Abstract
Aberrant protein aggregation is a dominant pathological feature in neurodegenerative diseases. Protein aggregates cannot be processed by the proteasome; instead, they are frequently concentrated to a perinuclear inclusion body, the aggresome, and subsequently removed by autophagy. Paradoxically, proteasomes are also concentrated at aggresomes and other related inclusion bodies prevalent in neurodegenerative disease. Here, we show that proteasomes are crucial components in aggresome clearance. The disassembly and disposal of aggresomes requires a proteasomal deubiquitinating enzyme, Poh1, which cleaves ubiquitinated proteins and releases ubiquitin chains. In Poh1-deficient cells, aggresome clearance is blocked. Remarkably, microinjection of free lysine (K) 63-linked ubiquitin chains restores aggresome degradation. We present evidence that free ubiquitin chains produced by Poh1 bind and activate the deacetylase, HDAC6, which in turn stimulates actinomyosin- and autophagy-dependent aggresome processing. Thus, unanchored ubiquitin chains are key signaling molecules that connect and coordinate the proteasome and autophagy to eliminate toxic protein aggregates.
Introduction
Misfolded proteins resulting from genetic mutations, defective protein maturation, or environmental stress are primarily poly-ubiquitinated and degraded by the proteasome system. When proteasome activity becomes inadequate, misfolded proteins accumulate and form toxic aggregates (Tran and Miller, 1999). Aggregated proteins cannot be properly unfolded to pass through the proteolytic barrel of the proteasome and can, in fact, inhibit proteasome activity. This results in further buildup and aggregation of misfolded proteins, a pathological feature common in neurodegenerative disease (Bennett et al., 2007; Pandey et al., 2007; Snyder et al., 2003). Aggregated proteins resistant to proteasomal degradation are instead processed by autophagy where they are sequestered by autophagosomes and delivered to lysosomes for clearance (Holmberg et al., 2004; Venkatraman et al., 2004; Webb et al., 2003). Thus, autophagy acts as a compensatory degradation system when the proteasome system is impaired. How these two complementary degradative systems communicate and coordinate to dispose of toxic protein aggregates remains poorly understood.
Protein aggregates are commonly concentrated by microtubule-dependent dynein motors to a perinuclear inclusion body, the aggresome, before they are processed by autophagy (Kopito, 2000; Yao, 2010). The formation of aggresomes, which are related to Lewy bodies prevalent in neurodegenerative disease, is proposed to facilitate the autophagic clearance of toxic protein aggregates at the microtubule organizing center (MTOC), where autophagosomes and lysosomes are concentrated (Iwata et al., 2005; Lee et al., 2010; McNaught et al., 2001). Curiously, despite their inability to degrade protein aggregates, proteasomes are also accumulated at aggresomes and Lewy bodies (McNaught et al., 2002; Wigley et al., 1999). Whether these inclusion body-associated proteasomes have any function is not known. The apparent convergence of autophagy and proteasomes at these unique inclusion bodies, however, raises an intriguing possibility of a functional interaction of these two degradative systems.
Aggresome formation requires several ubiquitin-binding proteins (Reviewed in (Yao, 2010)). In this ubiquitin-dependent machinery, the protein deacetylase, HDAC6, plays a central role by regulating both the concentration and autophagic clearance of protein aggregates (Iwata et al., 2005; Kawaguchi et al., 2003; Lee et al., 2010). In a Drosophila model, transgenic expression of HDAC6 suppresses toxicity caused by proteasome deficiency, suggesting that HDAC6 is involved in the compensatory autophagy when proteasomes are impaired (Pandey et al., 2007). How HDAC6 might connect the proteasome and autophagy machinery is not known. Interestingly, HDAC6 contains an unusual ubiquitin-binding domain, the BUZ finger, which is required for association with and clearance of ubiquitinated protein aggregates (Kawaguchi et al., 2003; Lee et al., 2010). Structural determination revealed that the BUZ finger binds to ubiquitin at its C-terminal Gly-Gly (GG) residues, unlike most other ubiquitin binding domains that interact with the hydrophobic core (Ouyang et al., 2011; Pai et al., 2007). This structural feature indicates that HDAC6 specifically binds free ubiquitin chains but not ubiquitinated proteins (Ouyang et al., 2011), raising an interesting possibility that unanchored ubiquitin chains could regulate HDAC6-dependent ubiquitinated protein aggregate processing.
In this report, we present a surprising finding that the proteasome system is an integral part of the autophagy-dependent degradative machinery for protein aggregates. We provide evidence that the proteasome, via its de-ubiquitinating enzyme subunit Poh1, stimulates aggresome clearance by producing unanchored free ubiquitin chains, which bind and activate HDAC6. HDAC6, in turn, induces an actinomyosin system that promotes the de-aggregation and autophagic clearance of the aggresome. These findings establish a critical signaling function of inclusion body-associated proteasomes and identify unanchored ubiquitin chains as the missing link that connects the proteasome and autophagy.
