Abstract
We have investigated whether the recently discovered transcription factor, zinc finger BED domain-containing protein 6 (ZBED6), is expressed in insulin-producing cells and, if so, to what extent it affects beta cell function. ZBED6 was translated from a ZC3H11A transcript in which the ZBED6-containing intron was retained. ZBED6 was present in mouse βTC-6 cells and human islets as a double nuclear band at 115/120 kDa and as a single cytoplasmic band at 95–100 kDa, which lacked N-terminal nuclear localization signals. We propose that ZBED6 supports proliferation and survival of beta cells, possibly at the expense of specialized beta cell function—i.e., insulin production—because (i) the nuclear ZBED6 were the predominant forms in rapidly proliferating βTC-6 cells, but not in human islet cells; (ii) down-regulation of ZBED6 in βTC-6 cells resulted in altered morphology, decreased proliferation, a partial S/G2 cell-cycle arrest, increased expression of beta cell-specific genes, and higher rates of apoptosis; (iii) silencing of ZBED6 in the human PANC-1 duct cell line reduced proliferation rates; and (iv) ZBED6 binding was preferentially to genes that control transcription, macromolecule biosynthesis, and apoptosis. Furthermore, it is possible that beta cells, by switching from full length to a truncated form of ZBED6, can decide the subcellular localization of ZBED6, thereby achieving differential ZBED6-mediated transcriptional regulation.
Zinc finger BED domain-containing protein 6 (ZBED6) is a recently discovered transcription factor restricted to and highly conserved among placental mammals (1). The intron-less ZBED6 has evolved from a domesticated DNA transposon and is located in one of the first introns of another gene, zinc finger CCCH-type containing 11A (ZC3H11A), with which it has no sequence homology. ZBED6 belongs to the CCCH zinc-finger BEAT and DREF (BED) domain-containing family and contains two DNA-binding BED domains and one hobo-Ac-Tam3 (hATC) dimerization domain. ZBED6 has a broad tissue distribution in mammals (1). In muscle tissue of pigs, it has been observed that ZBED6 negatively regulates insulin-like growth factor-2 (IGF2) transcription (1). A G-to-A transition in intron 3 of IGF2 abrogates the binding site for ZBED6 and leads to a threefold up-regulation of IGF2 mRNA expression in skeletal muscle. Mutant pigs lacking the ZBED6 binding site display increased muscle mass, bigger hearts, and less fat deposition (2). Silencing of ZBED6 in mouse C2C12 myoblast cells was associated with elevated IGF2 expression, increased cell proliferation, and a faster wound-healing process. Chromatin immunoprecipitation (ChIP) sequencing using C2C12 cells identified >2,000 ZBED6 binding sites in the genome. Genes associated with ZBED6 binding sites show a highly significant enrichment for certain Gene Ontology (GO) classifications, including development and transcriptional regulation (1).
It is possible that ZBED6 controls not only IGF2 expression in muscle, but also the function of insulin-producing beta cells. For example, genes coding for transcription factors crucial to the pancreatic beta cell—such as Neurog3, Nkx6.1, NeuroD2, and v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (MafA)—were found to be putative binding targets for ZBED6 in myoblast cells (1). ZBED6-mediated control of IGF2 may be relevant to beta cells because it has been observed that aberrant IGF2 production in embryonic pancreas preceded the subsequent beta cell mass anomaly that develops in diabetic Goto–Kakizaki rats (3). Furthermore, the human insulin gene is clustered closely together with IGF2 (4), pointing to the possibility that not only IGF2, but also insulin might be controlled by a common ZBED6-dependent regulatory mechanism. Thus, it is conceivable that the putative expression of ZBED6 in beta cells influences or controls important events in beta cell development and function and, as a consequence, might be pertinent to the pathogenesis of various types of diabetes mellitus. The aim of the present investigation was therefore to study ZBED6 expression in insulin-producing cells and to determine whether ZBED6 knockdown affected basal beta cell functions such as proliferation, cell death, and insulin production.
Results
Differential Expression Pattern of ZBED6 in βTC-6 Cells and Human Islets and Stable Silencing of ZBED6 in βTC-6 Cells.
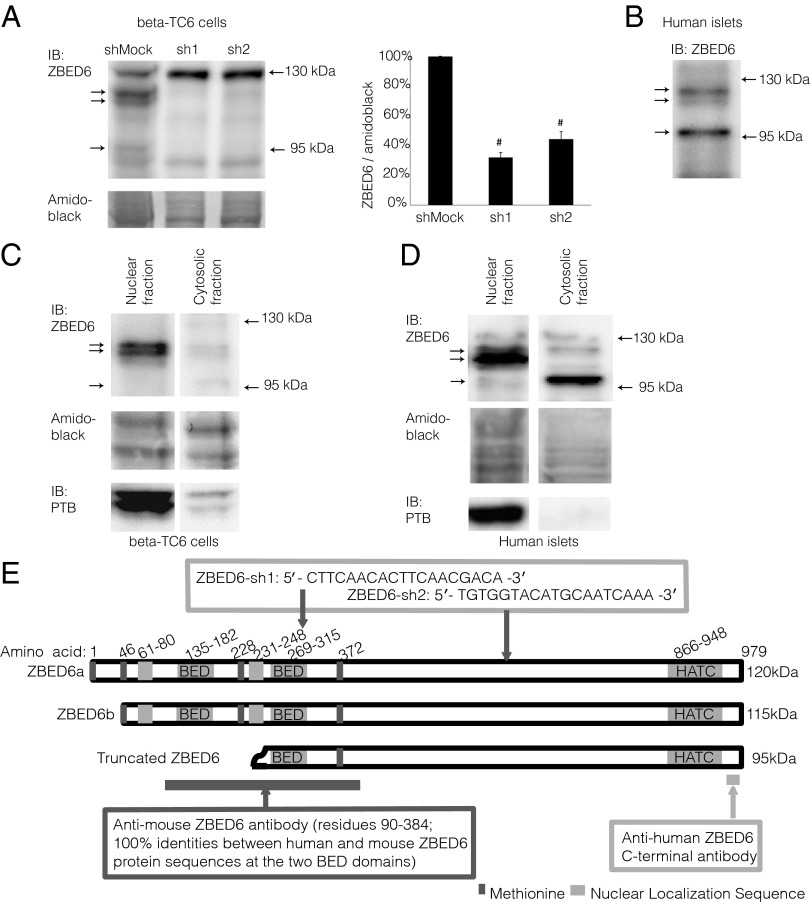
