Abstract
Recessive mutations in MEGF10 are known to cause a congenital myopathy in humans. Two mutations in the extracellular EGF-like domains of MEGF10, C326R and C774R, were associated with decreased tyrosine phosphorylation of MEGF10 in vitro. Y1030 was identified to be the major tyrosine phosphorylation site in MEGF10 and is phosphorylated at least in part by c-Src. Overexpression of wild-type MEGF10 enhanced C2C12 myoblast proliferation, while overexpression of Y1030F mutated MEGF10 did not. We conclude that MEGF10-mediated signaling via tyrosine phosphorylation helps to regulate myoblast proliferation. Defects in this signaling pathway may contribute to the disease mechanism of MEGF10 myopathy.
Keywords: MEGF10, myopathy, tyrosine phosphorylation
1. Introduction
Mutations in MEGF10 were recently found to cause an autosomal recessive skeletal muscle disease [1–3]. MEGF10 myopathy patients experience progressive congenital muscle weakness and respiratory failure, without any signs of central nervous system dysfunction [2,3]. MEGF10 is expressed in satellite cells of skeletal muscle [4]. Satellite cells are muscle stem cells that play a key role in muscle growth and regeneration. Satellite cells normally remain quiescent, but activate upon muscle injury or exercise and undergo asymmetrical division, leading to self-renewal of the stem cell population and production of myogenic cells that differentiate into new muscle fibers. Satellite cells were found to be depleted in the skeletal muscle tissue of a MEGF10 myopathy patient [2]. Concordantly, it was reported that MEGF10 overexpression in C2C12 myoblasts enhances cell proliferation and that knockdown of MEGF10 in muscle fibers leads to a reduction of satellite cells due to premature differentiation [4]. MEGF10 is also highly expressed in the central nervous system [5]. In neuronal tissue, MEGF10 has been reported to play roles in neuronal cell engulfment [6,7], amyloid-β protein uptake [8] and retinal neuron spacing [9].
MEGF10 is a single transmembrane protein that has 17 EGF-like domains in the extracellular N-terminus and a C-terminal cytoplasmic domain with 13 tyrosine residues that may be involved in signal transduction [10]. MEGF10 has two mammalian homologues, MEGF11 and MEGF12 (also known as PEAR1 and Jedi-1), and one orthologue in Drosophila melanogaster, draper. Tyrosine phosphorylation of MEGF11 and MEGF12 has been reported to contribute to platelet activation [11,12] and glial phagocytosis of apoptotic neurons [6,7,13], and tyrosine phosphorylation signaling of draper is important for phagocytic activity [13,14]. Recently, it has also been reported that MEGF10 and MEGF12 are tyrosine phosphorylated and regulate phagocytosis of apoptotic neurons via the Src family kinase-Syk pathway [7]; similar phosphorylation and regulation patterns have been reported for draper [11,14]. We hypothesized that MEGF10 participates in a signaling pathway that is important for satellite cell function. However, there have been no studies of MEGF10 signaling in muscle and little is known about the mechanism of disease for MEGF10 myopathy.
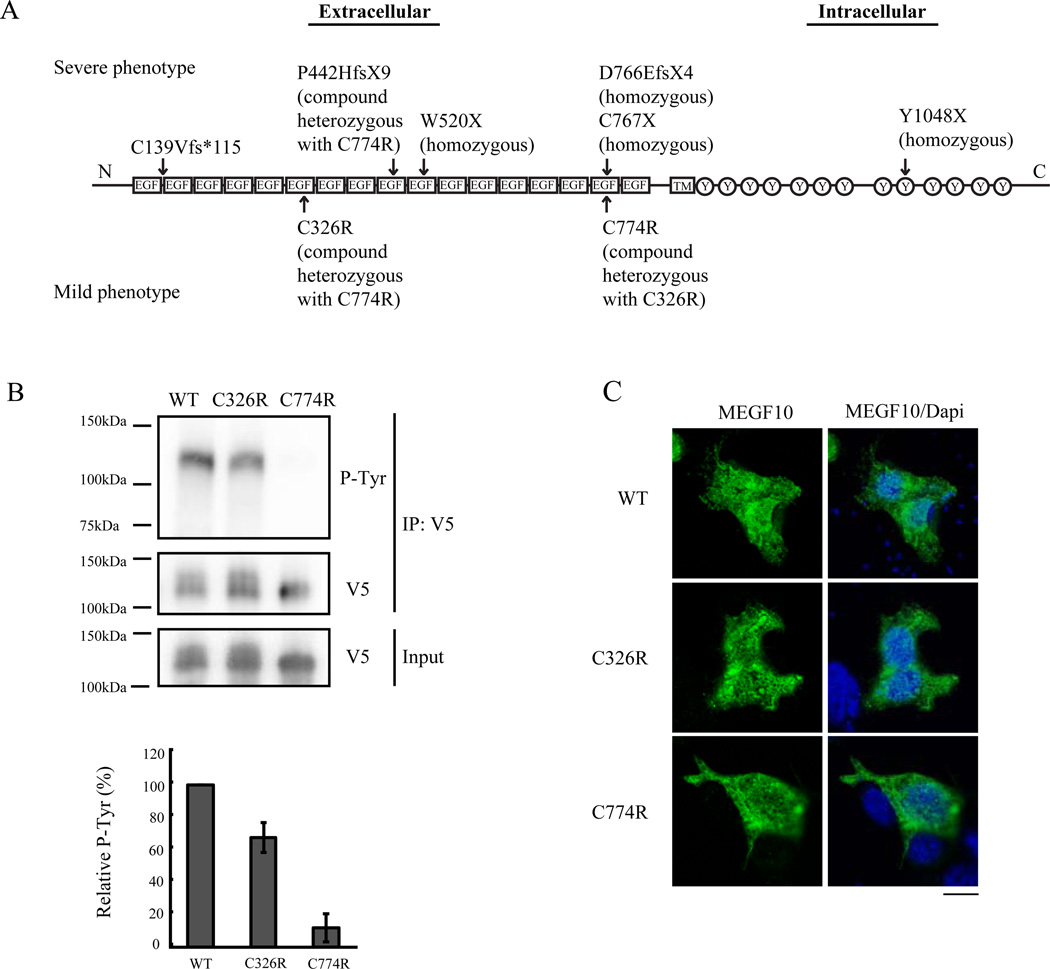