Results
HDAC6 is required for aggresome disassembly and clearance
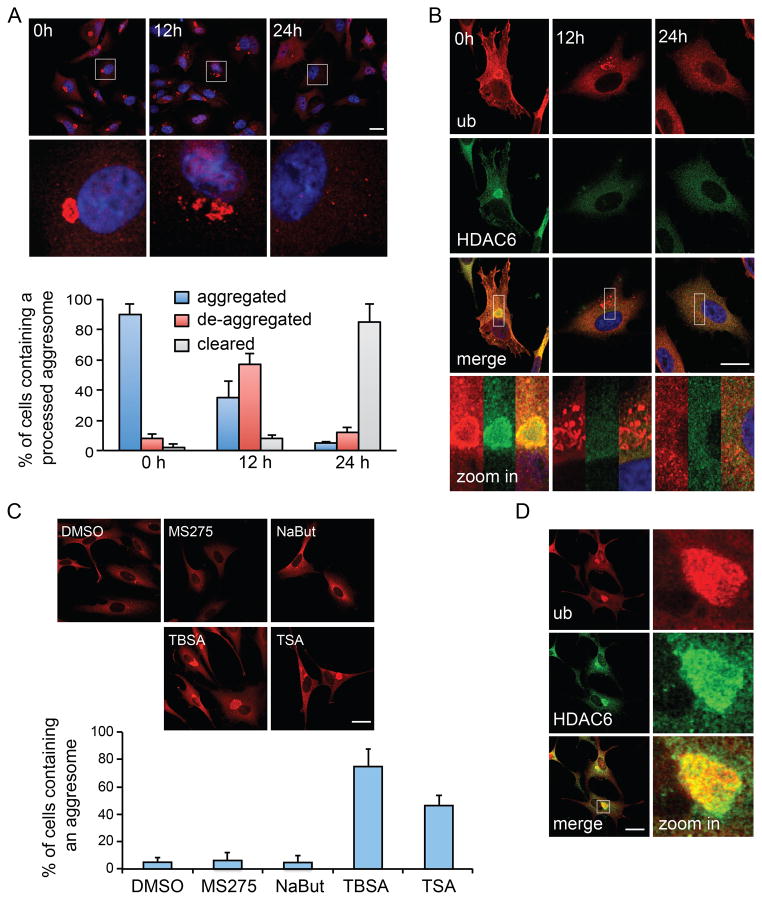
While aggresome biogenesis has been extensively characterized, how a large inclusion body is processed and becomes accessible for degradation by autophagy remains largely unknown. To investigate how aggresomes are processed, we treated cells with a proteasome inhibitor (MG132), which induced aggresome formation in >80% of cells. We then imaged aggresomes at different time points after MG132 was removed. As shown in Fig 1A, 24 hours after MG132 washout, the ubiquitin-positive aggresomes were no longer visible in the majority of cells. Staining for additional markers of protein aggregates, including p62 and α-synuclein, confirmed the loss of aggresomes (Supplemental Fig S1A–B). Further analysis indicated that aggresomes were first broken into several large fragments and then smaller micro-aggregates before they were eliminated from cells (Fig 1A and supplemental Fig S1A–B, 12h), suggesting a “de-aggregation” step that precedes final clearance by autophagy. Indeed, inhibition of autophagy by 3-methyladenine (3-MA) or bafilomycin A1 (BFA) prevented aggresome clearance and resulted in the accumulation of micro-aggregates at the perinuclear region (Supplemental Fig S1C).
Figure 1. HDAC6 is required for aggresome de-aggregation and clearance.
A549 cells were treated with MG132 (5 μM) for 24h to induce aggresome formation, followed by MG132-free medium for the indicated times. Aggresomes were identified by staining with an antibody against ubiquitin (ub, clone FK1). (A) Ubiquitin immuno-staining (red) indicates that the large aggresomes (0h) were de-aggregated (12h) before final clearance (24h). Bottom panels show zoomed areas (white squares). Nuclei were stained with DAPI (blue). The status of aggresomes at different time points after MG132 washout was quantified in the histogram. (B) Representative images of HDAC6 (green) in relationship to the aggresome (red) by double-immuno-staining during MG132 washout. Bottom panels show zoomed areas (white squares). Note that protein aggregates are no longer positive for HDAC6 after 12h washout (middle panels). (C) A549 cells were treated with an HDAC6-selective inhibitor, Tubastatin A (TBSA, 10 μM), pan-HDAC inhibitor, Trichostatin A (TSA, 1 μM), or class I HDAC inhibitors, MS275 (10 μM) or sodium butyrate (NaBut, 1mM) during MG132 washout. The presence of aggresome was analyzed and quantified as described in (A). (D) Representative images of HDAC6 and the aggresome (ub) in TBSA-treated cells. For (A) and (C), three independent experiments were quantified. Error bars show ± S.E.M. Scale bar = 25 μm. See also Figure S1.
Interestingly, while HDAC6 was initially concentrated at the aggresome, as previously reported (Kawaguchi et al., 2003), it was no longer detectable at the ubiquitin-positive structures once aggresomes de-aggregated (Fig 1B). As HDAC6 protein levels remained stable over this time course (data not shown), this finding indicates that HDAC6 dissociates from the aggresome rather than being degraded upon de-aggregation. We used a highly selective HDAC6 inhibitor, Tubastatin A (TBSA) (Butler et al., 2010), to probe the potential role of HDAC6 in aggresome processing. As shown in Fig 1C, TBSA treatment completely inhibited the de-aggregation and clearance of the aggresome. Aggresome clearance was also significantly inhibited by Trichostatin A (TSA), a pan-HDAC inhibitor that inactivates HDAC6 (Hubbert et al., 2002), while inhibitors targeting other HDAC members (MS275, Sodium Butyrate) had little effect. Interestingly, TBSA treatment also prevented the dissociation of HDAC6 from protein aggregates (Fig 1D), suggesting that HDAC6 is released from protein aggregates after its activation. These results show that HDAC6 activity is required for the disassembly and clearance of the aggresome.