Immunoblot analysis of murine βTC-6 cell lysates using an antibody directed against the N-terminal ZBED6 BED domains revealed several bands (Fig. 1A). To identify immunoreactivity specific for ZBED6, we down-regulated ZBED6 protein levels by transduction with lentiviral vectors expressing shRNA that specifically targets ZBED6 mRNA. Two shRNA sequences targeted to different sites of ZBED6 mRNA were used, and the transduced βTC-6 cells were named ZBED6–sh1 and –sh2 (Fig. 1E). A lentiviral vector carrying a scrambled shRNA sequence was used to create transduced control cells (shMock). Down-regulation of ZBED6 was confirmed by real-time RT-PCR showing reductions of ZBED6 mRNA levels by 75% (sh1) and 59% (sh2) (Table 1). When comparing shMock-treated cells with sh1 and sh2, we observed ZBED6-specific immunoreactivity as a double band with the approximate molecular masses of 115/120 kDa and a single, weaker 95-kDa band (Fig. 1A). In sh1 and sh2 cells, these bands were clearly weakened, resulting in a 60–70% decrease in ZBED6 intensity when expressed per total amidoblack staining (Fig. 1A). Other bands were not affected by the shRNA, indicating some nonspecific reactivity of the antibody.
Fig. 1.
ZBED6 expression, knockdown, and subcellular localization in βTC-6 cells and human islets. (A) Expression of ZBED6 protein in βTC-6 cells after stable transduction with ZBED6-targeting shRNA. ZBED6 immunoreactivity was obtained by using the anti-mouse ZBED6 antibody. Arrows indicate the positions of the three ZBED6 immunoreactive bands. The positions of the 130- and 95-kDa molecular-mass markers are given. Results are means ± SEM for three observations. #P < 0.01. (B) Expression of ZBED6 proteins in human islets detected by using the anti-human ZBED6 antibody. (C) βTC-6 cytosolic and nuclear fractions were analyzed by immunoblotting using the anti-mouse ZBED6 antibody. The subcellular fractionation procedure was verified by immunoblot analysis of polypyrimidine tract binding protein, a nuclear RNA-binding protein. (D) Human islet nuclear and cytosolic fractions were analyzed by immunoblotting using the anti-human ZBED6 antibody. Results are representative for three observations. (E) Schematic presentation of three human ZBED6 forms with molecular mass of 120, 115, and 95 kDa. Putative translation start sites (methionine at residues 1, 46, 228, and 372 in the ZBED6a isoform) are indicated together with the localization of the BED1 (residues 135–182), BED2 (residues 269–315), and HATC (residues 866–948) domains, and putative nuclear localization signals (NLS) at residues 61–80 and 231–248; the two NLS sequences were identified in mouse C2C12 cells (1), but the second NLS in human ZBED6 (KKHDKSASDALRAERGRFL) differs at two positions from the mouse sequence (KKRDKSASDALRAKRGRFL), and it is therefore uncertain whether the sequence is active as an NLS in human ZBED6. Epitope regions for mouse and human ZBED6 antibodies as well as shRNA target sites are depicted.
Table 1.
Real-time RT-PCR results from βTC-6 cell lines
| Cell lines |
|||
| Gene | shMock, % | ZBED6–sh1, % | ZBED6–sh2, % |
| Zbed6 | 100 | 25 ± 3* | 41 ± 5* |
| Igf2 | 100 | 165 ± 8* | 115 ± 23 |
| Pdx1 | 100 | 265 ± 43* | 186 ± 22* |
| Glucokinase | 100 | 263 ± 69* | 121 ± 28 |
| MafA | 100 | 388 ± 99* | 224 ± 56 |
| Insulin | 100 | 326 ± 45* | 155 ± 12* |
Results were normalized to GAPDH levels and expressed in percent of shMock. Results are means ± SEM three to five independent observations.
P < 0.05 vs. shMock using Student’s t test.
To visualize ZBED6 expression in human islets, we used an anti-human ZBED6 C-terminal antibody. Analysis of islet lysates revealed three bands with anticipated molecular masses (Fig. 1B). The upper two corresponded well in size with the 115/120-kDa bands, and the third was slightly larger (100 kDa) than the 95-kDa ZBED6 band observed in βTC-6 cells. Interestingly, the 100-kDa band was more prominent in human islet cells (Fig. 1B), whereas the 115/120-kDa bands were the most prominent in the βTC-6 cell line (Fig. 1A). We attempted to down-regulate ZBED6 in human islet cells using the lentiviral ZBED6–sh1 vector, but we could not achieve a significant silencing of ZBED6 mRNA, possibly due to a high cell death rate after the lentiviral transduction.
ZBED6 Subcellular Localization.
βTC-6 cells were separated into nuclear and cytoplasmic fractions. Immunoblot analysis revealed that the large, 115/120-kDa ZBED6 forms were mainly localized to the nuclear fraction, whereas the small, 95-kDa form was instead cytosolic (Fig. 1C). Also, in human islet cells the 115/120-kDa forms were predominantly located to the nuclei, and the smaller, 100-kDa form was mainly cytoplasmic (Fig. 1D).
Immunostaining of ZBED6 in βTC-6 Cells, Mouse Pancreas, and Human Islets.