To date, MEGF10 myopathy has been reported in 7 families from different populations (Fig 1A) [2,3,15]. First, five families were reported to possess various homozygous nonsense mutations or compound heterozygous nonsense and missense mutations, all showing severe phenotypes [2]. In one of these severely-affected families, a heterozygous C774R mutation was paired with a heterozygous P442HfsX9 nonsense mutation. Three of the patients died in infancy due to respiratory complications. Later, we reported a sixth family with a milder phenotype who harbored the compound heterozygous missense mutations C326R and C774R, located at the second cysteine of the 6th EGF-like domain and the 4th cysteine of the 16th EGF-like domain, respectively [3]. The three affected individuals in this family are now in their third decade, and one is still ambulatory. They have milder respiratory complications than the initial cohort of 5 families, only requiring non-invasive nocturnal ventilatory support. These observations suggest that the C774R mutation is as deleterious as a nonsense mutation, while the C326R mutation may be less consequential. Recently, a seventh family with a homozygous frameshift deletion of exon 7 in MEGF10 was reported to display a severe phenotype [15].
Figure 1. Tyrosine phosphorylation in C326R and C774R mutant MEGF10.
(A) Diagram of the MEGF10 protein domains with arrows indicating previously reported human patients’ mutations [2,3,15]. Upper mutations are from individuals with severe disease phenotypes. Lower mutations are from three patients in one family with a mild phenotype. Note that the C774R mutation has been associated with both severe and mild phenotypes. EGF: EGF-like domain, TM: transmembrane domain, Y: tyrosine residue. (B) HEK293T cells were transfected with wild type, C326R mutant, and C774R mutant MEGF10 tagged with V5. Cell lysates were immunoprecipitated with anti-V5 antibody, then subjected to immunoblotting with anti-phosphotyrosine (P-Tyr) antibody or anti-V5 antibody. The C774R mutant shows a greater defect in tyrosine phosphorylation than the C326R mutant. IP V5: V5 tagged immunoprecipitated lysates. Western blot densitometry of the bands shows decreased tyrosine phosphorylation in C326R and C774R. Quantitative analysis was normalized against intensities of immunoprecipitated V5 bands (data are relative to wild type MEGF10, n=5). (C) Wild type, C326R mutant, and C774R mutant MEGF10 constructs were transfected into 293T cells. One day after transfection, cells were fixed and stained with anti-V5 antibody (green). Representative cells are shown. The left column shows MEGF10 staining with anti-V5 antibody. The right column shows merged images of DAPI-labeled nuclei (blue) with the images on the left. Mutant proteins show the same subcellular localization pattern as the wild type proteins. Scale bar: 10µm.
The current study investigates the tyrosine phosphorylation signaling of MEGF10, using constructs representing the C326R and C774R mutations and tyrosine phosphorylation deficient mutations. MEGF10 tyrosine phosphorylation contributes to MEGF10-induced muscle cell proliferation, potentially explaining the loss-of-function mechanism of this disease.
2. Material and Methods
2.1. Construction of expression vectors for MEGF10 and mutants
The V5-tag sequence was inserted into XhoI- and XbaI-sites in pCS2(+). The human MEGF10 cDNA was cloned by PCR using the TOPO TA Cloning Kit (Life Technologies Corporation) then subcloned into pCS2(+)-V5. Human c-Src cDNA was generously provided by Professor Shoichi Ishiura (The University of Tokyo, Tokyo, Japan). The Myc-tagged c-Src was generated by subcloning c-Src into pcDNA3.1-Myc vector (Life Technologies Corporation). Various MEGF10 mutants, including C326R, C774R, Y1030F and Y1030D, deletion mutants, and the kinase-inactive dominant negative variant of c-Src (K298R) [16] were generated by site-directed mutagenesis as previously described [17]. Primers used in this study are listed in Supplemental Table 1.