Aggresome clearance requires the proteasomal deubiquitinating enzyme Poh1
Using an in vitro binding assay, we confirmed that HDAC6 purified from 293T cells binds to free ubiquitin chains but not to those conjugated to a protein (Fig 2A). This finding led us to investigate whether unanchored ubiquitin chains play a role in aggresome processing. Recently, an ubiquitin E3 ligase, TRAF6, was shown to produce unanchored ubiquitin chains (Xia et al., 2009). Knockdown of TRAF6 by a specific siRNA, however, did not have an appreciable effect on aggresome clearance (data not shown). In principle, free ubiquitin chains can also be produced by Poh1 (RPN11), a JAMM/MPN+-domain containing deubiquitinating enzyme that cleaves off ubiquitin chains en bloc from substrates targeted to the proteasome (Verma et al., 2002; Yao and Cohen, 2002). Poh1 is a subunit of the regulatory 19S proteasome, which along with the proteolytic core (20S proteasome), are concentrated at the aggresome (Supplemental Fig S2A, (Wigley et al., 1999)). We tested if Poh1 is required for aggresome clearance by siRNA-mediated knockdown. Knockdown (KD) of Poh1 (by ~ 70%) led to an accumulation of ubiquitinated proteins indicative of impaired proteasome activity (Supplemental Fig S2E); however, it did not cause aggresome formation in the absence of MG132 (Fig 2B). Importantly, the inactivation of Poh1 by four different sets of siRNA all dramatically inhibited de-aggregation and clearance of MG132-induced aggresomes (Fig 2B and Supplemental Fig S2B–D). In contrast, aggresomes were cleared normally upon knockdown of two related but non-proteasomal JAMM/MPN+ deubiquitinating enzymes, Amsh and Brcc36, or proteasomal deubiquitinating enzymes Usp14 and Uch37, which trim ubiquitin from the distal end (Supplemental Fig S3). Furthermore, the buildup of aggresomes in Poh1 KD cells was not affected by inhibiting microtubule-dependent delivery of nascent cytoplasmic aggregates to the MTOC by nocodazole during the MG132 washout period (Fig 2C). Thus, the aggresome accumulation in Poh1 KD cells is mainly due to a defect in aggresome clearance rather than an increase in aggregate formation. Importantly, the aggresome clearance defect in Poh1 KD cells was effectively reversed by the re-introduction of wild type but not a catalytically inactive (H113A/H115A) mutant Poh1 expressed at the similar level (Fig 2D, Supplemental Fig S2F) (Gallery et al., 2007). It should be pointed out that Poh1 is a stoichiometric subunit of 19S proteasomes required for proper 26S proteasome assembly and function (Verma et al., 2002). As Poh1 (H113A/H115A) mutant can fully support proteasome assembly (Verma et al., 2002), its failure to restore aggresome clearance in Poh1 KD cells indicates that deubiquitinating enzyme activity of Poh1 is required for aggresome clearance.
Figure 2. Poh1 facilitates aggresome clearance by producing unanchored ubiquitin chains.
(A) Immuno-purified FLAG-HDAC6 produced in 293T cells was incubated with unanchored ubiquitin chains (K63-linked Ub 1–7) or ubiquitinated protein (ub-protein, See Methods for details). The input and bound fractions were analyzed by immuno-blotting with the indicated antibodies. (B) A549 cells were transfected with control or Poh1-specific siRNA and treated with MG132 (5 μM, 24 hr) to induce aggresome or followed by MG132 washout as indicated. Aggresomes (marked by arrows) were detected and quantified as described in Fig 1a. Values represent the mean ± S.E.M. n = 3. (C) Poh1 knockdown (KD) cells were treated with MG132 to induce aggresomes followed by MG132 washout in the presence of DMSO or 2.5 μM of nocodazole (Noc.) for 24h. Cells containing aggresomes were quantified and averaged from three independent experiments in the histogram. Note that nocodazole treatment disrupted microtubule networks (α-tubulin, green) but had no effect on aggresome clearance. (D) A549 cells stably expressing shRNA-resistant wild type (wt) or catalytically inactive (CI) H113/115A mutant Poh1 were infected with Poh1-shRNA lentivirus. The percentage of cells containing an aggresome after MG132 washout was analyzed and quantified as described in (B). (E) Poh1 KD cells were pre-treated with MG132 to induce aggresome formation. 3h after MG132 was removed, cells were microinjected with indicated ubiquitin species or BSA mixed with fluorescence-conjugated dextran. The presence of aggresomes was analyzed 21h post-injection. Injected cells were identified by dextran (green, top panels) and marked by white dotted lines (bottom panels). Aggresomes were identified by staining with an antibody specific for K48-linked ubiquitin (red, clone Apu2) and marked by white arrowheads in injected cells. Note that only Poh1 KD cells injected with the K63-linked ubiquitin chains do not contain aggresomes. Non-injected cells retained aggresomes under all conditions (yellow arrowheads) and served as an internal control. Right panel shows the quantification from three independent experiments. 50 to 100 injected cells were scored in each experiment. *, p < 0.01. Error bars indicate ± S.E.M. Scale bar = 25 μm. (F) Control and Poh1 KD A549 cells were treated with 5 μM MG132 for 24h, or MG132 followed by 24h washout in the presence or absence of 10 mM 3-MA as indicated. Detergent insoluble fractions were isolated from whole cell lysates and resolved by SDS-PAGE, followed by immuno-blotting with a K48- or K63- specific ubiquitin antibodies as indicated. Actin is used as a loading control. See also Figure S2, S3, S4 and S8.
Poh1 activates aggresome clearance by producing unanchored K63 ubiquitin chains
If Poh1 promotes aggresome processing by producing unanchored ubiquitin chains, it would be expected that introduction of exogenous free ubiquitin chains should restore aggresome clearance in Poh1 KD cells. To test this hypothesis, we first treated Poh1 KD cells with MG132 to induce aggresome formation. After removal of MG132, we microinjected the cells with either K63- or K48- linked free ubiquitin chains, as both types of ubiquitination were associated with detergent-insoluble protein aggregates (Supplemental Fig S4A). As a control, we also injected ubiquitinated protein conjugated with K63-linked ubiquitins, which do not bind HDAC6 in vitro (Fig 2A). As shown in Fig 2E, after 24h of washout, non-injected Poh1 KD cells retained aggresomes, as expected (yellow arrowheads). Remarkably, aggresomes arrested in Poh1 KD cells were efficiently cleared after microinjection of K63-linked free ubiquitin chains. In contrast, K48-linked free ubiquitin chains did not promote aggresome clearance. Injection of proteins conjugated with K63-linked ubiquitins also failed to induce aggresome clearance, as expected. To ensure that aggresome clearance stimulated by K63-linked ubiquitin chains is not an indirect effect of supplying mono-ubiquitin upon their disassembly, we injected mono-ubiquitin in Poh1 KD cells. We found that mono-ubiquitin injection modestly increased aggresome clearance although this activity was much weaker when compared to K63-linked ubiquitin chains (Fig 2E). These results support the conclusion that Poh1 promotes aggresome clearance primarily by producing specific unanchored ubiquitin chains.