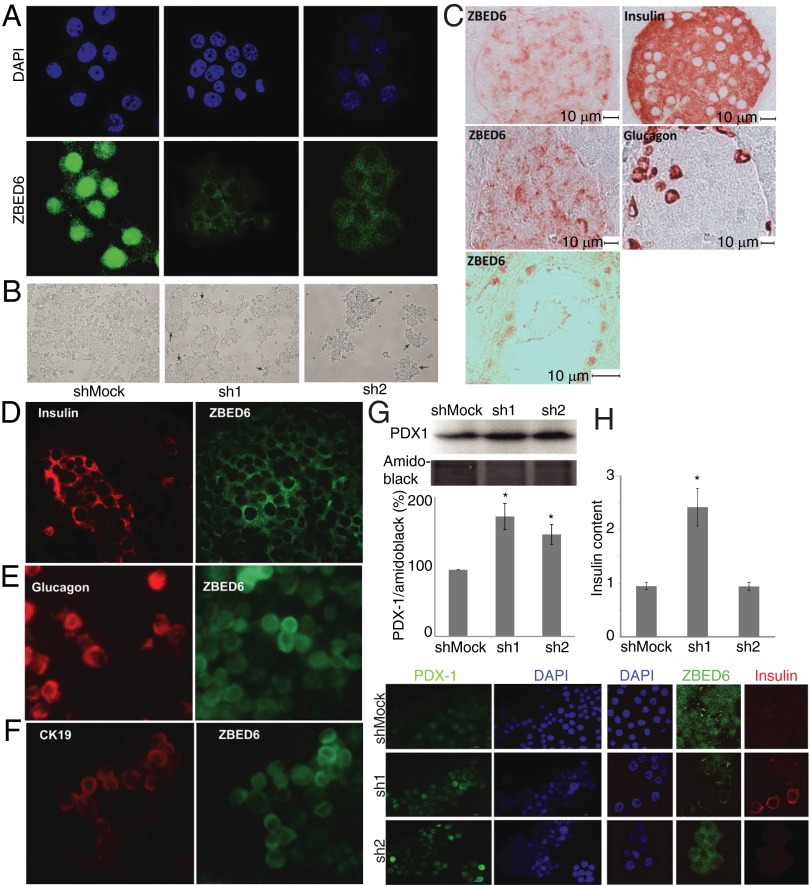
Immunostaining with the anti-mouse ZBED6 antibody revealed a strong nuclear and weak cytoplasmic staining in βTC-6 cells (Fig. 2A). In sh1 and sh2 βTC-6 cells, ZBED6 nuclear immunostaining was dramatically decreased (Fig. 2A). This finding corroborates our immunoblotting results showing that βTC-6 cells express high levels of nuclear ZBED6 and that knockdown affects mainly this form of ZBED6.
Fig. 2.
ZBED6 expression in islet cells and changes in morphology and gene expression after ZBED6 knockdown. (A) Immunofluorescence staining of βTC-6 cells with anti-mouse ZBED6 antibody (green) and DAPI (blue). ShMock, sh1, and sh2 cells were analyzed in parallel. Results are representative for three independent experiments. (B) Effects of ZBED6 knockdown on βTC-6 cell morphology. Cells were photographed in a phase contrast microscope after 3 d of in vitro culture on a standard NUNC plastic support. Pictures show low-magnification overview of cell cultures, and arrows show position of cells growing in 3D structures that resemble islets. (C) Immunohistochemical staining of 6.5-wk-old C57BL/6 mouse pancreas. Consecutive (5 μm) sections were stained for ZBED6 and insulin and for ZBED6 and glucagon (brown). The sections were briefly counterstained with hematoxylin (blue). Scale bars are given. Bottom shows a pancreatic duct stained for ZBED6. (D–F) Pairwise immunofluorescence costaining of human islet cells for ZBED6 and insulin (D), glucagon (E), and cytokeratin 19 (CK19) (F). (G and H) PDX1 and insulin expression are increased in ZBED6 silenced βTC-6 cells. (G) PDX1 levels based on five independent immunoblottings; results are means ± SEM. (Bottom) Immunofluorescence staining with PDX1 antibody and DAPI is shown. (H) Insulin contents (picograms of insulin per nanograms of DNA) based on three independent experiments; results are means ± SEM. *P < 0.05 using Student's t test. (Lower) Immunofluorescence staining with ZBED6 antibody, insulin antibody and DAPI.
In mouse pancreas ZBED6, immunohistochemical staining was stronger in islets than in surrounding exocrine tissue (Fig. 2C). The signal was mainly cytoplasmic and was present in both insulin- and glucagon-positive cells, as assessed by staining in consecutive sections. In pancreatic duct cells, however, ZBED6 staining was mainly nuclear (Fig. 2C).
We also stained isolated human islets, partially fragmented by trypsin digestion. Again, ZBED6 immunoreactivity was mainly cytoplasmic in insulin-positive islet cells (Fig. 2D). In addition, glucagon-positive cells displayed a rather weak cytoplasmic ZBED6 signal (Fig. 2E). Some islet-like cell clusters also contained duct cells, as indicated by cytokeratin 19 staining (Fig. 2F). These cells displayed stronger ZBED6 immunoreactivity than the endocrine cells, and in some cases clear nuclear ZBED6 staining was observed.
RNA Sequencing of ZBED6 and ZC3H11A mRNA in Human Islets.
