Abstract
The nature of influenza virus to randomly mutate and evolve into new types is an important challenge in the control of influenza infection. It is necessary to monitor virus evolution for a better understanding of the pandemic risk posed by certain variants as evidenced by the highly pathogenic avian influenza (HPAI) viruses. This has been clearly recognized in Egypt following the notification of the first HPAI H5N1 outbreak. The continuous circulation of the virus and the mass vaccination programme undertaken in poultry have resulted in a progressive genetic evolution and a significant antigenic drift near the major antigenic sites. In order to establish if vaccination is sufficient to provide significant intra- and interclade cross-protection, lentiviral pseudotypes derived from H5N1 HPAI viruses (A/Vietnam/1194/04, A/chicken/Egypt-1709-01/2007) and an antigenic drift variant (A/chicken/Egypt-1709-06-2008) were constructed and used in pseudotype-based neutralization assays (pp-NT). pp-NT data obtained was confirmed and correlated with HI and MN assays. A panel of pseudotypes belonging to influenza Groups 1 and 2, with a combination of reporter systems, was also employed for testing avian sera in order to support further application of pp-NT as an alternative valid assay that can improve avian vaccination efficacy testing, vaccine virus selection, and the reliability of reference sera.
1. Background
Egypt faced its first H5N1 outbreak in 2006 where a highly pathogenic avian influenza (HPAI) virus was detected in poultry [1]. The strategy used by the Egyptian authorities to mitigate this relied on vaccinating poultry, depopulating infected areas, and increasing awareness and biosecurity levels. Despite these efforts, by 2008, the H5N1 virus had become endemic, and vaccine-escape variants have emerged despite commercial poultry vaccines exhibiting protection in laboratory settings [2]. For each year, from 2009 through 2012, Egypt has had more laboratory-confirmed human cases reported to the WHO than any other country, and global concern regarding Egyptian H5N1 influenza viruses is currently high, as some isolates have been reported to possess at least two mutations, of the 4 (or 5) needed to confer ferret-to-ferret airborne transmissibility [3]. Despite the mass vaccination program undertaken in poultry, the continuous circulation of the virus has resulted in a progressive genetic evolution and a significant antigenic drift with multiple mutations near the major antigenic sites [4]. To date, the WHO has identified 12 new H5N1 clades, and the Egyptian clade 2.2.1 was further split into a new subclade 2.2.1.1 [5, 6]. The past experience in Egypt has proved that controlling avian influenza in poultry is the primary method to reduce the human risk from infection and monitoring virus evolution can be extremely important for understanding the pandemic risk posed by certain subtypes, especially those prone to antigenic drift mechanisms as evidenced by the genetic and antigenic divergence of H5N1 HPAI viruses in Egypt [7–9].
Furthermore, it has been highlighted as a priority to combine vaccination with the implementation of specific systems to detect early infection with low pathogenicity avian influenza (LPAI) viruses and to study naturally acquired or vaccine-induced immunity in avian species via appropriate diagnostic tools and serological surveillance [10, 11]. Recent studies have stressed the need of reinforcing serological tests as an auxiliary tool to evaluate the potency of commercial vaccines and monitor vaccine-driven evolution of emerging variants and consequent choice of seed viruses [2]. This has been clearly recognized when the inactivated vaccine containing an H5 virus belonging to a different lineage to the Eurasian H5N1 (H5N2/Mexico) is being actively used in order to control the HPAI outbreak in Egypt from 2006 [12–14].
As shown by our earlier study, the emergence of an Egypt H5N1 drift variant (circulating one year later from the first H5N1 outbreak) exhibited significantly decreased cross-reactivity by haemagglutination inhibition (HI) and microneutralization (MN) assays against the Mexican vaccine seed strain [15]. This evidence, together with previous observations, has raised the important question of the mechanism of antigenic drift under vaccine pressure. Additionally, the key role of an active serological postvaccination surveillance for the assessment of vaccine efficacy and evaluation of cross-neutralizing capability of the vaccine concurrent with incremental virus escape from neutralizing antibodies is important [16].
There is currently a wide range of serological assays available for influenza; the choice is mainly based on the viral protein targeted, the level of specificity required (subtype specific or nonsubtype specific tests), and the laboratory facilities needed for certain strains [17]. Despite the complexity of the antibody response against influenza viruses, the standard serological tests such as HI and MN are routinely employed in avian influenza reference laboratories as promoted by the FAO/OIE Network of expertise on animal influenza (http://www.offlu.net/) and WHO [18]. More recently, due to their wide applicability and sensitivity, pseudotype-based neutralization (pp-NT) assays have been shown to be valid alternatives to these established methods for studying the serological profiles of highly pathogenic influenza viruses, vaccine-induced immunogenicity, and serological cross-reactivity of haemagglutinins (HAs) from different clades [19–21]. Moreover, recent studies [22, 23] have revealed that broadly cross-neutralizing antibodies binding to the stalk region of HA can be indirectly measured by HA pp-NT assays and to a lesser extent by MN but not by HI which only measures those antibodies that bind to the globular head and interfere with receptor binding [24–27]. This study reports on the screening of avian sera for antibodies elicited by LPAI and HPAI viruses and proposes a new perspective for the widening application and validation of pp-NT serological assays especially with the potential to streamline the screening of large sample sets collated from in-field seroepidemiology studies and vaccination programmes [17]. Towards this aim, we have firstly constructed HA pseudotypes from an HPAI Egyptian H5N1 virus and its closely related antigenic drift variant for a comparative serological framework to study cross-strain immunity induced by an LPAI H5N2 vaccine. Subsequently, in this study, the HA-pseudotype panel has been expanded in order to demonstrate their unique versatility (via the use of alternative reporter systems), reliability (by testing sera from naturally infected birds and reference sera) and propose them as powerful tools to support in-field and laboratory-based avian serology.