2.2. Cell culture
Human embryonic kidney (HEK) 293T cells (GenHunter Corporation) and mouse myoblast C2C12 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies Corporation) supplemented with 10% and 20% fetal bovine serum (FBS, Atlanta Biologicals) respectively, penicillin (100 units/ml, Sigma), and streptomycin (100 µg/ml, Sigma). All cells were maintained in a 5% CO2 incubator at 37°C.
2.3. Immunoprecipitation
The constructs were transfected into HEK293T cells or C2C12 cells using Lipofectamine 2000 reagent (Life Technologies Corporation) and Fugene HD (Promega), respectively, according to the manufacturers’ protocols. Cells were collected 24 hours after transfection and lysed in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM Na3VO4) containing a Complete Mini EDTA-free Protease Inhibitor Cocktail tablet (Roche). Protein was immunoprecipitated by either anti-V5 monoclonal antibody (Life Technologies Corporation) or anti-Myc monoclonal antibody (Life Technologies Corporation) using Protein G Sepharose 4 Fast Flow (GE Healthcare Life Science). Immunoprecipitants were eluted with SDS-PAGE sample buffer (125 mM Tris-HCl pH 6.8, 5% 2-mercaptoethanol, 2% SDS, 10% glycerol, 0.01% bromophenol blue), boiled at 100°C for 5 minutes and subjected to western blot analysis. NuPage Tris-acetate 8–10% gels (Life Technologies Corporation) were used for the assays. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore), then blocked with 5% Non-fat dry milk (Labscientific) or 2.5% bovine serum albumin in Tris-buffered saline (25 mM Tris-HCl pH 7.4, 137 mM NaCl, 2.7 mM KCl) containing 0.05% Tween 20 (Sigma). For detection of the V5 and Myc tags, the proteins were incubated overnight at 4°C using anti-V5 and anti-Myc antibodies at a 1:2000 dilution. Tyrosine phosphorylation of MEGF10 was detected by incubation overnight at 4°C using PY20 antibody (BD Bioscience) at a 1:2000 dilution. After incubation for 1 hour at room temperature with horseradish peroxidase (HRP, Life Technologies Corporation) conjugated anti-mouse IgG antibody, the bands were visualized by Immobilon Western Chemiluminescence HRP Substrate (Millipore) and analyzed by Universal Hood II Gel Imager (Bio-Rad Laboratories).
2.4 Immunofluorescence analysis
C2C12 and 293T cells were grown on plastic slide chambers and transfected with wild type MEGF10, C326R mutants, or C774R mutants using Lipofectamine 2000 (Life Technologies Corporation). The cells were washed with phosphate-buffered saline (PBS), fixed for 10 min in 4% paraformaldehyde at room temperature, then permeabilized with 0.25% Triton-X in PBS. After blocking with 5% goat serum in 2% BSA, the cells were incubated at room temperature with anti-V5 antibody for 1 hour. After 4 washes with PBS, the cells were incubated with Alexa-488 conjugated goat anti-mouse antibody with 4’ 6-diamidino-2-phenylindole (DAPI). Confocal images were obtained using an LSM 700 laser scanning confocal microscope (Carl Zeiss) with a 63× 1.4 NA objective using Zen image acquisition software (Carl Zeiss). Images were processed using Image J software and Adobe Photoshop (Adobe Systems).
2.5. Lentiviral vectors
V5-tagged wild type MEGF10, Y1030F mutant MEGF10, and Y1030D mutant MEGF10 were subcloned from pCS2(+)-V5 vectors into lentiviral expression vectors (pCDH-CuO-MCS-EF1-IRES-GFP; System Biosciences Inc.) using XbaI and NotI sites. Lentiviral vector production and infection were performed as previously described [18]. Briefly, lentiviral vectors along with third-generation lentiviral packaging plasmids (MDL/RRE, Rev and VSV-G) were transfected into HEK293T cells that were grown in 100 mm dishes in DMEM containing 10% FBS using Lipofectamine 2000 reagent (Life Technologies Corporation) according to the manufacturer’s protocol. Lentivirus-containing media were filtered through 45 µm filters and concentrated by 40% PEG solution, then pelleted by centrifugation at 2600 g for 45 minutes at 4°C. Lentiviral infection was done at a viral titer of more than 1×109 TU/ml.