Because insulin-producing cells expressed ZBED6 proteins with different sizes (Fig. 1), we next analyzed ZBED6 gene transcription and alternative splicing by RNA sequencing of six human islet samples from two separate donors. Because the ZBED6 coding sequence is located in intron 2 of ZC3H11A, we analyzed the obtained sequencing reads originating from the entire ZC3H11A gene. Essentially no reads were present upstream of the first exon of ZC3H11A (Fig. S1), whereas numerous reads were uniformly distributed from the start of exon 1 to the end of exon 4 of ZC3H11A, consistent with frequent retention of introns 1–3. Few reads covering the first splice site (intron 1) were found (Fig. S1), indicating incomplete splicing of intron 1. No reads indicated alternative splicing in intron 2 or 3 (Fig. S1) that could explain the presence of the three isoforms of ZBED6 detected in this study. This result suggests that intron 2, containing ZBED6, is retained in most ZC3H11A transcripts in human islets and that there is only one major transcription start site (TSS) for the genes in these cells. Remaining introns (4–19) of the ZC3H11A transcript were efficiently spliced because many reads covering the subsequent splice sites were detected in all human islet samples.
Mass Spectrometry Analysis of ZBED6 in βTC-6 Cells.
To further characterize the observed ZBED6 proteins with the molecular masses 120, 115, and 95 kDa (Fig. 1), we immunoprecipitated ZBED6 from βTC-6 cells using the anti-mouse ZBED6 antibody. Specific bands, which did not appear in the nonspecific IgG control, were visualized by silver staining (Fig. S2). Five bands with molecular masses between 125 and 70 kDa were cut out and analyzed by mass spectrometry. The sizes of the bands did not exactly match those observed in the immunoblotting experiments described above (Fig. 1), but the gel electrophoresis systems were different in the two experimental setups, which could explain the minor differences. The upper band, 125 kDa, was identified with high confidence as ZBED6 (e value = 1.8 × 10−4). Fingerprinting identified nine peptide fragments of ZBED6 starting at position 76 and reaching to 887. This finding supports the hypothesis that the 120- to 125-kDa band represents full-length ZBED6 (predicted molecular mass: 109 kDa) and that unknown posttranslational modifications enhanced the apparent molecular mass with ∼11–16 kDa. In addition, the second band (110 kDa) was identified with high confidence as ZBED6 (e value = 2.0 × 10−4). The same peptide fragments were found in this band as in the 125-kDa band. These two bands were further verified by tandem mass spectrometry (MS/MS) by analyzing the peptide FLQIVAPDYR (439–448, score 42). The explanation for the smaller size of the second band is not known, but the decreased size may correspond to an alternative translation start site at the second methionine at amino acid 46, which generates a predicted 5-kDa smaller protein (1). The third band (100-kDa double band) was identified with lower probability as ZBED6 using the GPMAW (General Protein/Mass Analysis for Windows) software based on four peptides starting at amino acid 257 and ending at 659. MS/MS analysis of the AVPQLYDSVR (459–468) peptide also gave some support for the ZBED6 identity. No peptides from the ZBED6 N-terminal region were observed in this band. The 95- to 100-kDa size of the third band therefore corresponds to an N-terminal truncation somewhere before amino acid 257. This truncation would generate a predicted 80- to 85-kDa protein that migrates somewhat more slowly due to posttranslational modifications. Such an N-terminal–truncated form of ZBED6 fits well with the observed cytoplasmic localization of the 95-kDa band because the ZBED6 NLS are located at amino acids 61–80 and 231–248 (1). Interestingly, the third band was not pure because the DDHD domain-containing 2 (DDHD2) protein also was identified in the same double band. It is not clear whether DDHD2 immunoprecipitated with ZBED6 or whether the ZBED6 antibody cross-reacted with DDHD2. Bands 4 and 5 did not generate any ZBED6 protein identification.
ChIP-Sequencing Analysis of ZBED6 in Human Islets.
We used the anti-mouse ZBED6 antibody for Illumina paired-end ChIP-sequencing (ChIP-seq) experiments on human islet cells and called 1,658 peaks using a threshold of 12 overlapping ChIP fragments. Most of the peaks (63%) had a match to the ZBED6 binding sequence GCTCGC in the peak center (Table S1a and Fig. S3). The IGF2 site was bound in islet cells, but the MafA, pancreatic and duodenal homeobox 1 (PDX1), and glucokinase genes, which also had an elevated expression in the sh1 and sh2 cells (Table 1), did not have significant ZBED6 signals. ZBED6 binding sites were mainly found in CpG islands (88%) with a preference for locations immediately downstream of TSS. The enrichment at TSS was higher than seen in the previous C2C12 experiment, with 62% of all islet peaks located within 1 kb of TSS. This enrichment was even more pronounced (73%) for sites containing the motif, indicating that ZBED6 mainly affects gene expression by binding to proximal regulatory elements. For GO analysis, we used the DAVID program on genes with strong ZBED6 enrichment (at least 19 overlapping fragments) within 10 kb of the TSS. This process gave a list of 351 genes (Table S1b), for which GO analysis revealed enrichment of transcription, transcription regulation, apoptosis, positive regulation of cellular biosynthetic process, and transmembrane genes (Table S2).
Effect of ZBED6 Knockdown on βTC-6 Cell Morphology.
ZBED6 knockdown and control cells were seeded to Nunclon Surface (NUNC) culture dishes at the same cell density and then cultured in parallel at identical conditions. After 3 d of culture, morphological changes were observed in both sh1 and sh2 βTC-6 cells compared with the Mock cells. The ZBED6 knockdown cells rounded up and formed 3D islet-like clusters, whereas the control cells to a higher degree spread out and covered the surface as a monolayer (Fig. 2B).
Effects of ZBED6 Knockdown on βTC-6 Cell Gene Expression and PDX1/Insulin Levels.
βTC-6 cells, with or without knockdown of ZBED6, were analyzed by semiquantitative real-time RT-PCR for the beta cell markers insulin, PDX1, glucokinase, and MafA (Table 1). In addition, the known ZBED6 target gene Igf2 was analyzed, and lowered ZBED6 mRNA levels were paralleled by increased Igf2 expression (Table 1). This result is in line with a previous study reporting that ZBED6 acts as an Igf2 repressor (1). The mRNA levels of insulin, PDX1, glucokinase, and MafA were all significantly increased in sh1 cells (Table 1). A similar pattern was observed in sh2 cells, but the effect was not as pronounced as in sh1 cells, indicating that the sh2 sequence is less efficient in knocking down ZBED6. To experimentally verify the observed alterations in mRNA levels, we analyzed PDX1 protein levels by immunostaining and immunoblot analysis. Again, PDX1 protein levels were increased more potently in sh1 cells than in sh2 cells, compared with control cells (Fig. 2G). Also insulin contents were potently increased in sh1 cells, but not in sh2 cells, compared with control cells (Fig. 2H).