2. Materials and Methods
2.1. Plasmids and Pseudotype Virus Production
Lentiviral pseudotypes with HA envelope glycoproteins derived from the HPAI viruses H5N1 (A/Vietnam/1194/04, A/chicken/Egypt/1709-1/2007 and A/chicken/Egypt/1709-6/2008) and H7 (A/chicken/Italy/13474/99) were constructed as described previously, with exogenous soluble neuraminidase (NA) (1 Unit/plate; Sigma) being added after transfection in order to induce the release of HA-pseudotypes from the surface of producer cells [28–30]. H5 and H7 pseudotypes were produced by cotransfection of HEK-293T cells with a complex comprising HA-expression plasmids (pl. 18-HA), the HIV type 1 gag-pol (pCMV-Δ8.91) and the firefly luciferase reporter constructs (pCSFLW) using Fugene 6 (Roche) that facilitates highly efficient DNA transport into cells [31–33]. Additionally green fluorescent protein (GFP) pseudotypes bearing H5 glycoproteins from A/Vietnam/1194/04 and A/chicken/Egypt 1709-1/2007 strains were generated by incorporation of GFP retroviral construct (pCSGW) as reporter [34–38]. Concurrently, a no-HA control was generated by co-transfection of producer cell lines with two plasmids, gag-pol pCMV-Δ8.91 and pCSFLW.
2.2. Serum Samples
Five panels of sera were evaluated in this study and were all provided by the FAO-OIE and National Reference Laboratory for Newcastle disease and Avian influenza, Istituto Zooprofilattico delle Venezie. Panel 1 consisted of 10 sera positive for antibodies to the LPAI H5N2 vaccine strain (A/chicken/Mexico/232/94/CPA) obtained from chickens vaccinated at 21 days of age and boosted after 3 weeks with a commercially available inactivated vaccine, which has been used in previous studies [12, 15]. Panel 2 consisted of 10 sera positive for H7 collected from turkeys during an Italian outbreak caused by an LPAI virus H7N3. Panel 3 consisted of 10 sera positive for H5 with stratified incremental HI titers (ranging from 1 : 4 to 1 : 2048) collected from chickens vaccinated with an inactivated adjuvanted H5N2 vaccine and were used for comparative firefly luciferase and GFP-pseudotype neutralization assays. In order to test for influenza HA group-specific virus neutralization, panel 4, consisting of 16 reference hyperimmune sera produced against 16 influenza subtypes (from Group 1 and Group 2), was also provided. These antisera (H1N1, H2N3, H3N8, H4N8, H5N1, H6N2, H7N3, H8N4, H9N2, H10N1, H11N9, H12N5, H13N6, H14N5, H15N9, and H16N2) were produced in specific pathogen-free chickens by inoculation with viruses (inactivated by beta-propriolactone if HPAI viruses) as described previously [12]. A panel of 41 negative sera (panel 5) confirmed by agar gel immunodiffusion assay (AGID) and Enzyme-linked immunosorbent assay (ELISA) was also employed.
2.3. HI and MN Assays
All the sera collected from vaccinated chickens were evaluated using standard protocols for HI and MN assays using A/chicken/Egypt/1709-1/2007 and A/chicken/Egypt/1709-6/2008 field strain antigens. HI assays were also carried out for the H5- and H7-positive serum panels using the test antigens: H5N2 (homologous to the Mexican LPAI vaccine) and H7N1 (A/Starling/Africa/985/79), respectively. Standard protocols were followed for both assays as described previously [15, 39]. For the 41 negative sera a titer of 2 was assigned when tested by HI.
2.4. Firefly Luciferase pp-NT Assay
For this assay, firefly luciferase pseudotypes bearing HAs from HPAI H5 (A/Vietnam/1194/04, A/chicken/Egypt/1709-1/2007, A/chicken/Egypt/1709-6/2008) and the HPAI H7 strain (A/chicken/Italy/13474/99) were used. Two-fold serial dilutions of serum samples were mixed with an equal volume of pseudotype virus resulting in 5 × 105 relative light units (RLUs) after 48 hr under standard conditions. After a 1-hour incubation at 37°C, 1 × 104 HEK-293T cells were added to each well of a 96-well flat-bottomed culture plate, and RLUs were evaluated after 48 hr of incubation with a luminometer (Promega Glo Max 96) using the Bright-Glo substrate (Promega). To measure neutralization activity, the 50% and/or 90% inhibitory dose (IC90) was determined as the serum dilution resulting in a 50% and/or 90% reduction of a single round of infection (reporter gene-mediated signal) [28, 29, 40]. All the results were compared to control wells containing virus alone, with the RLUs from cell-only wells subtracted from all the readings. Additionally, 41 negative sera were also tested by using firefly luciferase pseudotypes (A/Vietnam/1194/04, A/chicken/Egypt/1709-1/2007, and A/chicken/Egypt/1709-6/2008).
2.5. GFP pp-NT Assay
The pp-NT assay was additionally performed using pseudotypes bearing HA from HPAI A/Vietnam/1194/04 and A/chicken/Egypt 1709-1/2007 strains using a packaging construct with GFP reporter gene, and the GFP pp-NT assay was essentially performed as described previously [29, 31]. In order to determine, for each strain, the amount of HA-pseudotyped virus required for this assay, complete medium was dispensed into each well of a clear 96-well flat-bottomed plate, and 8 rows of 2-fold serial dilutions of the initial virus stock were prepared, followed by the addition of 50 μL of HEK-293T cells to each well. 3 days after infection, GFP expression was monitored using fluorescence microscopy. Normally 3 random fields of view are used to score the overall fraction of GFP-expressing cells, and the volume of HA-pseudotyped virus used for the assay was calculated by choosing the reciprocal pseudotype virus dilution that corresponds to the amount required to transduce 100 cells/well. The serum neutralization activity was estimated as the reduction of fluorescence expressed by the percentage of green cells in the presence of serum. Sera with no presence of neutralizing antibodies or negative sera were defined as 100% green cells or high GFP expression.