2.6.Cell proliferation assay
C2C12 myoblasts were infected 1 day after plating with lentiviral vectors expressing wild type MEGF10, Y1030F mutant MEGF10, Y1030D mutant MEGF10, or empty vector and subjected to cell proliferation assays. C2C12 myoblasts were seeded at 12,500 cells in 60 mm dishes. The number of cells over an approximately 1 cm2 growth area was counted in 4 randomly selected areas at 24, 48 and 72 hours after plating. To avoid the effect of contact inhibition, cell counts were performed prior to confluence. Numbers of viable cells were also assessed 48 hours after plating by the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer’s protocol.
2.8. Statistical analysis
Data are presented as mean ± standard deviation. Mean differences were statistically analyzed by t-test using R software version 2.11.0.
3. Results
3.1. The C774R mutation is associated with greater impairment of MEGF10 tyrosine phosphorylation than the C326R mutation
Most of the disease-causing mutations in MEGF10 reported to date are nonsense mutations [2,15], but we have also reported two missense mutations, C326R and C774R, in the extracellular domain of MEGF10 (Fig. 1A) [3]. To determine whether tyrosine phosphorylation status of MEGF10 is affected by these mutations, the C326R and C774R mutants were overexpressed in HEK293T cells and C2C12 cells. Mutant proteins were immunoprecipitated with anti-V5 antibodies and subjected to Western blot analysis with anti-phosphotyrosine antibodies (PY20) to assess tyrosine phosphorylation status. In both cell lines, wild type MEGF10 protein was tyrosine-phosphorylated. Tyrosine phosphorylation was severely decreased in the C774R mutant and mildly decreased in the C326R mutant (Fig. 1B, Supplementary Fig. 1A). To test for the possibility that this tyrosine phosphorylation involves a different protein that interacts with MEGF10 and has a similar molecular weight, we performed reciprocal immunoprecipitation using anti-phosphotyrosine (PY20) antibody. MEGF10 was immunoprecipitated with anti-phosphotyrosine antibody, confirming that MEGF10 is the protein that is tyrosine phosphorylated (Supplementary Fig. 2).
Wild type MEGF10 showed a broad band in a 3–8% gradient gel, suggesting the presence of glycosylation (Fig. 1B). C774R mutant MEGF10 showed a thinner band compared to the wild type protein, suggesting that the defective tyrosine phosphorylation associated with the C774R mutation may be related to reduced glycosylation. Overexpressed wild type and C774R mutant MEGF10 were treated with N-glycosidase. The treated proteins showed similarly sized bands, migrating at ~100kDa, also suggesting that C774R has less N-glycosylation (Supplementary Fig. 3).
We analyzed the subcellular localization of MEGF10 in HEK293T and C2C12 cells to evaluate the possibility that either the C326R or C774R mutation alters protein localization. MEGF10 has been shown to localize to the plasma membrane [19]. Immunofluorescent staining of V5 tagged-MEGF10 shows a lattice-like pattern at the adherent surface between the cells and the tissue culture dish, in concurrence with a previous report [19]. Mutant C326R and C774R MEGF10 display the same subcellular localization patterns as wild type MEGF10 in both HEK293T and C2C12 cells, suggesting that the phosphorylation defects of these mutant MEGF10 proteins is not due to aberrant subcellular localization (Fig. 1C, Supplementary Fig.1B).
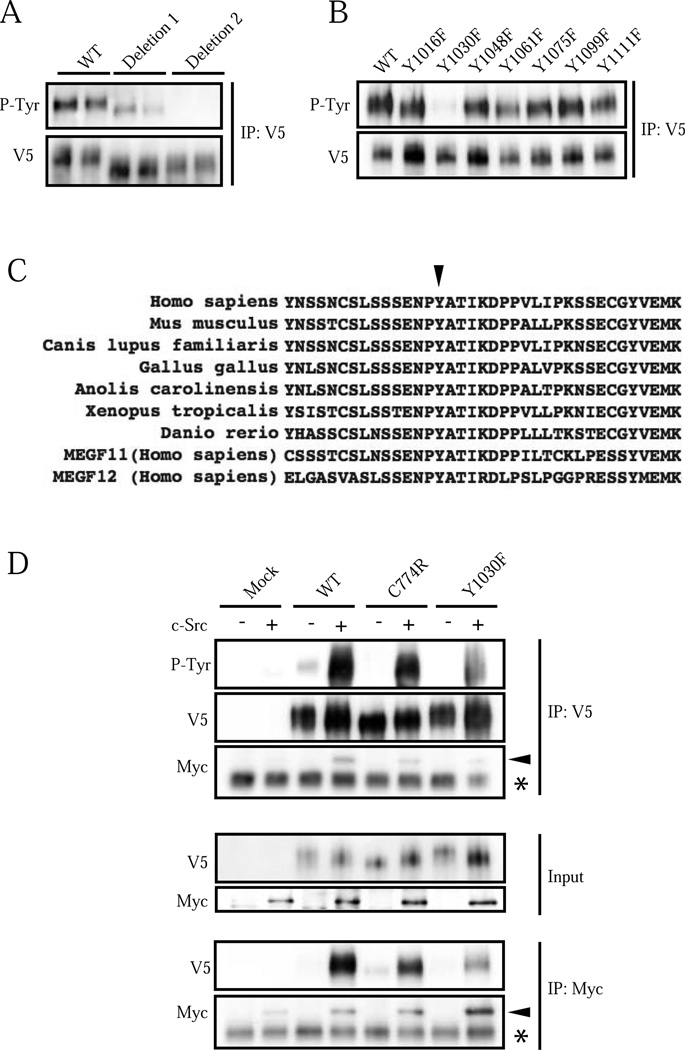
3.2. MEGF10 is tyrosine phosphorylated at Y1030 by c-Src