Effects of ZBED6 Knockdown on βTC-6 Cell Proliferation and Cell-Cycle Distribution.
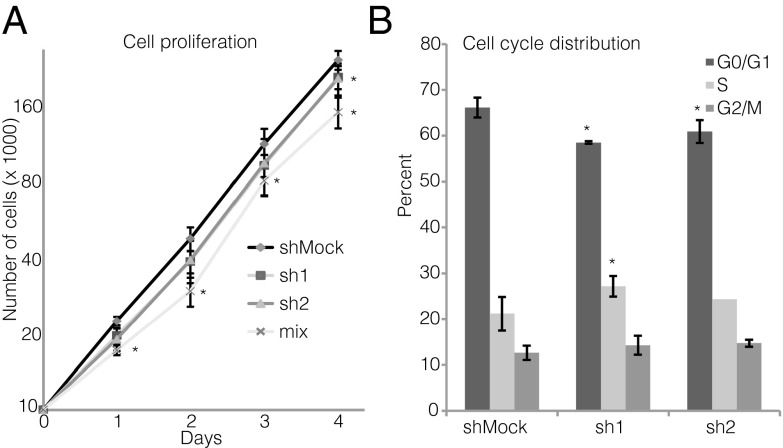
In view of the increased insulin content of ZBED6–sh1-knockdown cells, we next investigated βTC-6 cell proliferation rates. We observed that sh1 and sh2 βTC-6 cells proliferated at a lower rate than controls and that the cells transduced with a combination of sh1 and sh2 proliferated the slowest (Fig. 3A). The sh1 and sh2 both exhibited a lowered fraction of G1 cells (Fig. 3B). In addition, sh1 cells had an increased percentage of S-phase cells (Fig. 3B). These results indicate that ZBED6-knockdown cells proliferate at a lower rate due to a partial inhibition of the S/G2 phase of the cell cycle.
Fig. 3.
Effects of ZBED6 knockdown on βTC-6 cell proliferation (A) and cell-cycle distribution (B). Cell numbers on days 1–5 were determined by flow cytometry, and cell-cycle distribution was analyzed by propidium iodide staining and flow cytometry. Results are means ± SEM for three or four observations. *P < 0.05 (comparing silenced vs. corresponding Mock cells).
Effects of ZBED6 Knockdown on βTC-6 Cell Viability.
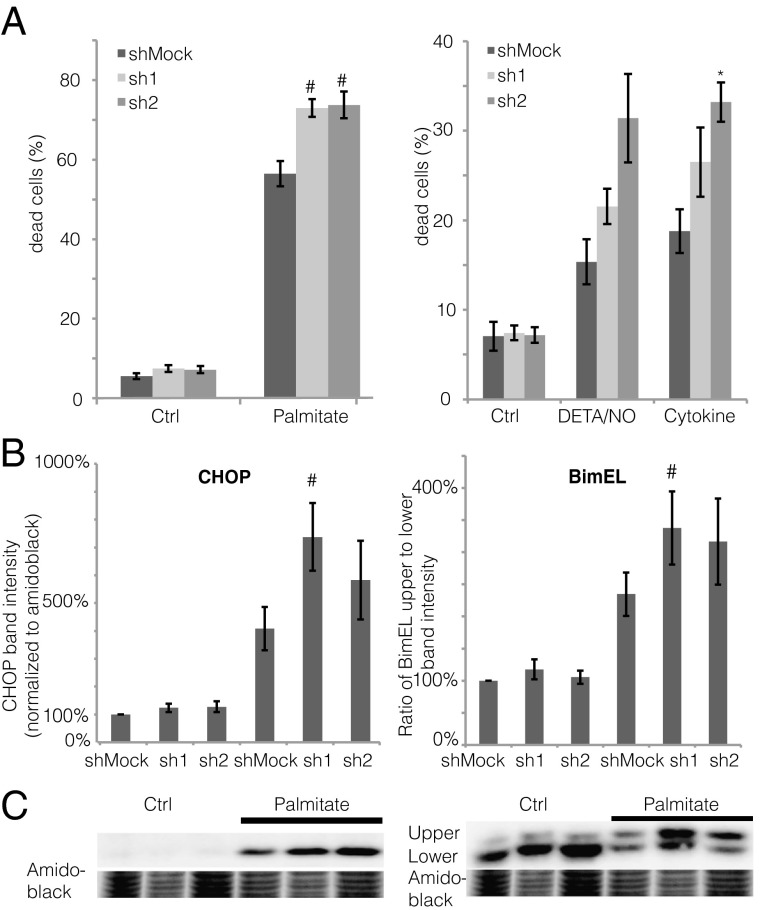
We next studied effects of ZBED6 knockdown on cell death rates in response to sodium palmitate, the nitric oxide (NO) donor diethylenetriamine (DETA)/NO, and the cytokines IL-1β and IFN-γ. At basal conditions, sh1 and sh2 βTC-6 cells displayed similar cell death rates to control cells (Fig. 4A). When stressed by overnight exposure to sodium palmitate, however, ZBED6 knockdown resulted in augmented cell death rates compared with Mock cells (Fig. 4A). In addition, in response to DETA/NO and cytokines, an increased cell death was observed, although statistical significance was in some cases not obtained (Fig. 4A). The sh1 knockdown-induced sensitivity to palmitate was paralleled by an augmented level of the proapoptotic protein CHOP (DDIT3) (Fig. 4B). We also observed an increased phosphorylation of the proapoptotic Bcl-2 family protein Bim (BCL2L11) in palmitate-treated sh1 cells, as assessed by calculating the ratio of the upper Bim band (phosphorylated) to the lower Bim band (nonphosphorylated) (Fig. 4B).