2.6. Statistical Analysis
The estimation of pseudotype transduction titers was performed using Excel software where pseudotype titers obtained at each of a range of dilution points were expressed as RLU/mL, and the arithmetic mean was calculated by GraphPad Prism (version 5, GraphPad Software, San Diego, CA, USA). Statistical analyses were also undertaken for the analysis of pp-NT assays using GraphPad. Pp-NT titers were normalized, and IC50 and IC90 values were calculated by dose-response inhibition analysis. In order to assess correlation between pp-NT, HI, and MN, antibody titers were log10 transformed, and Pearson's correlation analysis was used.
3. Results
The initial aim of the present study was to study, via a comparative serological approach, the profile, described in our earlier study [15] of influenza H5N1 subclade 2.2.1 A/chicken/Egypt/1709-1/07 virus and its antigenic drift variant belonging to subclade 2.2.1.1 A/chicken/Egypt/1709-6/08 in order to confirm the reliability of pp-NT results when employed in parallel with standard HI and MN tests. Subsequently, we investigated whether pseudotypes bearing HPAI H5 and H7 are accurately able to accurately detect neutralizing antibody responses elicited by LPAI H5 and H7 avian influenza viruses with the flexibility of using different reporter genes expressed by lentiviral vectors pseudotyped with influenza HA glycoproteins. In order to show the validity and robustness of the pp-NT method for its application to large-scale serological analyses, the results obtained by pp-NT assays were compared with HI and MN tests.
3.1. Panel H5 Positive (Collected from LPAI H5N2 A/chicken/Mexico/232/94 Vaccine Trial)
Neutralizing antibodies were measured using firefly luciferase HPAI H5 influenza pseudotypes bearing HA glycoproteins derived from the HPAI viruses, clade 1 A/Vietnam/1194/04, A/chicken/Egypt 1709-1/2007, and A/chicken/Egypt/1709-6/2008. These pseudotypes were used in a neutralization assay for the detection of antibodies in a panel of 10 sera collected from chickens vaccinated with an LPAI strain belonging to a different lineage: A/chicken/Mexico/232/94/CPA (H5N2). Mexican-derived A/H5N2 inactivated vaccines were commonly used for vaccination programs in poultry farms, as undertaken in Egypt, where the samples used for this study were collected [12, 41]. A broad range of IC90-neutralizing antibody titers was observed in these sera, tested in duplicate against A/Vietnam/1194/04, A/chicken/Egypt 1709-1/2007, and the drift variant A/chicken/Egypt/1709-6/2008 (Table 1). 41 negative sera previously tested AI antibody free by ELISA and AGID assays (data not shown) were also found negative by H5N1 A/Vietnam/1194/04, A/chicken/Egypt 1709-1/2007, and A/chicken/Egypt/1709-6/2008 HA pp-NT.
Table 1.
IC90-neutralizing antibody titres tested by pseudotype-based neutralization assays for chickens immunized with the Mexican-derived H5N2 strain.
| IC90-neutralizing antibody titres | |||
|---|---|---|---|
| Sera no. | A/Vietnam/1194/04 | A/ck/Egypt 1709-1/2007 | A/ck/Egypt 1709-6/2008 |
| 4822/V09-1 | 2560–5120 | >81920 | 160–320 |
| 4822/V09-3 | 2560–5120 | 20480–40960 | 80–160 |
| 4822/V09-4 | 320–640 | 1280–2560 | 40–80 |
| 4822/V09-5 | 2560–5120 | 10240–20480 | 80–160 |
| 4822/V09-6 | 2560–5120 | 20480–40960 | 320–640 |
| 4822/V09-7 | 2560–5120 | >81920 | 640–1280 |
| 4822/V09-8 | 1280–2560 | 40960–81920 | 80–160 |
| 4822/V09-9 | 1280–2560 | >81920 | 320–640 |
| 4822/V09-10 | 2560–5120 | 5120–10240 | 80–160 |
| 4822/V09-12 | 2560–5120 | 40960–81920 | 40–80 |
|
| |||
| Range of titres observed | (320–5120) | (1280–81920) | (40–1280) |
In order to assess whether the results obtained with pp-NT assay mirrored those obtained with conventional assays (HI and MN) extensively used for influenza serology, a regression analysis on paired datasets was performed in order to measure the significance of correlation. The results of this analysis were supported by a highly statistically significant correlation (P < 0.001) between antibody titers obtained by HI, MN, or pp-NT. As shown in the scatterplots, titers obtained via HI correlated strongly with titers obtained using clade 2.2.1 A/chicken/Egypt 1709-1/2007 (r 2 = 0.6291) (Figure 1) and the drift variant A/chicken/Egypt/1709-6/2008 (r 2 = 0.7972) (Figure 2). Similar levels of correlation were observed between pp-NT titres and MN titres for both Egyptian strains (Figure 3).
Figure 1.
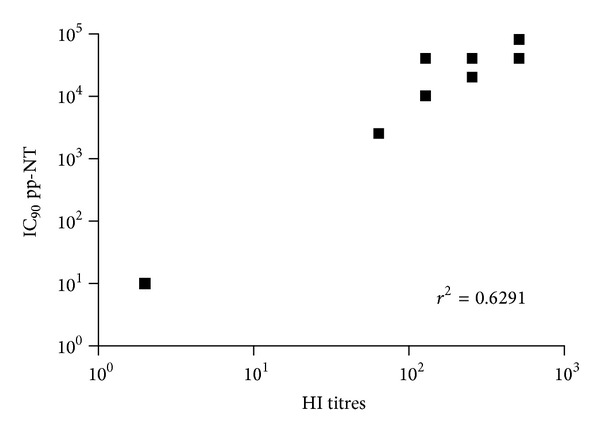
Comparison of pp-NT with HI antibody titer. Scatterplots showing the correlation of antibody logarithmic titers measured by pp-NT (using A/chicken/Egypt/1709-1/2007) versus HI (tested against A/chicken/Egypt/1709-1/2007). Correlation gave a P value < 0.0001.
Figure 2.
