. 2012;109(43):17436–17441. doi: 10.1073/pnas.1215175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ori A, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Annibale P, Scarselli M, Greco M, Radenovic A (2012) Identification of the factors affecting co-localization precision for quantitative multicolor localization microscopy. Optical Nanoscopy 1(9), 10.1186/2192-2853-1-9.
- 30.Durisic N, et al. Stoichiometry of the human glycine receptor revealed by direct subunit counting. J Neurosci. 2012;32(37):12915–12920. doi: 10.1523/JNEUROSCI.2050-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4(4):319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6(2):131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sengupta P, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8(11):969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2(1):157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 35.De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol. 2004;6(6):487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 36.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 37.Lo SY, et al. Intrinsic tethering activity of endosomal Rab proteins. Nat Struct Mol Biol. 2012;19(1):40–47. doi: 10.1038/nsmb.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veatch SL, et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE. 2012;7(2):e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lando D, et al. Quantitative single-molecule microscopy reveals that CENP-A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol. 2012;2(7):120078. doi: 10.1098/rsob.120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Endesfelder U, et al. Chemically induced photoswitching of fluorescent probes—A general concept for super-resolution microscopy. Molecules. 2011;16(4):3106–3118. doi: 10.3390/molecules16043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20(1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu FT, et al. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270(22):13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 43.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12(5):739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 44.McBride HM, et al. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98(3):377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 45.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 46.Sönnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149(4):901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mennella V, et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol. 2012;14(11):1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.