Fig. 4.
(A and B) Effects of ZBED6 knockdown on βTC-6 cell death in response to sodium palmitate, DETA/NO, and cytokines (A), and palmitate-induced CHOP (DDIT3) induction and Bim (BCL2L11) phosphorylation (B). BimEL is the longest splice form (23 kDa) of Bim. Results are means ± SEM for six experiments. *P < 0.05; #P < 0.01. (C) Representative immunoblot images for CHOP (Left) and Bim (Right).
Confirmation of the Specificity of ZBED6 Silencing in βTC-6 Cells.
Reexpression of ZBED6 reversed sh1-induced alterations in proliferation rates and PDX1 levels in βTC-6 cells (Fig. S4). We also confirmed that the lentiviral shRNA sequences targeted Zbed6 and did not affect Zc3h11a expression using mouse BAC clones expressing GFP-tagged ZBED6 or ZC3H11A (Figs. S5 and S6).
Effects of ZBED6 Knockdown on PANC-1 Cell Proliferation.
It has been suggested that beta cells in some cases are generated through transdifferentiation of pancreatic duct cells (5). To establish whether ZBED6, which is expressed in nuclei of pancreatic duct cells (Fig. 2 C–F), controls proliferation and differentiation of these putative beta cell precursors, we silenced ZBED6 in human pancreatic cancer cell line PANC-1 cells using the same approach as with βTC-6 cells (Fig. S7A). We observed that PANC-1 cells express high contents of the nuclear ZBED6 form and low contents of the cytosolic form (Fig. S7B). Both forms were efficiently knocked down by the lentiviral sh1 and sh2 vectors. Interestingly, the proliferation rates were markedly lower in the sh1 and sh2 cells than in the shMock cells (Fig. S7C). This finding is in agreement with the βTC-6 cell results, indicating a proliferative role of ZBED6.
Discussion
Here we report that the newly discovered transcription factor ZBED6 is expressed in an insulin-producing cell line, mouse islets, and human beta cells. ZBED6 appeared both in nuclei, as the larger, 115/120-kDa double band, and in cytosol, as the smaller, 95/100-kDa form. In a recent study, it was observed that expression of ZBED6 in mouse myoblasts, driven by a BAC containing the entire Zbed6 gene, resulted in a full-length nuclear protein with the approximate molecular mass of 115 kDa (6). In murine stem cells, however, the same overexpression strategy resulted in a truncated ZBED6 protein, which lacked the N-terminal located NLS sequence and the DNA-binding domains (6). Given the similarity of the ZBED6 molecular masses presently observed with those observed in the other two cell types (6), it is possible that insulin-producing cells express three forms of ZBED6: two full-length or close to full-length forms that enter the nuclei and act as transcription factors and one N-terminal truncated form (devoid of one of the BED DNA binding domains and NLS) that remains in the cytosol to perform hitherto-unknown functions. This notion is supported by our mass spectrometry analysis of the three different ZBED6 bands showing that the shorter, 95/100-kDa band lacked N-terminal peptide sequences.
ZBED6 originates from a DNA transposon that integrated into one of the first introns of ZC3H11A in a common ancestor of all placental mammals, an event that must have happened >200 million years ago (1). Available data indicate that ZC3H11A and ZBED6 are transcribed from a common promoter located upstream of ZC3H11A exon 1. This interpretation is supported by ENCODE data showing one major POL2 ChIP peak in the vicinity of ZC3H11A exon 1, as well as the distribution of 5′ ends of capped analysis of gene expression mRNAs (CAGE tags) (Fig. S1C). ZBED6 expression is thus dependent on the retention of the intron containing the ZBED6 coding sequence in ZC3H11A transcripts, and an unknown mechanism of alternative splicing must determine whether ZC3H11A transcripts with or without the ZBED6 sequence are produced. To explain the origin of the different ZBED6 forms in islet cells, we analyzed ZBED6 transcription and mRNA splicing in human islet cells. We did not observe multiple TSS or alternative splicing generating mRNAs with different ZBED6 reading frames, but instead near complete retention of the ZBED6-containing intron in transcripts from the ZC3H11A/ZBED6 locus. Because the ZBED6 ORF is located upstream of the translation start site for ZC3H11A, ZBED6 intron retention would result in translation of ZBED6 and not of ZC3H11A. Bioinformatic analysis of the ZC3H11A sequence using the Viral IRES Prediction System software (http://140.135.61.250/vips/) did not reveal any potential internal ribosome entry site downstream of ZBED6 that could possibly drive ZC3H11A translation from the bicistronic transcript, and we can also conclude that it is not possible to translate a hybrid ZBED6–ZC3H11A protein because of the many stop codons in the sequence between the ZBED6 and ZC3H11A ORF in the bicistronic ZBED6/ZC3H11A transcript (Fig. S1D). Furthermore, the interpretation that the ZC3H11A protein is only translated from the transcript lacking introns 1–3—and thus ZBED6—is strongly supported by our data showing that silencing of ZBED6 in mouse C2C12 myoblast cells, expressing ZBED6 and ZC3H11A at a similar level, does not affect ZC3H11A expression (Fig. S5). Thus, we conclude that islet cells predominantly express the bicistronic ZBED6/ZC3H11A transcript that results in the translation of the ZBED6 protein and that ZC3H11A and ZBED6 are produced from separate mRNA molecules.