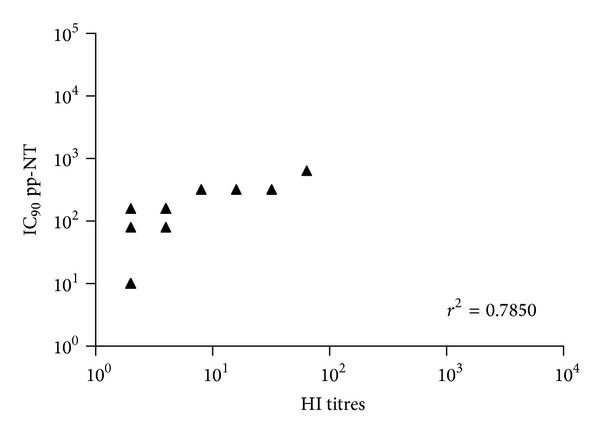
Comparison of pp-NT with HI antibody titers. Scatterplots showing the correlation of antibody logarithmic titers measured by pp-NT (using A/chicken/Egypt/1709-6/2008) versus HI (tested against A/chicken/Egypt/1709-6/2008). Correlation P value < 0.0001.
Figure 3.
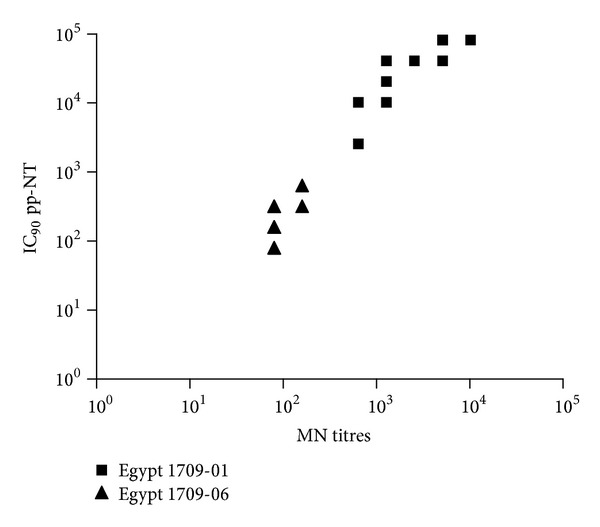
Comparison of pp-NT with MN antibody titers. Scatterplots showing the correlation of antibody logarithmic titers measured by pp-NT (using A/chicken/Egypt/1709-1/2007 and A/chicken/Egypt/1709-6/2008) versus MN (tested against A/chicken/Egypt/1709-1/2007 and A/chicken/Egypt/1709-6/2008).
Similar correlation parameters were observed between HI titers and clade 1 A/Vietnam/1194/04 pseudotype (Figure 4).
Figure 4.
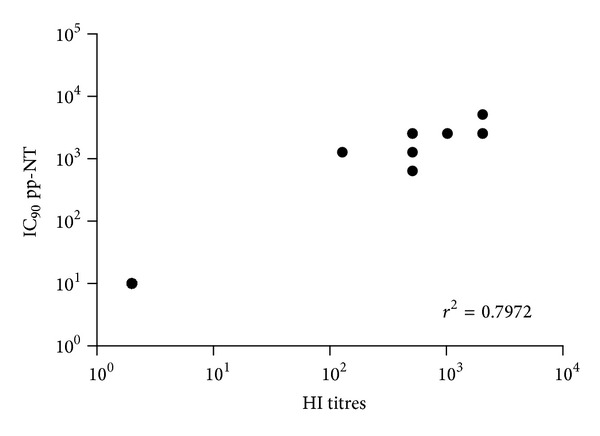
Comparison of pp-NT with HI antibody titers. Scatterplots showing the correlation of IC90 pp measured by pp-NT (using A/Vietnam/1194/04) versus HI (tested against A/chicken/Hidalgo/28159-232/1994). Correlation gave a P value < 0.0001.
3.1.1. Incremental HI Positive H5 Serum Panel
Measurement of Neutralizing Antibodies Using GFP and Firefly Luciferase HPAI H5 Pseudotypes. In order to determine the reliability and the applicability of the pp-NT assay using different reporter systems, H5 A/Vietnam/1194/04 and A/chicken/Egypt 1709-1/2007 pseudotypes carrying the GFP were tested against a panel of sera positive by HI with incremental titers ranging from 1 : 8 to 1 : 2048. Three sera (3929-1, 3929-9, and 3929-6) were scored as 100% neutralization activity with pp titers >1 : 1280 (no GFP expression was observed) and sera 3930-19, 3931-26, and 3930-20 showed 50% neutralization activity at 1 : 80, 1 : 160, and 1 : 320 when tested against A/Vietnam/1194/04. IC50 values corresponding to titers around ≤1 : 40 were obtained for sera with HI titers lower than 1 : 32. For A/chicken/Egypt 1709-1/2007 pseudotypes, a similar pattern was observed: 4 sera (3929-1, 3929-9, 3929-6, and 3930-19) have shown complete neutralization; for 3 sera (3931-26, 3930-20, and 3933-41) 50% neutralization activity was scored between 1 : 640 and 1 : 1280. For 2 sera (3933-42 and 3933-50) percentage values of 50% lay between 1 : 80 and 1 : 320.
In order to support the quantitative results obtained using GFP-pseudotypes, the panel of sera was tested in parallel against firefly luciferase HA pseudotype. pp-NT results were found to correlate strongly with HI showing a similar neutralization profile; however, for sera with an HI titre lower than 1 : 32, it has not been possible to determine the respective pp-NT neutralization values when H5 A/Vietnam/1194/04 pseudotypes have been used (Table 2).
Table 2.
