Abstract
Asthma is a complex genetic disorder with a heterogeneous phenotype, largely attributed to the interactions among many genes and between these genes and the environment. Numerous loci and candidate genes have been reported to show linkage and association to asthma and atopy. Although some studies reporting these observations are compelling, no gene has been mapped that confers a sufficiently high risk of asthma to meet the stringent criteria for genomewide significance. Using 175 extended Icelandic families that included 596 patients with asthma, we performed a genomewide scan with 976 microsatellite markers. The families were identified by cross-matching a list of patients with asthma from the Department of Allergy/Pulmonary Medicine of the National University Hospital of Iceland with a genealogy database of the entire Icelandic nation. We detected linkage of asthma to chromosome 14q24, with an allele-sharing LOD score of 2.66. After we increased the marker density within the locus to an average of one microsatellite every 0.2 cM, the LOD score rose to 4.00. We designate this locus “asthma locus one” (AS1). Taken together, these results provide evidence of a novel susceptibility gene for asthma on chromosome 14q24.
Introduction
Bronchial asthma [MIM 600807], the most common chronic disease affecting children and young adults, is a complex genetic disorder with several overlapping phenotypes (Cookson and Moffatt 2000; Weiss 2001). There is strong evidence of a genetic component in asthma (Bleecker et al. 1997; Kauffmann et al. 2002). Multiple environmental factors are also known to modulate the clinical expression of asthma as well as the asthma-associated phenotypes: bronchial hyperresponsiveness, atopy, and elevated immunoglobulin E (IgE) (Cookson 1999; Holloway et al. 1999; Koppelman and Postma 1999). It is a commonly held view that asthma is caused by multiple interacting genes, some having a protective effect and others contributing to the disease pathogenesis, with each gene having its own tendency to be influenced by the environment (Cookson 1999; Holloway et al. 1999; Koppelman and Postma 1999). Thus, the complex nature of the asthma phenotype, together with substantial locus heterogeneity and environmental influence, has made it difficult to uncover genetic factors that underlie asthma. Although multiple linkage and association studies have implicated >500 atopy and asthma loci or candidate genes throughout the genome (Collings et al. 2000), only one linkage study (Lee et al. 2000) has yielded an atopy locus that, without doubt, meets criteria for genomewide significance (Lander and Kruglyak 1995), and only a few of the associations reported qualify for possible significance. The regions that have been most consistently reported to be in linkage or association with asthma include the cytokine gene cluster on chromosome 5 (including interleukin [IL]-3, IL-4, IL-5, IL-9, and IL-13), chromosome 11 (the β chain of FcεRI), chromosome 12 (stem cell factor, interferon γ, insulin-like growth factor, and Stat-6), and chromosome 16 (IL-4R), suggesting that these regions may harbor genes that contribute to the development of asthma and allergies. In addition, these data support the hypothesis that genes involved in immunological processes play a critical role in the pathogenesis of asthma. Because so few of these studies show significance or map genes with strong, or even moderate, effect, it has been difficult to interpret many of these results (Green et al. 1998; Wjst and Immervoll 1998; Unoki et al. 2000; Hakonarson et al. 2001; Hakonarson and Wjst 2001). In the present study, we report significant linkage to chromosome 14q24, with a LOD score of 4.00 (P=8.70×10-6), in a genomewide linkage study of 596 patients with asthma who are members of 175 Icelandic families.
Material and Methods
Patients
The original patient list contained the names of >7,000 patients who attended the private clinics or outpatient clinics of allergists practicing at the Allergy/Pulmonary Departments of the National University Hospital of Iceland during the years 1977–2001 (European Community Respiratory Health Survey Group 1997). For this study, we selected patients with physician-diagnosed asthma who were being treated with asthma drugs and who were related to at least one other patient within and including six meiotic events (six meiotic events separate second cousins), as revealed by a computerized genealogy database. The patients were 12–70 years of age (mean 39.3 years), and 62% were female. Information was gathered regarding age at diagnosis, medications, hospital admissions, and family history of atopy and asthma. The diagnoses of asthma and atopy were clinically reconfirmed in the study by means of a new physical examination, measurements of skin test reactivity to 12 aeroallergens (including birch, grass, Rumex crispus, cat, dog, horse, Cladosporium, Mucor, Alternaria, Dermatophagoides pteronyssinus, D. farinae, and Lepidoglyphus destructor), total IgE levels, and pulmonary function tests. Unless a baseline forced expiratory volume in 1 s (FEV1) was ⩽70% of predicted value (based on sex, height, and race), a methacholine (MCh)-challenge test was performed. The phenotype assessments, PFTs, and MCh tests were performed according to guidelines of the American Thoracic Society (ATS) (Cockcroft et al. 1977; Palmquist et al. 1988). Patients were considered atopic if their skin-prick–test reaction was positive (i.e., ⩾3 mm or ⩾50% of the histamine-positive control response). The diagnosis of asthma in Iceland is based on the diagnostic criteria outlined by the U.S. National Heart, Lung, and Blood Institute and the ATS (American Thoracic Society 1995; National Institutes of Health 1997) and includes any of the following measures:
-
1.