The mechanisms by which the three ZBED6 forms are generated in insulin-producing cells remain unknown, but the different variants may originate from proteolytical processing or the use of alternative translation start sites. Indeed, the first three methionines of the ZBED6 ORF (Fig. 1E) correspond well to the sizes of the three ZBED6 forms, supporting the notion that the three forms may represent alternative translational start sites. Alternatively, differentiated beta cells may express specific proteases that cleave ZBED6 at specific sites so that the N-terminal region is removed. For example, by using the GPS-UUP software (Version 1.0; http://ccd.biocuckoo.org), a high-score calpain proteolytical site was predicted at amino acid 241. Calpains are calcium-activated proteases, and a previous study reported that Calpain-3 removes the nuclear localization signal peptide from the enzyme dUTPase (7), thereby controlling the subcellular localization of the enzyme. Further studies are required to unravel how insulin-producing cells control ZBED6 size and thereby most likely also its subcellular localization and effects on transcriptional regulation.
ZBED6 acts as an IGF2 repressor in pig skeletal muscle by binding to the GCTCG consensus motif present in the CpG island in intron 3 (1). As assessed by ChIP-seq in myoblasts, ZBED6 target sites appear to exist in many genes other than IGF2 (1), suggesting that ZBED6 might transcriptionally regulate numerous pathways and functions. The results of the present investigation support this notion because ChIP-sequencing of human islet cells revealed ZBED6 binding to 351 genes, using 19 overlapping reads as threshold. The 351 genes were significantly clustered into categories dealing with transcriptional regulation, apoptosis, biosynthetic processes, and transmembrane proteins. To investigate this finding more closely, we down-regulated ZBED6 in βTC-6 cells using a lentiviral shRNA approach. Because βTC-6 cells mainly express the nuclear 115/120-kDa ZBED6 forms, it is likely that our silencing approach mainly addressed the role of ZBED6 transcription factor activity. In addition, in this study IGF2 expression was augmented by ZBED6 knockdown, suggesting that ZBED6 acts as an Igf2 repressor in mouse beta cells. This finding was further supported by our ChIP-seq data showing ZBED6 binding to the IGF2 site in human islets. The mRNA for insulin was similarly up-regulated in response to ZBED6 down-regulation in mouse βTC-6. Because the genes for insulin and IGF2 are closely located in the human genome (4), it is possible that ZBED6 binding to the IGF2 site also affects insulin transcription. Interestingly, the mRNA levels for MafA, a transcription factor that functions as a beta cell differentiation factor during development and a beta cell function-maintaining factor in adult life (8), was increased in ZBED6 knockdown cells. ZBED6 binding to the MafA gene was found in myoblasts (1), but was not presently observed in human islet cells. In addition to MafA, ZBED6 bound also the genes of the beta cell transcription factors Neurog3 and NeuroD2 in muscle cells (1), but not in islet cells. Thus, it may be that ZBED6 acts as a repressor of these genes in non-beta cells and that ZBED6 binding to the same genes is abolished in beta cells. A lack of ZBED6 binding to MafA, Neurog3, and NeuroD2 might result, as a secondary effect, in the presently observed rise in PDX1 and glucokinase mRNA levels.
It is tempting to speculate that the full-length ZBED6 protein—similarly to ZBED1 in HeLa cells (9) and ZBED3 in Caenorhabditis elegans (10)—promotes or maintains proliferation in beta cells and in parallel represses differentiation and specialized cell function, in this case the machinery for glucose sensing and insulin release. In support for this notion are the findings that (i) nuclear ZBED6 immunoreactivity was higher in the rapidly proliferating/low-insulin-producing βTC-6 cells than in slowly proliferating/high-insulin-producing mouse and human islet cells; (ii) knockdown of ZBED6 in βTC-6 cells resulted in decreased proliferation, a partial S/G2 cell-cycle arrest, and the morphological appearance of differentiated islet-like clusters rather than nondifferentiated monolayers; (iii) the insulin and PDX1 contents were enhanced in ZBED6–sh1 cells; and (iv) ZBED6 binding sites were present close to human genes involved in transcriptional regulation and macromolecule biosynthesis in islet cells. In addition, our finding that silencing of nuclear ZBED6 expression in the human pancreatic duct cell line PANC-1 resulted in a reduced proliferation also supports a proliferative role of this transcription factor. It has been reported that PANC-1 cells undergo partial transdifferentiation into an endocrine-like phenotype when induced by neurogenin 3/cell cycle and apoptosis regulator 1 (11), insulinoma-associated antigen-1 (12), or c-Kit activation (13). To test whether down-regulation of ZBED6 also promoted PANC-1 cell transdifferentiation, we analyzed PDX1 immunoreactivity in the sh1– and sh2–PANC-1 cells. However, we observed no signs of endocrine differentiation. Thus, it is likely that silencing of ZBED6 is not sufficient for PANC-1 endocrine differentiation and that other transcription factors initiate the process. This interpretation does not exclude the possibility that ZBED6 plays a more decisive role in primary duct cells in vivo or that ZBED6, rather than being an initiator, instead modulates duct cell transdifferentiation synergistically with other factors. In this context, it appears of great interest to study whether forced ZBED6 down-regulation potentiates neurogenin-3–induced expression of beta cell markers in pancreatic duct cells.
ZBED6-silenced βTC-6 cells also displayed an enhanced sensitivity to the cytotoxic actions of sodium palmitate, a saturated free fatty acid that is increased in type 2 diabetes and that is known to negatively affect beta cell function and survival (14). Furthermore, there was also a trend to increased sensitivity to nitrosative stress and proinflammatory cytokines, indicating a general shift in the apoptotic machinery in favor of enhanced cell death rates. This notion is supported by the finding that the Bcl-2 family protein Bim was hyperphosphorylated in ZBED6 knockdown cells, an event associated with JNK activation and increased rates of apoptosis (15), and that the induction of the proapoptotic transcription factor CHOP (16) was similarly potentiated. A possible explanation for these findings may be that altered expression of ZBED6-controlled apoptosis and cell-cycle arrest genes, occurring in continuously proliferating beta cell lines, promotes the activation of cell cycle-dependent proapoptotic signals and that such events synergize with external cell death signals. Conversely, it is also possible that cells with a more pronounced beta cell phenotype, induced by ZBED6 silencing, are more susceptible to proapoptotic stimuli than nondifferentiated cells.