Comparison between pp-NT assays using different reporter systems (GFP and CSFLW luciferase) and HI tests. IC50-neutralizing antibody titres tested by A/Vietnam/1194/04 and A/Egypt 1709-01/07.
| Sera no. | HI | IC50
A/Vietnam/1194/04 GFP-pp |
IC50
A/Vietnam/1194/04 Luc-pp |
IC50
A/Egypt 1709-01/07 GFP-pp |
IC50
A/Egypt 1709-01/07 Luc-pp |
|---|---|---|---|---|---|
| 3933-42 | 1 : 8 | <40 | 80–160 | 80–160 | 80–160 |
| 3933-50 | 1 : 16 | <40 | 640–1280 | 160–320 | 80–160 |
| 3933-41 | 1 : 32 | <40 | 640–1280 | 320–640 | 1280–2560 |
| 3930-20 | 1 : 64 | 160–320 | 2560–5120 | 640–1280 | 2560–5120 |
| 3931-26 | 1 : 128 | 80–160 | 640–1280 | 1280 | 5120–10240 |
| 3930-19 | 1 : 256 | 40–80 | 2560–5120 | >1280 | 5120–10240 |
| 3929-6 | 1 : 512 | >1280 | >10240 | >1280 | 2560–5120 |
| 3929-9 | 1 : 1024 | >1280 | 5120–10240 | >1280 | >10240 |
| 3929-1 | 1 : 2048 | >1280 | >10240 | >1280 | >10240 |
3.2. Panel H7 Positive (Collected from an LPAI H7 Outbreak in Italy)
A panel of 10 sera collected from turkeys during an Italian epizootic caused by an LPAI H7 virus was tested by A/chicken/Italy/13474/99 HA-pseudotype assay and by HI using H7N1 (A/Starling/Africa/985/79) as antigen. All sera were positive by HI showing a panel of different titers and 10/10 closely correlated with titers obtained by pp-NT as shown in Figure 5. 41 chicken sera confirmed AI antibody free by ELISA and AGID assays were also found negative when tested by H7 A/chicken/Italy/13474/99 pseudotype-based assay.
Figure 5.
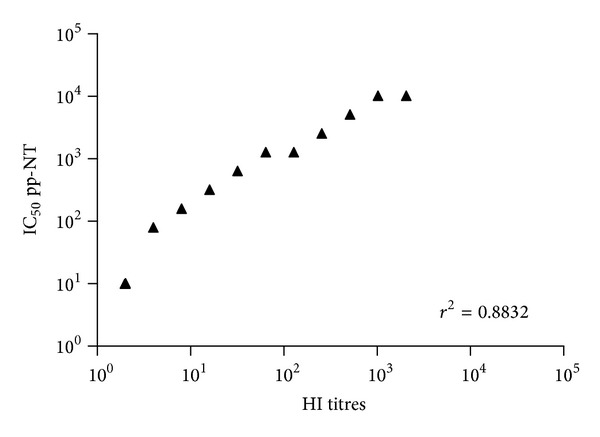
Comparison of pp-NT with HI antibody titers. Scatterplots showing the correlation of antibody logarithmic titers measured by pp-NT (using A/chicken/Italy/13474/99) versus HI (tested against A/starling/Africa/985/79 (H7N1)). Correlation gave a P value < 0.0001.
3.3. Cross-Reactivity of Influenza HA Groups 1 and 2 Pseudotypes Using a Panel of Avian Reference Sera against All 16 HA Subtypes
In order to determine the extent of HA-group-specific “heterosubtypic” cross-reactivity, we tested the ability of reference hyperimmune avian sera (raised against H1N1, H2N3, H3N8, H4N8, H5N1, H6N2, H7N3, H8N4, H9N2, H10N1, H11N9, H12N5, H13N6, H14N5, H15N9, and H16N2) to neutralize pseudotypes produced with the Group 1 viruses belonging to different clades: H5 A/chicken/Egypt 1709-1/2007 and A/chicken/Egypt/1709-6/2008 and Group 2 virus: H7 A/chicken/Italy/13474/99. The quantity of H5 and H7 pseudotypes was chosen in order to have a virus input around 1 × 105 RLUs, and the cross-neutralization activity for both subtypes was determined as the serum dilution resulting in 50% reduction of luciferase signal. Sera with IC50 titers equal to or below 1 × 101 were considered not cross-reactive as shown in Figure 6. It is notable that HA-influenza pseudotypes are able to detect cross-specific neutralization within Groups 1 and 2, and, with some variation observable, H5 pseudotypes show similar patterns of cross-reactivity. Both Group 1 H5 pseudotypes (A/chicken/Egypt/1709-1/2008 and A/chicken/Egypt/1709-6/2008) exhibited cross-reactivity with sera generated against the subtypes H1N1, H2N3, H6N2, and H8N4. The Group 2 H7 pseudotype (A/chicken/Italy/13474/99) exhibited cross-reactivity with sera generated against H3N8, H4N8, H10N1, and H15N9. As additional control, H5 and H7 pseudotypes were also tested against reference sera H5N1 and H7N3, respectively, in order to provide evidence of cross-reactivity between Group 1 and Group 2 (Figure 6).
Figure 6.
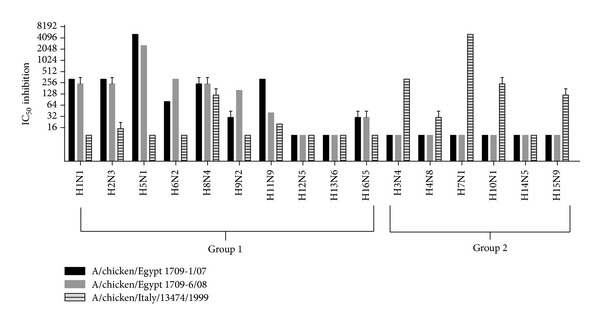
pp-NT assay showing the presence of cross-reactivity in avian reference sera between 1 and 2 influenza groups tested by H5 A/chicken/Egypt/1709-1/2008 and A/chicken/Egypt/1709-6/2008 and H7 A/chicken/13474/Italy/1999. Values corresponding to 50% neutralization (IC50) and with threshold serum dilution of ≥10 were considered positive.