Both K63- and K48-linked ubiquitin chains associated with protein aggregates were markedly reduced after MG132 washout, concomitant with aggresome clearance (Fig 2F, compare Lane 1 and 2). We reasoned if Poh1 cleaves K63-linked ubiquitin chains to activate autophagic aggresome clearance, the decrease in K63-linked ubiquitins on protein aggregates would depend on Poh1 but not autophagy. Indeed, the loss of K63-linked ubiquitination was inhibited by Poh1 knockdown but not by the autophagy inhibitor, 3-MA (Fig 2F, Upper Panel, and Supplemental Fig S4B). In contrast, the loss of K48-linked ubiquitins was largely suppressed by 3-MA, indicating that they were degraded along with protein aggregates by autophagy (Fig 2F, Bottom Panel, and Supplemental Fig S4B). Supporting this proposition, K48-linked ubiquitins were retained in Poh1 KD cells, which are defective in aggregate clearance (Bottom Panel, Lane 4–6). Altogether, these findings indicate that Poh1 cleaves K63-linked ubiquitin chains from protein aggregates prior to their autophagic degradation.
Unanchored ubiquitin chains bind and activate HDAC6 activity
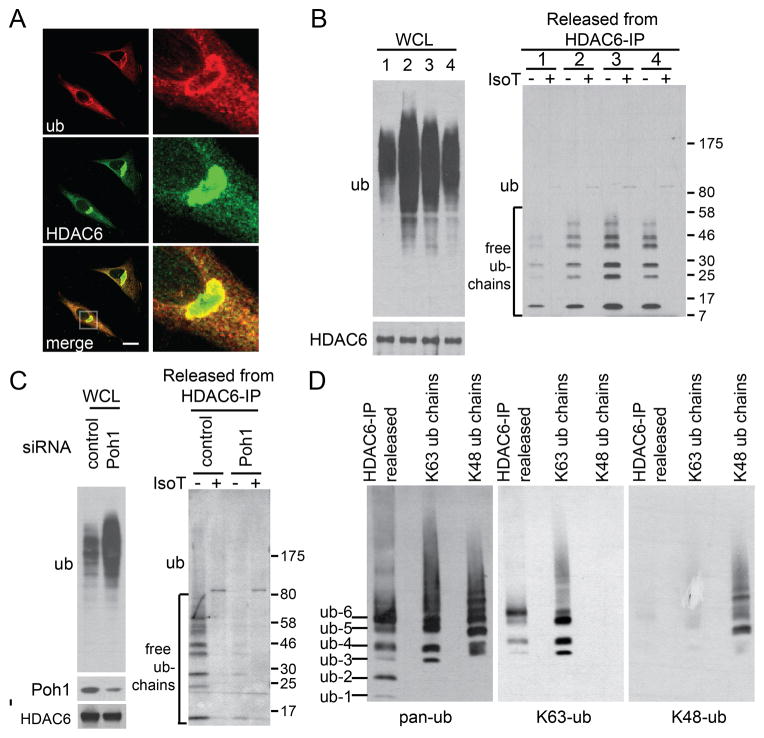
Similar to the effect of TBSA, HDAC6 did not dissociate from aggresomes in Poh1 KD cells after MG132 washout, suggesting that Poh1 is required for HDAC6 activation (Fig 3A). As HDAC6 binds unanchored ubiquitin chains in vitro (Fig 2A), we investigated if HDAC6 is a regulatory target of Poh1. We first ascertained whether HDAC6 binds endogenous free ubiquitin chains during aggresome clearance. We modified a binding assay that takes advantage of two unique properties of free ubiquitin chains (Zeng et al., 2010): resistance to heat and sensitivity to isopeptidase T (IsoT), which digests unanchored poly-Ub chains by binding to the C-terminal tail of the ubiquitin (Reyes-Turcu et al., 2006). As shown in Fig 3B, immuno-precipitation of HDAC6 indeed pulled down heat-resistant and IsoT-sensitive ubiquitin species, which are particularly prominent after MG132 washout (Fig 3B and Supplemental Fig S5, compare group 1 and 3). Most importantly, the free ubiquitin chains associated with HDAC6 were greatly diminished in Poh1 KD cells (Fig 3C), indicating that Poh1 is required for the production of free ubiquitin chains bound by HDAC6. As Poh1-dependent aggresome clearance specifically involves the generation of K63-linked ubiquitin chains (Fig 2E–F), we investigated the nature of HDAC6-associated unanchored ubiquitin chains by linkage-specific ubiquitin antibodies. As shown in Fig 3D, unanchored ubiquitin chains pulled down by HDAC6 were positive for the K63-linkage but not the K48-linkage. These findings support the idea that HDAC6 binds K63-linkage containing free ubiquitin chains produced by Poh1 during aggresome clearance.
Figure 3. HDAC6 binds unanchored ubiquitin chains during aggresome clearance in a Poh1-dependent manner.
(A) Representative image of HDAC6 and aggresome (ub) in Poh1 KD cells 24h after MG132 washout. (B) FLAG-HDAC6 was immuno-precipitated under the following conditions: 1) no treatment, 2) 5μM MG132 for 24h, 3) 12h MG132 washout, 4) 24h MG132 washout. The immune complexes were subjected to heating, and the eluates were treated with Isopeptidase T (IsoT) as indicated (see Methods for details). Samples were analyzed by immuno-blotting with antibodies for ubiquitin or HDAC6. Note that MG132 washout (conditions 2 to 3) led to a decrease in total ubiquitinated protein levels in whole cell lysate (WCL), but an increase in HDAC6-associated free ubiquitin chains. (C) Free ubiquitin chains associated with HDAC6 in wild type and Poh1 KD cells were assessed 12h after MG132 washout. Note that Poh1 KD led to the accumulation of total ubiquitinated proteins (left panel) but a reduction of HDAC6-associated unanchored ubiquitin chains (right panel). (D) Free ubiquitins chains released from HDAC6-IP components were immuno-blotted using a pan-ubiquitin antibody (pan-ub), K63-specific ubiquitin antibody (K63-ub) and K48-specific ubiquitin antibody (K48-ub) sequentially. Recombinant K63- or K48-linked poly-ubiquitin chains were loaded onto the same gel to validate the specificity of linkage-specific ubiquitin antibodies. See also Figure S5.