MEGF10 has multiple potential tyrosine phosphorylation sites in its cytoplasmic domain. It is not fully understood which tyrosine residues may be involved in phosphorylation signaling. To identify the phosphorylation site(s), two MEGF10 deletion mutants transfected into HEK293T cells were initially screened. The MEGF10-deletion 1 mutant with a deletion from the Y879 to Y1002 amino acid residues showed slightly decreased tyrosine phosphorylation as compared to wild type MEGF10. On the contrary, the MEGF10-deletion mutant 2 with a deletion of the Y1016 to Y1099 amino acid residues showed complete loss of tyrosine phosphorylation (Fig. 2A). This indicated that MEGF10 has an important tyrosine phosphorylation site between Y1016 and Y1099. A series of tyrosine-to-phenylalanine mutants in this range were generated and then overexpressed in HEK293T cells. Among these, only the Y1030F mutant lost tyrosine phosphorylation, suggesting that Y1030 is a major tyrosine phosphorylation site for MEGF10 (Fig. 2B). MEGF10 phosphorylation was also ablated when the Y1030F mutant was expressed in C2C12 myoblasts (Supplementary Fig. 1A). Y1030 is well conserved in many vertebrate species and has comparable residues in MEGF11 (Y944) and MEGF12/PEAR1/Jedi-1 (Y925) (Fig. 2C).
Figure 2. MEGF10 is tyrosine phosphorylated at Y1030 and c-Src phosphorylates MEGF10.
(A) MEGF10 deletion mutant 1 (deleted from Y879 to Y1002) and deletion mutant 2 (deleted from Y1016 to Y1099) were transfected into HEK293T cells. Cell lysates were immunoprecipitated with anti-V5 antibody, then subjected to immunodetection with anti-P-Tyr or anti-V5 antibodies. Experiments were run and are shown in duplicate. (B) Tyrosine-to-phenylalanine mutants were transfected into HEK293T cells. The Y1030F mutant shows marked decrease of tyrosine phosphorylation while the other mutants displayed normal patterns, suggesting that Y1030 is the major tyrosine phosphorylation site of MEGF10. IP V5: V5 tagged immunoprecipitated lysates. (C) Y1030 is highly conserved in vertebrates (arrowhead). This tyrosine residue is also conserved in human MEGF11 and MEGF12/PEAR1/Jedi-1 protein sequences. (D) Wild type MEGF10, C774R, Y1030F or pCS2(+)-V5 (mock) were co-transfected with Myc-tagged c-Src or pcDNA3.1-Myc (c-Src negative) into HEK293T cells, and then immunoprecipitated with anti-V5 or anti-Myc antibodies. The C774R mutant shows weaker tyrosine phosphorylation in the setting of c-Src overexpression compared to wild type MEGF10. The Y1030F mutant MEGF10 shows less tyrosine phosphorylation compared to wild type or C774R mutant MEGF10 when co-expressed with c-Src. Myc-tagged c-Src is co-immunoprecipitated with MEGF10. Note the decreased binding in the C774R and Y1030F mutants (arrowhead indicates Myc-tagged c-Src). IP V5: V5 tagged immunoprecipitated lysates. WT: wild type MEGF10. *: IgG.
The MEGF10 homologue MEGF12/PEAR1/Jedi-1 is phosphorylated by c-Src at Y925 [11]. To further characterize the MEGF10 tyrosine phosphorylation signaling mechanism, we co-transfected wild type MEGF10 with Myc-tagged c-Src, dominant negative kinase-dead c-Src (c-Src K298R) or empty vector control into HEK293T cells. Wild type MEGF10 co-expressed with c-Src showed a marked increase in tyrosine phosphorylation compared to wild type MEGF10 co-expressed with empty vector (Fig. 2D, Supplementary Fig. 4). In addition, co-expression of wild type MEGF10 with dominant negative c-Src demonstrated a loss of tyrosine phosphorylation compared to wild type MEGF10 co-expressed with empty vector control (Supplementary Fig. 4). These results indicate that overexpressed MEGF10 is phosphorylated by c-Src.