4. Discussion and Conclusions
Up to 1995, there had been only three reports of avian influenza viruses infecting humans, in 1959, 1977, and 1981. However, since 1996 there have been regular reports of natural infections of humans with avian influenza viruses [42]. Although these infections seem to have been limiting, with very little human to human transmission, the potential emergence of a virus capable of spread in the human population could occur via different mechanisms such as avian and human virus reassortment, recirculation of existing subtypes, and/or gradual adaptation of animal viruses to human transmission. The emergence of influenza viruses highlighted the ability of H5 and H7 subtypes to mutate from low to the highly pathogenic variant after introduction into domestic poultry [43–45]. It follows that all HPAI viruses should have an LPAI progenitor, and these incidents have raised concern about potential pandemics caused by viruses of the H5 and H7 subtypes or by any other avian influenza viruses with the potential to be transmitted to a variety of nonavian hosts including humans [44, 46–48]. In some cases mutation seems to have taken place rapidly after introduction from the wild bird reservoir; in others the LPAI virus has circulated in poultry for months prior to mutating. The factors responsible and the mechanism by which LPAI virus mutates into HPAI virus remain unclear. However, it is reasonable to assume that the wider the circulation of LPAI in poultry, the higher the chance that mutation to HPAI will occur [42]. The recent implementation of active surveillance and vaccination policies with administration of appropriate vaccines in domesticated poultry has facilitated eradication of HPAI in many countries [49]. The control of infection in poultry and the validation of more sensitive and specific assays for detecting antibodies to avian influenza viruses in avian and non-avian species represent some of the main objectives for influenza experts from the animal and public health sectors [50]. The measurement of neutralizing antibody responses is critical for influenza serodiagnosis, for the evaluation of novel vaccines and their effectiveness against drift variants arising as a consequence of vaccine pressure. The pp-NT assay represents a reliable and safe test to determine neutralizing antibody responses to all subtypes of influenza viruses [28, 51]. This neutralization assay has shown high sensitivity and specificity when compared with the established serological tests, HI and MN, and has demonstrated wide applicability for antiviral and therapeutic antibody screening and for the evaluation of vaccine efficacy. Moreover, all these methods together can be used to evaluate how well the circulating isolates match the AI vaccine formulations in order to update the vaccine by using criteria similar to those used for human influenza vaccines [52]. Exploiting the inherent sensitivity of this assay, the aim of this study was to determine the levels of antibody response against H5 and H7 pseudotypes carrying the polybasic cleavage site, in sera obtained from avian species vaccinated with commercially available inactivated vaccine that has been used in poultry farms or naturally infected with LPAI influenza viruses, and to show the correlation between pp-NT and the classical serological assays: HI and MN.
A panel of H5-positive sera obtained from chickens vaccinated with an H5N2 A/chicken/Mexico/232/94/CPA strain was tested previously by us against the Egyptian H5N1 challenge strains (A/chicken/Egypt/1709-1/2007, A/chicken/Egypt/1709-6/2008), and significant differences between these strains have been shown by HI and MN assays, most likely due to antigenic drift driven by the implementation of vaccination in poultry [15]. In parallel, pseudotypes bearing HPAI HAs were constructed (A/Vietnam/1194/04, A/chicken/Egypt 1709-1/2007 and A/chicken/Egypt/1709-6/2008) [15, 29]. Titers obtained via HI and MN correlated strongly (Figures 1–5) with those obtained using H5 pseudotypes (for A/chicken/Egypt/1709-1/2007: r 2 = 0.62 and for A/chicken/Egypt/1709-6/2008: r 2 = 0.78). When A/Vietnam/1194/04 pseudotypes were used the correlation was r 2 = 0.79 despite the fact that the HA used in this pp-NT assay was not antigenically matched as it belongs to a different clade. The rank of ordered neutralizing values obtained by pseudotypes mirrored the HI and MN assays. Interestingly, compared with HI and MN, the pp-NT overall gives higher numerical titers and appears to be more sensitive than MN. Recent studies have raised the possibility that the lower incorporation of HA spikes into retroviral pseudotypes, compared to the wild-type virus, makes pseudotypes more sensitive allowing the binding of antibodies not only on antigenic sites of HA surface but also on the HA stalk as shown in previous studies [24, 53].
Similar results were obtained when a control panel of sera positive by HI against H7 was tested against A/chicken/Italy/13474/99 HA pseudotypes showing not only the presence of a neutralizing antibody response against HPAI H7 in sera from chickens infected by an LPAI virus but also a profile of neutralization that strongly correlates with HI. The pp-NT assay has the potential to be used in resource-limited countries where the cost-benefit of this assay could be increased by the availability of different reporter systems, for example, the use of GFP reporter instead of firefly luciferase. Additionally for laboratories lacking fluorescence or luciferase detection capability, β-galactosidase reporters could be used [31]. The results from this study revealed that the neutralization profile for pp-NT using a GFP reporter does not show as clearly as firefly luciferase pp-NT the titer stratification (especially for sera that give low responses by HI). A comparative analysis of results obtained using the two different reporters on the same set of sera was performed and shows a clear correlation and a strong neutralizing profile although no correlate of protection has been yet established for pp-NT assay (Table 2) [28].
Results for cross-reactivity analyses of Groups 1 and 2 HA influenza pseudotypes against antisera from all 16 HA subtypes shed new light on the performance of the pp-NT assay using HI standards. Firstly, the specificity that can be gained by the use of influenza pseudotypes considering that the cut-off for negative sera was assigned for IC50 values equivalent or below 1 × 101, and H5 and H7 pseudotypes showed some degree of cross-reactivity with sera generated from viruses belonging to the same HA groups and strong reactivity when H5 and H7 pseudotypes were tested against H5-H7 hyperimmune sera as shown in Figure 6.
Furthermore, reactivity observed validates the reliability and the quality of OIE-FAO reference sera that represents a prerequisite for the improvement of sero-diagnosis and can help to evaluate the effectiveness of vaccine strategies bearing in mind that an extensive library of reference sera for all influenza strains is an essential aspect for pandemic influenza preparedness [54]. It is likely that a new panel of reference sera will need to be prepared for use with pseudotype-based assays as they become more widely used in the future.