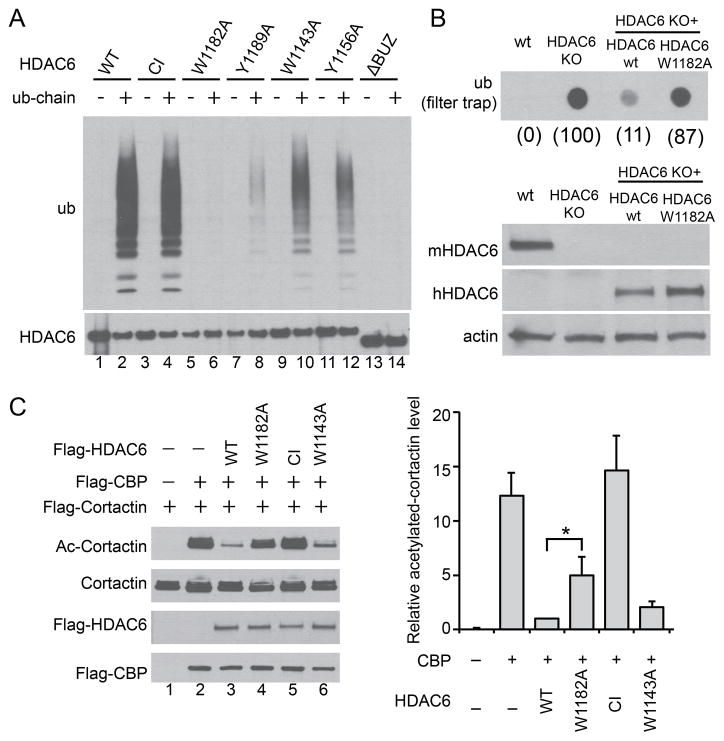
We next investigated whether ubiquitin chain binding affects HDAC6 activity. To generate a ubiquitin-chain binding deficient mutant of HDAC6, we mutated several residues in the BUZ finger that were predicted to make contacts with ubiquitin (Ouyang et al., 2011). As shown in Figure 4A, the W1182A point mutation markedly disrupted the ability of HDAC6 to bind free ubiquitin chains (Fig 4A, Lane 6). We then asked if the W1182A mutant can restore protein aggregate clearance in HDAC6 knockout fibroblasts (KO MEF) by a filter-trap assay (Lee et al., 2010). As shown in Fig 4B, HDAC6 KO MEF reconstituted with HDAC6-W1182A mutant, in contrast to those with wild type HDAC6 (wt), failed to suppress the accumulation of ubiquitinated protein aggregates. This result indicates that binding of free ubiquitin chains is important for HDAC6 to promote protein aggregate clearance. Interestingly, the level of acetylated tubulin, a key substrate of HDAC6 (Hubbert et al., 2002), was comparable between the HDAC6-W1182A mutant and wild type HDAC6 reconstituted KO MEFs (Supplemental Fig S6A). Thus microtubule deacetylation and protein aggregate clearance can be functionally uncoupled. Consistent with this observation, disruption of the microtubule network by nocodazole, which prevents aggresome formation (Johnston et al., 1998), had little effect on aggresome clearance (Supplemental Fig S6B).
Figure 4. Free ubiquitin chains regulate HDAC6 activity.
(A) Immuno-purified FLAG-HDAC6 wild type (WT), H215A/H610A catalytic inactive (CI), and various point mutants were incubated with free ubiquitin chains as indicated. The bound fractions were detected by immuno-blotting with an ubiquitin antibody. (B) Upper panel, filter-trap analysis of SDS-insoluble ubiquitinated aggregates accumulated in wild-type (wt), HDAC6 KO, and KO MEFs stably expressing human wild type HDAC6 (wt) or W1182A mutant subject to 24h MG132 washout. The relative ubiquitin signal intensity was quantified and presented in parenthesis under each genotype where HDAC6 KO MEFs was set at 100. Lower panel, whole-cell extracts from indicated cell lines were immuno-blotted using antibodies for human HDAC6 (hHDAC6), mouse HDAC6 (mHDAC6) and actin. (C) Cortactin, the acetyltransferase CBP, and wild type or HDAC6 mutant plasmids were co-transfected into 293T cells as indicated. Cortactin was immuno-precipitated followed by immuno-blotting with an antibody for acetylated (Ac) cortactin or total cortactin. The relative cortactin-acetylation level (Ac-cortactin/cortactin) was quantified by scanning densitometry and presented in the right panel. Error bars indicate ± SD (n=3). The statistical significance was assessed using two-way ANOVA analysis with Dunnett’s test. *, p < 0.05. See also Figure S6.
In addition to microtubules, HDAC6 also regulates the actin cytoskeleton (Gao et al., 2007; Zhang et al., 2007). HDAC6 promotes autophagy-dependent protein aggregate clearance, at least in part, by deacetylating and activating cortactin, an accessory factor for actin polymerization (Lee et al., 2010). We therefore investigated whether free ubiquitin chain binding modulates HDAC6 deacetylase activity toward cortactin. As shown in figure 4C, the ubiquitin-binding deficient W1182A-HDAC6 mutant is less efficient at promoting cortactin deacetylation than wild type HDAC6 in cells (Fig 4C, compare Lane 3 and 4). In contrast, the W1143A HDAC6 mutant, which retains significant ubiquitin binding activity (Fig 4A, Lane 10), possesses a near wild type cortactin deacetylase activity (Fig 4C, Lane 6). Altogether, these results indicate that full HDAC6 deacetylase activity towards cortactin requires the binding of free ubiquitin chains.