Wild type, C774R mutant, or Y1030F mutant MEGF10 was then co-transfected with Myc-tagged c-Src into HEK293T cells. Co-expression of human c-Src showed markedly enhanced phosphorylation of wild type MEGF10 and weakly enhanced phosphorylation of the C774R mutant. The Y1030F mutant showed reduced but not absent phosphorylation in the presence of c-Src compared to wild type or C774R MEGF10, indicating that c-Src primarily phosphorylates Y1030, but may also phosphorylate other tyrosine residues when overexpressed (Fig. 2D). These other tyrosine residues may not be phosphorylated under physiological normal conditions, as the Y1030F mutant shows marked decreased of phosphorylation without overexpression of c-Src (Fig. 2B&D). It is also possible that Y1030 needs to be phosphorylated first, initiating a signaling event that leads to the other residues being phosphorylated. In addition, overexpressed c-Src was able to co-immunoprecipitate with MEGF10, suggesting that c-Src associates with MEGF10 (Fig 2D, Supplementary Fig. 4). This association was decreased when kinase-dead c-Src was overexpressed with wild type MEGF10 (Supplementary Fig. 4) or when c-Src was overexpressed with C774R or Y1030F mutant MEGF10 (Fig. 2D), suggesting that c-Src binding to MEGF10 correlates with tyrosine phosphorylation of this protein.
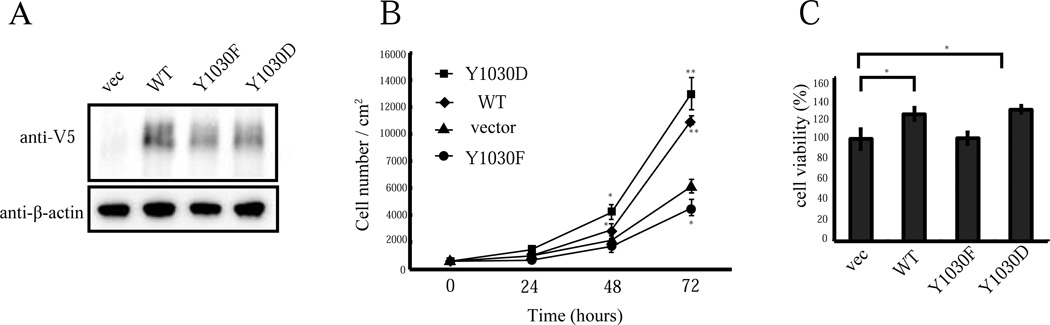
3.3. MEGF10 tyrosine phosphorylation is important for muscle cell proliferation
Overexpression of MEGF10 increases C2C12 cell proliferation [4]. To evaluate whether MEGF10 phosphorylation at Y1030 is involved in muscle cell proliferation signaling, we overexpressed wild type, Y1030F mutant, or Y1030D mutant MEGF10, with an empty vector control, in C2C12 myoblasts using lentiviral vectors (Fig. 3A). Overexpression of wild type MEGF10 increased cell proliferation, in concordance with a previous report [4]. This effect was nullified in the Y1030F mutant overexpressing myoblasts. The phosphorylation-mimicking mutant Y1030D and wild type MEGF10 showed similar increased cell proliferation when overexpressed in myoblasts, suggesting that wild type MEGF10 is phosphorylated in proliferating myoblasts (Fig. 3B). A cell viability assay and a proliferation marker Ki-67 positive cell count confirmed that overexpression of wild type MEGF10 and the Y1030D mutant increases cell proliferation in C2C12 myoblasts, whereas the overexpressed Y1030F mutant did not (Fig. 3C, Supplementary Fig. 5). These results suggest that MEGF10 tyrosine phosphorylation at Y1030 signaling enhances myoblast cell proliferation.
Figure 3. MEGF10 phosphorylation is important for myoblast proliferation.
(A) C2C12 myoblasts were infected with lentiviral vectors to introduce either wild type MEGF10, Y1030F mutant MEGF10, Y1030D mutant MEGF10, or empty vector. The expression of wild type MEGF10 and mutant MEGF10 proteins are shown by immunoblots. β-actin was used as an internal control. (B) Proliferation of C2C12 cells expressing wild type (WT), Y1030F mutant, or Y1030D mutant MEGF10. Lentivirus-infected myoblasts were cultured in growth medium and cell counts were performed at 24, 48 and 72 hours after plating. C2C12 myoblasts expressing wild type MEGF10 or Y1030D mutant MEGF10 showed increased proliferation, whereas C2C12 myoblasts expressing the Y1030F mutant do not. (Error bars represent standard deviation, n=4, *p<0.05, **p<0.001) (C) The numbers of viable cells are increased in C2C12 cells expressing wild type MEGF10 (WT) and Y1030D mutant MEGF10 compared to control C2C12 cells and those expressing Y1030F mutant MEGF10 (Error bars represent standard deviation, n=4, *p<0.005).