The pp-NT assay is a valid surrogate for the more complex and time-consuming MN and for HI. Influenza pseudotypes can be employed to screen antibody responses on the particle surface due to the fact that HA is the major antigen of the virus against which neutralizing antibodies are produced [55]. It will permit HA subtyping, antigenic tracking of virus evolution, and help to improve both the evaluation of vaccine effectiveness and vaccine virus strain selection.
References
- 1.Joannis T, Lombin LH, De Benedictis P, Cattoli G, Capua I. Confirmation of H5N1 avian influenza in Africa. Veterinary Record. 2006;158(9):309–310. doi: 10.1136/vr.158.9.309-b. [DOI] [PubMed] [Google Scholar]
- 2.Kayali G, Kandeil A, El-Shesheny R, et al. Do commercial avian influenza H5 vaccines induce cross-reactive antibodies against contemporary H5N1 viruses in Egypt? Poultry Science. 2013;92:114–118. doi: 10.3382/ps.2012-02637. [DOI] [PubMed] [Google Scholar]
- 3.Neumann G, Macken CA, Karasin AI, Fouchier RA, Kawaoka Y. Egyptian H5N1 influenza viruses-cause for concern? PLOS Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002932.e1002932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grund C, Abdelwhab E-SM, Arafa A-S, et al. Highly pathogenic avian influenza virus H5N1 from Egypt escapes vaccine-induced immunity but confers clinical protection against a heterologous clade 2.2.1 Egyptian isolate. Vaccine. 2011;29(33):5567–5573. doi: 10.1016/j.vaccine.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Donis RO, Smith G, Brown IH, et al. Continuing progress towards a unified nomenclature for the highly pathogenic H5N1 avian influenza viruses: divergence of clade 2·2 viruses. Influenza and other Respiratory Viruses. 2009;3(2):59–62. doi: 10.1111/j.1750-2659.2009.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith GJD, Donis RO. Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza and other Respiratory Viruses. 2012;6(1):1–5. doi: 10.1111/j.1750-2659.2011.00298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balish AL, Davis CT, Saad MD, et al. Antigenic and genetic diversity of highly pathogenic avian influenza a (H5N1) viruses isolated in Egypt. Avian Diseases. 2010;54(1):329–334. doi: 10.1637/8903-042909-Reg.1. [DOI] [PubMed] [Google Scholar]
- 8.Cattoli G, Monne I, Fusaro A, et al. Highly pathogenic avian influenza virus subtype H5N1 in Africa: a comprehensive phylogenetic analysis and molecular characterization of isolates. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004842.e4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusaro A, Joannis T, Monne I, et al. Introduction into Nigeria of a distinct genotype of avian influenza virus (H5N1) Emerging Infectious Diseases. 2009;15(3):445–447. doi: 10.3201/eid1503.081161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capua I, Marangon S. Control and prevention of avian influenza in an evolving scenario. Vaccine. 2007;25(30):5645–5652. doi: 10.1016/j.vaccine.2006.10.053. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson KG, Colegate AE, Podda A, et al. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet. 2001;357(9272):1937–1943. doi: 10.1016/S0140-6736(00)05066-2. [DOI] [PubMed] [Google Scholar]
- 12.Terregino C, Toffan A, Cilloni F, et al. Evaluation of the protection induced by avian influenza vaccines containing a 1994 Mexican H5N2 LPAI seed strain against a 2008 Egyptian H5N1 HPAI virus belonging to clade 2.2.1 by means of serological and in vivo tests. Avian Pathology. 2010;39(3):215–222. doi: 10.1080/03079451003781858. [DOI] [PubMed] [Google Scholar]
- 13.Lee C-W, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. Journal of Virology. 2004;78(15):8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abdelwhab EM, Grund C, Aly MM, Beer M, Harder TC, Hafez HM. Multiple dose vaccination with heterologous H5N2 vaccine: immune response and protection against variant clade 2.2.1 highly pathogenic avian influenza H5N1 in broiler breeder chickens. Vaccine. 2011;29(37):6219–6225. doi: 10.1016/j.vaccine.2011.06.090. [DOI] [PubMed] [Google Scholar]
- 15.Cattoli G, Milani A, Temperton N, et al. Antigenic drift in H5N1 avian influenza virus in poultry is driven by mutations in major antigenic sites of the hemagglutinin molecule analogous to those for human influenza virus. Journal of Virology. 2011;85(17):8718–8724. doi: 10.1128/JVI.02403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen GL, Subbarao K. Attacking the flu: neutralizing antibodies may lead to “universal” vaccine. Nature Medicine. 2009;15(11):1251–1252. doi: 10.1038/nm1109-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desvaux S, Garcia JM, Nguyen TD, et al. Evaluation of serological tests for H5N1 avian influenza on field samples from domestic poultry populations in Vietnam: consequences for surveillance. Veterinary Microbiology. 2012;156(3-4):277–284. doi: 10.1016/j.vetmic.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Comin A, Toft N, Stegeman A, Klinkenberg D, Marangon S. Serological diagnosis of avian influenza in poultry: is the haemagglutination inhibition test really the “gold standard”? Influenza and Other Respiratory Viruses. 2013;7:257–264. doi: 10.1111/j.1750-2659.2012.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB. Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP) Morbidity and Mortality Weekly Report. 2005;54(8):1–40. [PubMed] [Google Scholar]
- 20.Katz JM, Lim W, Bridges CB, et al. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. Journal of Infectious Diseases. 1999;180(6):1763–1770. doi: 10.1086/315137. [DOI] [PubMed] [Google Scholar]