In summary, our ZBED6 knockdown experiments point to a role of nuclear ZBED6 as an inhibitor of functional beta cell differentiation and as an activator of the cell cycle. Future experimentation is warranted to clarify whether ZBED6 controls embryonic beta cell differentiation and proliferation. Finally, the putative function of the cytoplasmic form of ZBED6 is not understood, but this truncated ZBED6 protein may participate in cytoplasmic functions of the fully mature and nonproliferating beta cell.
Materials and Methods
The materials and methods are described in detail in SI Materials and Methods, which includes the following subjects: cell culture, generation of stable ZBED6–shRNA βTC-6 and PANC-1 cell lines, RNA isolation and cDNA synthesis, human islet RNA sequencing, ZBED6 antibodies, ChIP-seq analysis, semiquantitative real-time RT-PCR, preparation of cytosolic and nuclear fractions, immunoblot analysis, immunoprecipitation of ZBED6 from βTC-6 cells, identification of proteins by mass spectrometry, total insulin content measurement, determination of cell viability by flow cytometry, cell proliferation and cell-cycle analysis, immunohistochemistry, immunofluorescence and confocal microscopy, reexpression of ZBED6 in shMock, sh1 and sh2 βTC6 cells, BrdU incorporation and flow cytometry analysis, silencing of ZBED6 in GFP–ZBED6 and GFP–ZC3H11A overexpressing C2C12 cells, and statistical analysis.
Supplementary Material
Acknowledgments
We thank Göran Hjälm for the development of ZBED6 antibodies and the construction of a GFP–ZBED6 BAC clone. We thank the Nordic Network for Clinical Islet Transplantation, supported by the Swedish national strategic research initiative EXODIAB (Excellence Of Diabetes Research in Sweden), and the Juvenile Diabetes Research Foundation (JDRF) for supply of human islets (JDRF Award 31-2008-413 and the European Consortium for Islet Transplantation/Islet for Basic Research program). Science for Life Laboratory–Uppsala, the Swedish National Infrastructure for Large-Scale DNA Sequencing, and Uppmax provided assistance in massive parallel sequencing. This work was supported in part by Swedish Research Council Grants 2010-11564-15-3 and 67-14643; the Swedish Diabetes Association; the Family Ernfors Fund; the Knut and Alice Wallenberg Foundation; and the Novo–Nordisk Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303625110/-/DCSupplemental.
References
- 1.Markljung E, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7(12):e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Laere AS, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425(6960):832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- 3.Calderari S, et al. Defective IGF2 and IGF1R protein production in embryonic pancreas precedes beta cell mass anomaly in the Goto-Kakizaki rat model of type 2 diabetes. Diabetologia. 2007;50(7):1463–1471. doi: 10.1007/s00125-007-0676-2. [DOI] [PubMed] [Google Scholar]
- 4.Mutskov V, Felsenfeld G. The human insulin gene is part of a large open chromatin domain specific for human islets. Proc Natl Acad Sci USA. 2009;106(41):17419–17424. doi: 10.1073/pnas.0909288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swales N, et al. Plasticity of adult human pancreatic duct cells by neurogenin3-mediated reprogramming. PLoS ONE. 2012;7(5):e37055. doi: 10.1371/journal.pone.0037055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11(4):305–311. doi: 10.1038/embor.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozóky Z, et al. Calpain-catalyzed proteolysis of human dUTPase specifically removes the nuclear localization signal peptide. PLoS ONE. 2011;6(5):e19546. doi: 10.1371/journal.pone.0019546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardo AS, Hay CW, Docherty K. Pancreatic transcription factors and their role in the birth, life and survival of the pancreatic beta cell. Mol Cell Endocrinol. 2008;294(1-2):1–9. doi: 10.1016/j.mce.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Ohshima N, Takahashi M, Hirose F. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J Biol Chem. 2003;278(25):22928–22938. doi: 10.1074/jbc.M303109200. [DOI] [PubMed] [Google Scholar]
- 10.Inoue T, Sternberg PW. C. elegans BED domain transcription factor BED-3 controls lineage-specific cell proliferation during organogenesis. Dev Biol. 2010;338(2):226–236. doi: 10.1016/j.ydbio.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu CK, Lai YC, Lin YF, Chen HR, Chiang MK. CCAR1 is required for Ngn3-mediated endocrine differentiation. Biochem Biophys Res Commun. 2012;418(2):307–312. doi: 10.1016/j.bbrc.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T, Wang H, Saunee NA, Breslin MB, Lan MS. Insulinoma-associated antigen-1 zinc-finger transcription factor promotes pancreatic duct cell trans-differentiation. Endocrinology. 2010;151(5):2030–2039. doi: 10.1210/en.2009-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, et al. c-Kit and stem cell factor regulate PANC-1 cell differentiation into insulin- and glucagon-producing cells. Lab Invest. 2010;90(9):1373–1384. doi: 10.1038/labinvest.2010.106. [DOI] [PubMed] [Google Scholar]
- 14.Unger RH. Lipotoxicity in the pathogenesis of obesity-dependent NIDDM: Genetic and clinical implications. Diabetes. 1995;44(8):863–870. doi: 10.2337/diab.44.8.863. [DOI] [PubMed] [Google Scholar]
- 15.Santin I, et al. PTPN2, a candidate gene for type 1 diabetes, modulates pancreatic β-cell apoptosis via regulation of the BH3-only protein Bim. Diabetes. 2011;60(12):3279–3288. doi: 10.2337/db11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirot P, et al. Transcriptional regulation of the endoplasmic reticulum stress gene chop in pancreatic insulin-producing cells. Diabetes. 2007;56(4):1069–1077. doi: 10.2337/db06-1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.