Poh1 and HDAC6 promote de-aggregation and clearance of aggresomes via an actinomyosin system
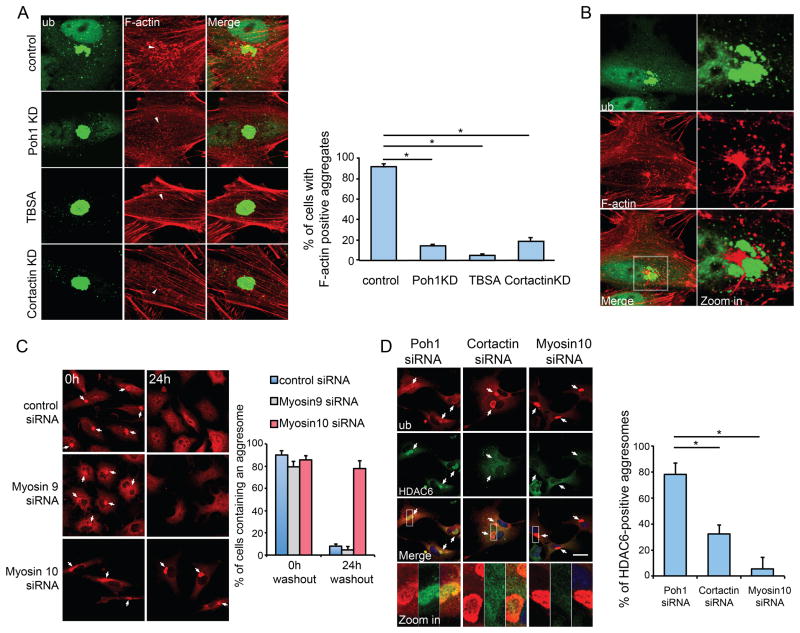
We have previously reported that HDAC6 promotes the formation of a cortactin-dependent actin network around the aggresome (Lee et al., 2010). Indeed, TBSA and cortactin siRNA treatment both inhibited F-actin network formation at the aggresome (Fig 5A). Importantly, Poh1 KD also suppressed these F-actin structures, phenocopying HDAC6 inactivation (Fig 5A). These results support the model wherein Poh1 activates HDAC6- and cortactin-dependent F-actin remodeling required for aggresome clearance. During aggresome de-aggregation, the F-actin network appeared to become interspersed among the protein aggregates (Fig 5B), raising the possibility that local actin cytoskeleton-dependent forces might be involved in the disassembly of the aggresome. Type II non-muscle myosin 9 (IIA) and 10 (IIB) are the main motors associated with the actin cytoskeleton (Vicente-Manzanares et al., 2009). We investigated whether these actin-dependent motors are involved in aggresome clearance by siRNA-mediated knockdown. As shown in Fig 5C (and Supplemental Fig S7A), knockdown of myosin 10, but not myosin 9, dramatically inhibited aggresome de-aggregation and clearance. Importantly, in contrast to Poh1 KD cells, aggresomes accumulated in myosin 10 KD cells are largely devoid of HDAC6 after MG132 washout. Similarly, HDAC6 was not retained on aggresomes in cortactin KD cells (Fig 5D). Collectively, these results are consistent with a model where HDAC6 acts as an effector downstream to Poh1 but upstream to the actinomyosin system. We conclude that proteasome-associated Poh1 activates HDAC6-dependent actinomyosin machinery to facilitate the de-aggregation and clearance of the aggresome.
Figure 5. De-aggregation of the aggresome requires the actinomyosin system.
A549 cells were pre-treated with MG132 (5μM) for 24h to induce aggresome formation. (A) Representative images of the aggresome (ubiquitin, green) and F-actin (red, Rhodamine-Phalloidin) 12h after MG132 washout in cells with indicated treatment. In the panels of F-actin, the location of the aggresome is marked by an arrowhead. Right Panel: Cells exhibiting F-actin punctae around the aggresome were quantified from three experiments. Error bars indicate ± S.E.M. *, p < 0.01. (B) Representative staining of F-actin and a de-aggregated aggresome 12h after MG132 washout. The right panel shows the zoomed areas (white squares). (C) A549 cells transfected with the indicated siNRA were imaged 0h or 24h after MG132 washout. Aggresomes (arrows) were detected with anti-ubiquitin antibody (red) and percentage of cells retained an aggresome was quantified from three independent experiments. Error bars show S.E.M. (D) Co-staining of HDAC6 (green) and ubiquitin (red) in cells expressing indicated siRNA 24h after MG132 washout. Arrows mark aggresomes. The right panel shows the quantification of aggresomes that are positive for HDAC6 (n=3). Error bars show ± S.E.M. *, p < 0.01. See also Figure S7.
Discussion
The paradoxical presence of proteasomes at the aggresome and pathological inclusion bodies has long been a mystery (McNaught et al., 2002; Wigley et al., 1999). In this report, we provided evidence that the proteasome system actively promotes de-aggregation and autophagy-dependent clearance of aggresome by producing unanchored ubiquitin chains. Our findings have uncovered a critical function of inclusion body-associated proteasomes and identified unanchored ubiquitin chains as signaling molecules that connect and coordinate the proteasome and autophagy to eliminate toxic protein aggregates.
Our studies identified the proteasomal deubiquitinating enzyme, Poh1, as a critical factor that controls the disassembly and degradation of the aggresome. In Poh1 deficient cells, aggresome clearance failed (Fig 2B). Although Poh1 is best characterized for its essential role in 26S proteasome-dependent proteolysis, several lines of evidence indicate that Poh1 activates aggresome clearance mainly by producing free K63 ubiquitin chains. First, although Poh1 inactivation likely affects multiple biological processes, microinjection of unanchored K63-linked chains was sufficient to restore aggresome clearance in Poh1 deficient cells. These results indicate that the production of unanchored K63-linked chains is key to Poh1-dependent aggresome clearance (Fig 2E). Second, proteasomal Poh1 possesses a K63-specific deubiquitinating enzyme activity (Cooper et al., 2009), which is required for activating aggresome clearance (Fig 2D). Poh1 can cleave K63-linkage, but not K48-linkage, within a ubiquitin chain containing both K63- and K48- linkages (Cooper et al., 2009). This unique endo-protease activity, in principle, could allow Poh1 to specifically cleave and release free K63-linked ubiquitin chains from protein aggregates, which are modified by both K48- and K63-linked ubiquitin chains (Fig S4A). Importantly, during aggresome clearance, protein aggregates indeed undergo Poh1-dependent K63-deubiquitination (Fig 2F), a process that could generate free ubiquitin chains. Lastly, consistent with this proposition, during aggresome clearance HDAC6 becomes associated with and activated by K63-linkage containing free ubiquitin chains, whose production depends upon Poh1 (Fig 3B–D). Although these data do not exclude the involvement of the canonical proteasomal function of Poh1, collectively they strongly indicate the production of unanchored ubiquitin chains is the main function of proteasomal Poh1 in promoting aggresome clearance. In this context, the proteasome does not function as classical proteolytic machinery; rather, it acts as a signaling complex that produces unanchored ubiquitin chains with regulatory activities.