4. Discussion
MEGF10 myopathy is the first human skeletal muscle disease caused by defects in a protein that is primarily expressed in muscle satellite cells [2–4]. Muscle satellite cells are progenitor cells that are necessary for repair of skeletal muscle fibers. In mice, satellite cell depletion has been shown to cause defective postnatal muscle growth, failure of muscle regeneration and also mild muscle atrophy over time in uninjured muscle [20–24]. Satellite cells were found to be absent in the muscle tissue of a patient with MEGF10 myopathy [2]. Consistent with these observations in human subjects and a previous report [4], we showed that MEGF10 overexpression increased C2C12 myoblast proliferation while a tyrosine phosphorylation defective mutant version of MEGF10 did not. In addition, tyrosine phosphorylation of C326R and C774R MEGF10 mutant proteins was reduced in patterns that are consistent with observations of genotype-phenotype correlations in the human disease. These results suggest that MEGF10 myopathy may be caused by a defect in muscle satellite cell proliferation due to impaired MEGF10 tyrosine phosphorylation signaling. Our data, in conjunction with reports that c-Src positively regulates C2C12 cell proliferation [25,26] and that overexpression of MEGF10 has been reported to increase C2C12 cell proliferation [4] suggest that c-Src may regulate muscle cell proliferation through the process of MEGF10 tyrosine phosphorylation.
MEGF10 has 17 EGF-like domains but little is known about the role of these domains in MEGF10 function. We hypothesized that the conformations of certain EGF-like domains of MEGF10 are important for transmitting extracellular signals. Both the C326R and C774R mutations are expected to disrupt the second disulfide bond of their respective EGF-like domains, but C326R results in a milder effect both biochemically and clinically than C774R, suggesting that the 16th EGF-like domain may be more important for recognizing extracellular stimuli and conducting signals than the 6th EGF-like domain. In addition, glycosylation of extracellular domains of EGFR is known to be important for receptor-ligand binding [27,28]. We observed that glycosylation is decreased in the C774R mutant compared to wild type and C326R MEGF10. MEGF10 glycosylation may also be involved in MEGF10 signal transduction.
Our experimental results indicate that MEGF10 transduces extracellular stimuli to the intracellular tyrosine-rich domain and participates in signal transduction pathways. Tyrosine phosphorylation of some immunoreceptors by Src family kinases (SFKs) results in recruitment of Syk kinase, which in turn mediates downstream signaling pathways [29]. MEGF10, MEGF12/PEAR1/Jedi-1, and their Drosophila orthologue draper participate in neuronal corpus engulfment signaling [7,13,14] via a similar tyrosine phosphorylation-SFKs-Syk cascade. We showed that c-Src associates with MEGF10 in a tyrosine-phosphorylation-dependent manner. Our data suggest that c-Src augments the phosphorylation of MEGF10 and helps form a signaling complex. It is not yet known what signaling molecules MEGF10 recruits or what ligand initiates MEGF10 signaling. Further studies will uncover further details regarding this signaling pathway that may play an important role in muscle cell proliferation.
We have demonstrated that among two disease-causing mutations in MEGF10, one is associated with more significant impairment of MEGF10 tyrosine phosphorylation than the other. Additionally, we have shown that MEGF10 tyrosine phosphorylation at Y1030 site is important for muscle cell proliferation. Our work suggests that defects in the MEGF10 signaling cascade may deplete muscle satellite cells, possibly contributing to the MEGF10 myopathy disease process. Further characterization of this pathway may enrich our understanding of muscle homeostasis. Elucidating the mechanism of MEGF10 myopathy may yield new molecular approaches for muscle cell proliferation which is important for muscle regeneration, paving the way for potential new therapies for a variety of muscle diseases, including MEGF10 myopathy itself and other muscle diseases.
Supplementary Material
Highlights.
Some MEGF10 mutations lead to impaired tyrosine phosphorylation of the MEGF10 protein.
MEGF10 has a major tyrosine phosphorylation site at tyrosine 1030 that is phosphorylated by c-Src.
Tyrosine phosphorylation of MEGF10 positively regulates muscle cell proliferation.