- 21.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature Structural and Molecular Biology. 2009;16(3):265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Anderson CM, de Feo CJ, et al. Cross-neutralizing antibodies to pandemic 2009 H1N1 and recent seasonal H1N1 influenza a strains influenced by a mutation in hemagglutinin subunit 2. PLoS Pathogens. 2011;7(6) doi: 10.1371/journal.ppat.1002081.e1002081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TT, Tan GS, Hai R, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathogens. 2010;6(2) doi: 10.1371/journal.ppat.1000796.e1000796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pica N, Hai R, Krammer F, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(7):2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steel J, Lowen AC, Wang TT, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1(1) doi: 10.1128/mBio.00018-10.e00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashem AM, Van Domselaar G, Li C, et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochemical and Biophysical Research Communications. 2010;403(2):247–251. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 27.Corti D, Suguitan AL, Jr., Pinna D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. Journal of Clinical Investigation. 2010;120(5):1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberini I, Del Tordello E, Fasolo A, et al. Pseudoparticle neutralization is a reliable assay to measure immunity and cross-reactivity to H5N1 influenza viruses. Vaccine. 2009;27(43):5998–6003. doi: 10.1016/j.vaccine.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 29.Temperton NJ, Hoschler K, Major D, et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza and Other Respiratory Viruses. 2007;1(3):105–112. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molesti E, Cattoli G, Ferrara F, Böttcher-Friebertshäuser E, Terregino C. The production and development of H7 Influenza virus pseudotypes for the study of humoral responses against avian viruses. Journal of Molecular and Genetic Medicine. 2012;7:315–320. [PMC free article] [PubMed] [Google Scholar]
- 31.Wright E, Temperton NJ, Marston DA, McElhinney LM, Fooks AR, Weiss RA. Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. Journal of General Virology. 2008;89(9):2204–2213. doi: 10.1099/vir.0.2008/000349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nature Biotechnology. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 33.Naldini L, Blömer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 34.Temperton NJ, Chan PK, Simmons G, et al. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerging Infectious Diseases. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright E, McNabb S, Goddard T, et al. A robust lentiviral pseudotype neutralisation assay for in-field serosurveillance of rabies and lyssaviruses in Africa. Vaccine. 2009;27(51):7178–7186. doi: 10.1016/j.vaccine.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demaison C, Parsley K, Brouns G, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human imunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Human Gene Therapy. 2002;13(7):803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 37.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 38.Besnier C, Takeuchi Y, Towers G. Restriction of lentivirus in monkeys. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowe T, Abernathy RA, Hu-Primmer J, et al. Detection of antibody to avian influenza a (H5N1) virus in human serum by using a combination of serologic assays. Journal of Clinical Microbiology. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temperton NJ. Molecular Biology of SARS-Coronavirus. Berlin, Germany: Springer; 2010. The use of retroviral pseudotypes for the measurement of antibody responses to SARS coronavirus; p. p. 9. [Google Scholar]
- 41.Kim J-K, Kayali G, Walker D, et al. Puzzling inefficiency of H5N1 influenza vaccines in Egyptian poultry. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(24):11044–11049. doi: 10.1073/pnas.1006419107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capua I, Alexander DJ. Animal and human health implications of avian influenza infections. Bioscience Reports. 2007;27(6):359–372. doi: 10.1007/s10540-007-9057-9. [DOI] [PubMed] [Google Scholar]
- 43.Banks J, Speidel ES, Moore E, et al. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Archives of Virology. 2001;146(5):963–973. doi: 10.1007/s007050170128. [DOI] [PubMed] [Google Scholar]
- 44.Banks J, Plowright L. Additional glycosylation at the receptor binding site of the hemagglutinin (HA) for H5 and H7 viruses may be an adaptation to poultry hosts, but does it influence pathogenicity? Avian Diseases. 2003;47:942–950. doi: 10.1637/0005-2086-47.s3.942. [DOI] [PubMed] [Google Scholar]
- 45.Webster RG. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Archives of Virology. 1997;1997(13):105–113. doi: 10.1007/978-3-7091-6534-8_11. [DOI] [PubMed] [Google Scholar]
- 46.Russell CJ, Webster RG. The genesis of a pandemic influenza virus. Cell. 2005;123(3):368–371. doi: 10.1016/j.cell.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 47.Horimoto T, Kawaoka Y. Pandemic threat posed by avian influenza A viruses. Clinical Microbiology Reviews. 2001;14(1):129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osterholm MT. Preparing for the next pandemic. New England Journal of Medicine. 2005;352(18):1839–1842. doi: 10.1056/NEJMp058068. [DOI] [PubMed] [Google Scholar]
- 49.Capua I, Alexander DJ. Avian influenza vaccines and vaccination in birds. Vaccine. 2008;26(4, supplement):D70–D73. doi: 10.1016/j.vaccine.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 50.Anderson T, Capua I, Dauphin G, et al. FAO-OIE-WHO joint technical consultation on avian influenza at the human-animal interface. Influenza and Other Respiratory Viruses. 2010;4:1–29. doi: 10.1111/j.1750-2659.2009.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferrara F, Molesti E, Böttcher-Friebertshäuser E, et al. The human transmembrane protease serine 2 is necessary for the production of Group 2 influenza A virus pseudotypes. Journal of Molecular and Genetic Medicine. 2012;7:309–314. [PMC free article] [PubMed] [Google Scholar]
- 52.Ada GL, Jones PD. The immune response to influenza infection. Current Topics in Microbiology and Immunology. 1986;128:1–54. doi: 10.1007/978-3-642-71272-2_1. [DOI] [PubMed] [Google Scholar]
- 53.Ding H, Tsai C, Zhou F, Buchy P, Deubel V, Zhou P. Heterosubtypic antibody response elicited with seasonal influenza vaccine correlates partial protection against highly pathogenic H5N1 virus. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0017821.e17821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vodeiko GM, Weir JP. Determination of H5N1 vaccine potency using reference antisera from heterologous strains of influenza. Influenza and other Respiratory Viruses. 2012;6(3):176–187. doi: 10.1111/j.1750-2659.2011.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee C-W, Senne DA, Suarez DL. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clinical and Vaccine Immunology. 2006;13(3):395–402. doi: 10.1128/CVI.13.3.395-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