We have identified HDAC6, a critical component of QC autophagy that degrades protein aggregates (Lee et al., 2010), as a key regulatory effector of Poh1-generated ubiquitin chains. HDAC6 binds unanchored K63-linked ubiquitin chain during aggresome clearance in wild type but not Poh1 deficient cells (Fig 3B–D). HDAC6 mutant with defective ubiquitin chain binding showed reduced activity in aggregate clearance and cortactin deacetylation, suggesting that binding to free ubiquitin chains stimulates HDAC6 activity (Fig 4). The regulation of HDAC6 by unanchored ubiquitin chains provides a simple model explaining how proteasome might communicate with autophagy machinery. We propose that proteasomal Poh1 serves to stimulate compensatory autophagy activity via HDAC6 when the proteasome activity is no longer adequate and challenged by protein aggregates. Indeed, Poh1 deficiency, similar to HDAC6 inhibition, leads to defects in aggresome de-aggregation and clearance (Fig 2) as well as autophagy maturation (Supplemental Fig S8). Interestingly, HDAC6 is also known to associate with another deubiquitinating enzyme, ataxin 3. Evidence suggests that ataxin 3 recruits HDAC6 to protein aggregates to facilitate aggresome formation (Burnett and Pittman, 2005; Ouyang et al., 2011). Indeed, in ataxin 3-deficient cells, HDAC6 no longer associates with ubiquitinated protein aggregates (Supplemental Fig S7B). Thus, HDAC6 appears to pair with two different deubiquitinating enzymes to control aggresome formation and clearance, respectively. Whether ataxin 3 produces unanchored ubiquitin chains and activates HDAC6 remains to be tested.
Unanchored ubiquitin chains were recently identified as key mediators that activate RIG-1 and TAK1 in response to cytokines (Xia et al., 2009; Zeng et al., 2010). The critical role of unanchored ubiquitin chains in aggresome clearance suggests that these unique ubiquitin species likely have a much broader regulatory function. Our data support K63-linked ubiquitin chains as the key mediators in activating HDAC6 and aggresome clearance although the involvement of other ubiquitin linkages cannot be excluded. We do not know how binding of K63-linked or other ubiquitin chains modulates HDAC6 activity. Structural study of HDAC6 with different ubiquitin chains would be needed to answer this question. Further studies would also be required to determine whether the length of the ubiquitin chain is critical. It is worth noting that ubiquitin chains with different linkage show differential activities in activating RIG-1 and TAK1 (Xia et al., 2009; Zeng et al., 2010). Thus, unanchored ubiquitin chains with different linkages could have distinct regulatory activities.
The unique dependence on K63-linked ubiquitin chains in aggresome clearance is intriguing, as this modification is not normally linked to proteasome-mediated degradation. However, we found that MG132 treatment led to a dramatic increase in K63-linked ubiquitination associated with protein aggregates (Supplemental Fig S4A). Thus, protein aggregates are likely tagged by mixed ubiquitin chains of both K48- and K63- linkage. These observations are of potential significance, as JAMM/MPN+-containing deubiquitinating enzymes, including Poh1, can cleave K63- but not K-48 linked ubiquitin chains in vitro (Cooper et al., 2009). We speculate that the addition of K63-linked ubiquitin chains to aggregated proteins creates substrates for Poh1, which subsequently cleaves and produces unanchored K63-linked ubiquitin chains that activate HDAC6 and autophagic-dependent aggresome clearance. Thus, ubiquitins associated with protein aggregates are not simply leftover marks on the misfolded proteins originally tagged for proteasomal degradation; rather, they might constitute a new ubiquitin code for aggresome-associated autophagy. Identifying the ligase responsible for K63-linked ubiquitination on protein aggregates and determining how Poh1 activity is regulated toward these unusual substrates are two critical issues to be addressed in the future.
It is widely assumed that inclusion bodies arise in neurodegenerative disease due to an impaired proteasomal system that fails to degrade misfolded proteins. Our study suggests that proteasome deficiency could also cause a failure to process and remove inclusion bodies. This dual mechanism could explain the prevalence of inclusion bodies in Parkinson’s disease, where proteasome deficiency has been documented (McNaught et al., 2001). Our study also shows that exogenous free ubiquitin chains can stimulate aggresome clearance in Poh1-deficient cells. The possibility that free ubiquitin chain-mimetics might activate inclusion body clearance offers a potential therapeutic solution for neurodegenerative disease.
Experimental Procedures
Cell fractionation
Cells were lysed in Triton buffer containing 50 mM Tris-Cl pH 7.4, 150 mM NaCl, 1% [v/v] Triton-X100, 1 mM EDTA, 1 mM DTT, 1mM Na3VO4, 0.1mM NaF and a cocktail of protease inhibitors by mixing for 20 min at 4 °C. Lysates were centrifuged at 12,000 g for 30 min. The supernatants were used as detergent-soluble fractions. The pellets were suspended in a buffer containing 50 mM Tris-Cl pH 7.4 plus 2% SDS, sonicated briefly, and analyzed as detergent-insoluble fractions.
Immuno-fluorescence and F-actin staining
Cells cultured on glass coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.2% [v/v] Triton-X100 in PBS, blocked with 5% BSA in PBS containing 0.1% Triton-X100, and incubated with antibodies, as described previously (Kawaguchi et al., 2003). F-actin staining was performed by incubating cells with Rhodamine-conjugated Phalloidin in PBS for 30 min (Gao et al., 2007). Images were taken using a Leica SP5 confocal microscope.
Microinjection
Microinjection was performed on an Olympus IX-70 inverted microscope equipped with an Eppendorf FemtoJet and TransferMan NK2 microinjection system. A549 cells transfected with Poh1 siRNA were cultured on poly-L-lysine coated glass coverslips, treated with 5 μM MG132 for 24 hours followed by three hours of MG132-free fresh medium prior to the injection. PBS solution containing 2 μg/μl of BSA control, K63-linked poly-ubiquitin chains, K63-ubiquitinylated protein, or K48-linked poly-ubiquitin chains was microinjected into the cytoplasm using holding pressure. About 0.2 pg proteins were injected per cell. Injected cells were incubated in fresh full growth medium at 37°C for another 21 hours before analysis. 70 to 100 cells were injected in each experiment with three independent experiments.