Acknowledgements
The authors thank Shoichi Ishiura for generously providing plasmids used in this study. We also thank Lane J. Mahoney, Elicia Estrella, Kyungah Cho, Motoyasu Satou, Chieko Aoyama, Megumu Ogawa, Norio Motohashi, Melissa Wu, Emanuela Gussoni, and Louis M. Kunkel for their helpful comments. SM is supported by the William Randolph Hearst Fund at Harvard Medical School. PBK is supported by Muscular Dystrophy Association (MDA) Research Grant 186796, a Pilot Grant at Boston Children’s Hospital, and NIH R01 NS080929. MSA is supported by Muscular Dystrophy Association (MDA) Development Grant MDA255059. Sanger DNA sequencing experiments were performed in the Molecular Genetics Core Facility at Children's Hospital Boston, supported by NIH P30 HD 18655 through the Intellectual and Developmental Disabilities Research Center (IDDRC) and NIH P50 NS40828 through the Neuromuscular Disease Project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartley L, et al. A congenital myopathy with diaphragmatic weakness not linked to the SMARD1 locus. Neuromuscul Disord. 2007;17:174–179. doi: 10.1016/j.nmd.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Logan CV, et al. Mutations in MEGF10, a regulator of satellite cell myogenesis, cause early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) Nat Genet. 2011;43:1189–1192. doi: 10.1038/ng.995. [DOI] [PubMed] [Google Scholar]
- 3.Boyden SE, et al. Mutations in the satellite cell gene MEGF10 cause a recessive congenital myopathy with minicores. Neurogenetics. 2012;13:115–124. doi: 10.1007/s10048-012-0315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holterman CE, Le Grand F, Kuang S, Seale P, Rudnicki MA. Megf10 regulates the progression of the satellite cell myogenic program. The Journal of cell biology. 2007;179:911–922. doi: 10.1083/jcb.200709083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki E, Nakayama M. MEGF10 is a mammalian ortholog of CED-1 that interacts with clathrin assembly protein complex 2 medium chain and induces large vacuole formation. Experimental cell research. 2007;313:3729–3742. doi: 10.1016/j.yexcr.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Wu HH, et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nature neuroscience. 2009;12:1534–1541. doi: 10.1038/nn.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheib JL, Sullivan CS, Carter BD. Jedi-1 and MEGF10 Signal Engulfment of Apoptotic Neurons through the Tyrosine Kinase Syk. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:13022–13031. doi: 10.1523/JNEUROSCI.6350-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh TD, Park SY, Bae JS, Yun Y, Bae YC, Park RW, Kim IS. MEGF10 functions as a receptor for the uptake of amyloid-beta. FEBS letters. 2010;584:3936–3942. doi: 10.1016/j.febslet.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 9.Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483:465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA research : an international journal for rapid publication of reports on genes and genomes. 2001;8:85–95. doi: 10.1093/dnares/8.2.85. [DOI] [PubMed] [Google Scholar]
- 11.Nanda N, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. The Journal of biological chemistry. 2005;280:24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 12.Kauskot A, Di Michele M, Loyen S, Freson K, Verhamme P, Hoylaerts MF. A novel mechanism of sustained platelet alphaIIbbeta3 activation via PEAR1. Blood. 2012;119:4056–4065. doi: 10.1182/blood-2011-11-392787. [DOI] [PubMed] [Google Scholar]
- 13.Logan MA, Hackett R, Doherty J, Sheehan A, Speese SD, Freeman MR. Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nature neuroscience. 2012;15:722–730. doi: 10.1038/nn.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, Freeman MR. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453:935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierson TM, et al. Novel SNP array analysis and exome sequencing detect a homozygous exon 7 deletion of MEGF10 causing early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD) Neuromuscular disorders : NMD. 2013 doi: 10.1016/j.nmd.2013.01.013. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fessart D, Simaan M, Zimmerman B, Comeau J, Hamdan FF, Wiseman PW, Bouvier M, Laporte SA. Src-dependent phosphorylation of beta2-adaptin dissociates the beta-arrestin-AP-2 complex. Journal of cell science. 2007;120:1723–1732. doi: 10.1242/jcs.03444. [DOI] [PubMed] [Google Scholar]
- 17.Mitsuhashi S, et al. A congenital muscular dystrophy with mitochondrial structural abnormalities caused by defective de novo phosphatidylcholine biosynthesis. American journal of human genetics. 2011;88:845–851. doi: 10.1016/j.ajhg.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander MS, et al. Regulation of DMD pathology by an ankyrin-encoded miRNA. Skeletal muscle. 2011;1:27. doi: 10.1186/2044-5040-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki E, Nakayama M. The mammalian Ced-1 ortholog MEGF10/KIAA1780 displays a novel adhesion pattern. Experimental cell research. 2007;313:2451–2464. doi: 10.1016/j.yexcr.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 20.Lepper C, Partridge TA, Fan CM. An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development. 2011;138:3639–3646. doi: 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy JJ, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung TH, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 24.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosoff WJ, Swope SL. Role for cellular Src kinase in myoblast proliferation. Journal of cellular physiology. 2002;193:328–339. doi: 10.1002/jcp.10182. [DOI] [PubMed] [Google Scholar]
- 26.Lim MJ, et al. Suppression of c-Src activity stimulates muscle differentiation via p38 MAPK activation. Archives of biochemistry and biophysics. 2007;465:197–208. doi: 10.1016/j.abb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi M, Yokoe S, Asahi M, Lee SH, Li W, Osumi D, Miyoshi E, Taniguchi N. N-glycan of ErbB family plays a crucial role in dimer formation and tumor promotion. Biochimica et biophysica acta. 2008;1780:520–524. doi: 10.1016/j.bbagen.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Liu YC, et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11332–11337. doi: 10.1073/pnas.1107385108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fodor S, Jakus Z, Mocsai A. ITAM-based signaling beyond the adaptive immune response. Immunology letters. 2006;104:29–37. doi: 10.1016/j.imlet.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.