Detection of HDAC6-associated free ubiquitin chains
The method to detect endogenous unanchored ubiquitin chains is modified from a previous report (Zeng et al., 2010). A549 cells stably expressing FLAG-HDAC6 were cultured in 15 cm dishes and treated under different conditions. Cellular confluence was allowed to reach ~85% prior to collection. The cells were lysed in NETN lysis buffer (170 mM NaCl, 20 mM Tris-Cl pH 8.0, 0.5 mM EDTA, 0.5 % [v/v] NP-40, 1 mM Na3VO4, 0.1 mM NaF) supplemented with 10 mM NEM, 20 μM MG132, protease inhibitor cocktail and a phosphatase inhibitor cocktail (Sigma-Aldrich). After centrifugation at 16000 g for 15 min, 5 mM DTT was added to the supernatant to quench the NEM. FLAG-HDAC6 and associated proteins were isolated using anti-FLAG M2 agarose (Sigma-Aldrich). The beads were washed three times with NETN buffer and then incubated in 40 μl Buffer F (20 mM HEPES-KOH at pH 7.0, 10% [v/v] glycerol, and 0.02% [w/v] CHAPS) at 77 °C for 5 min. Following centrifugation at 11000 g for 10 min, the supernatant was incubated with or without 100 nM IsoT at 30 °C for 30 min, resolved on a 4–20% gradient SDS-PAGE, transferred to nitrocellulose membranes and probed with an ubiquitin antibody. The quantification of the relative ubiquitin signal was obtained by scanning blot densitometry. To identify the linkage of HDAC6-bound ubiquitin chains, four 15 cm dishes of A549-FLAG-HDAC6 cells were collected after 12h MG132 washout and analyzed as described above with a few changes. The washed beads were incubated in 160 μl Buffer G (20 mM HEPES-KOH at pH 7.0, 2% [v/v] glycerol, and 0.02% [w/v] CHAPS) at 75 °C for 5 min to release unanchored ubiquitin chains. After centrifugation, 120 μl of supernatant was collected and lyophilized to ~15 μl followed by immune-blotting.
Ubiquitin binding assay
293T cells overexpressing FLAG-HDAC6 were lysed in RIPA buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% Sodium Deoxycholate, 1.0% [v/v] NP-40), and HDAC6 was immuno-precipitated using M2 beads. The beads were washed with RIPA buffer, resuspended in buffer B (100 mM NaCl, 20 mM Tris-Cl pH 7.4, 0.5 % [v/v] NP-40, 2% glycerol) and aliquoted into two parts, each containing ~10 pmol immobilized HDAC6 protein. The HDAC6-beads were incubated with 300 μl buffer B containing either ~ 70 pmol unanchored ubiquitin chains (Ub1–7) or ~ 70 pmol poly-ubiquitinylated protein (calculated with an estimated average molecular weight of 75 KD) at room temperature for 30 min. Unbound proteins were removed by centrifugation and washing. Proteins retained were eluted by electrophoresis sample buffer and tested by western blotting.
Cortactin deacetylation assays
For deacetylation in vivo, 293T cells were transfected with FLAG-tagged cortactin, acetyltransferase CBP and either wild type or mutant FLAG-HDAC6 expressing plasmids. The ratio of cortactin:CBP:HDAC6 plasmids was 8:8:1. Cell lysates were immuno-precipitated with FLAG-M2 beads, and the precipitated material was immuno-blotted with antibody against acetylated lysine and antibody against cortactin.
Supplementary Material
Highlights.
Proteasomes activate aggresome clearance by producing free ubiquitin chains
Proteasomal Poh1 produces free ubiquitin chains that bind and activate HDAC6
HDAC6 activates an actinomyosin system that facilitates aggresome clearance
Free ubiquitin chains functionally connect the proteasome and autophagy
Acknowledgments
We thank Drs. L. Sun, and J.Y. Lee and for technical advice on detecting unanchored ubiquitin chains and analyzing aggresomes, and Dr. Randall Pittman for providing ataxin 3 knockout fibroblasts. We thank Dr. F. Melandri for advice on ubiquitin chains. We thank Drs. X. Dong, B. Mathey-Prevot, D. Thiele, K.L. Norris and Ms. M. Woods and Mrs. A. McClure for critical reading of the manuscript. This work was supported by 2R01-NS054022 (NIH) to T.-P.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Pittman RN. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc Natl Acad Sci U S A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. Journal of the American Chemical Society. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallery M, Blank JL, Lin Y, Gutierrez JA, Pulido JC, Rappoli D, Badola S, Rolfe M, Macbeth KJ. The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Molecular cancer therapeutics. 2007;6:262–268. doi: 10.1158/1535-7163.MCT-06-0542. [DOI] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Lu J, Lee YS, Lee JY, Yao TP. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Molecular and cellular biology. 2007;27:8637–8647. doi: 10.1128/MCB.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. The Journal of biological chemistry. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat Rev Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Shashidharan P, Perl DP, Jenner P, Olanow CW. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16:2136–2148. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- Ouyang H, Ali YO, Ravichandran M, Dong A, Qiu W, MacKenzie F, Dhe-Paganon S, Arrowsmith CH, Zhai RG. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. The Journal of biological chemistry. 2011;287:2317–2327. doi: 10.1074/jbc.M111.273730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai MT, Tzeng SR, Kovacs JJ, Keaton MA, Li SS, Yao TP, Zhou P. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J Mol Biol. 2007;370:290–302. doi: 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. The Journal of biological chemistry. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- Tran PB, Miller RJ. Aggregates in neurodegenerative disease: crowds and power? Trends Neurosci. 1999;22:194–197. doi: 10.1016/s0166-2236(99)01409-5. [DOI] [PubMed] [Google Scholar]
- Venkatraman P, Wetzel R, Tanaka M, Nukina N, Goldberg AL. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, 3rd, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. The Journal of biological chemistry. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao TP. The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer. 2010;1:779–786. doi: 10.1177/1947601910383277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.