Patient has recurrent symptoms of cough and wheezing for >2 years and demonstrates clinical response to bronchodilator therapy (as measured by >15% increase in FEV1 after bronchodilator treatment).
-
2.
Patient has reduced FEV1 (i.e., <80%) at baseline prior to bronchodilator therapy and shows >15% improvement in FEV1 after bronchodilator therapy.
-
3.
Patient has recurrent symptoms of coughing and wheezing, and an MCh test, performed in accordance to ATS guidelines (American Thoracic Society 1995), shows >20% decrease in FEV1 at an MCh concentration ⩽8 mg/liter.
Severity of asthma was determined by a combination of signs and symptoms, including PFTs, MCh values, and requirements for therapy. On the basis of these criteria, ∼90% of the patients were scored as having mild-to-moderate asthma, and 10% were scored as having severe asthma. FEV1 >80% predicted was considered normal. Before blood samples were obtained, all patients included in the study were reexamined by the same two allergists who had confirmed the asthma phenotype and severity level and had supervised the measurements of total IgE levels, spirometry, and skin testing. In the present study, a reduction in FEV1 of ⩾20% at an MCh concentration of ⩽8 mg/liter is considered to be a positive result.
The patient participation rate for the study exceeded 90%. All patients gave signed informed consent, donated blood samples, completed a detailed medical questionnaire, and underwent all tests necessary for proper phenotyping. The study was approved by the Icelandic Data Protection Commission and the National Bioethics Committee. Personal identities of the patients and their family members were subsequently encrypted by the Data Protection Commission of Iceland (Gulcher et al. 2000). All blood and DNA samples were also coded in the same way. All participants were asked, by questionnaire, whether they had been diagnosed with asthma and/or atopy and whether they were receiving drugs to treat their condition. The names of the drugs were recorded and confirmed to be antiasthma and/or antiallergy medications. Blood was also collected from close relatives of the index patients, to increase the information available for the linkage analysis. The lung function of all participants was measured at the time the blood was drawn for the study.
Pedigrees
deCODE has built a computerized genealogy database with >650,000 names that includes all 285,000 living Icelanders and most of their ancestors (Gulcher and Stefansson 1998). The database has a connectivity >95% in the 20th century and >86% in the 19th century. Its maternal connections are 99.3% accurate, as measured by mitochondrial polymorphisms of maternally linked individuals (Helgason et al. 2000). The genealogy database was used to cluster the patients in pedigrees. The genealogy database is reversibly encrypted by the Data Protection Commission of Iceland before it is used in our laboratory (Gulcher and Stefansson 1998). Recursive algorithms are used with the encrypted personal identifiers to find all ancestors in the database who are related to any member of the patient list within a given number of preceding generations. The cluster function then identifies ancestors who are common to any two or more members of the patient list.
Genotyping
We genotyped 596 patients with asthma who belonged to 175 families, in which each patient was related to at least one other patient within and including six meiotic events. DNA samples from all 596 patients and 538 relatives were successfully genotyped, using 976 specific fluorescently labeled primers, with an initial genomewide average spacing of 3–4 cM. We have developed a microsatellite screening set, based, in part, on the ABI Linkage Marker (v. 2) screening and intercalating sets, in combination with >500 custom-made markers. All markers were extensively tested for multiplex PCR results. The PCR amplifications were set up and pooled using Cyberlab robots. The reaction volume was 5 μl, and, for each PCR, 20 ng of genomic DNA was amplified in the presence of 2 pmol of each primer, 0.25 U AmpliTaq Gold, 0.2 mmol/liter dNTPs, and 2.5 mmol/liter MgCl2. The PCR conditions were 95°C for 10 minutes, then 37 cycles of 15 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The PCR products were supplemented with the internal size standard, and the pools were separated and detected on an Applied Biosystems model 3700 sequencer, using Genescan (v. 3.0) peak-calling software. Alleles were called automatically with the DAC program (Fjalldal et al. 2001), and the program DecodeGT was used to fractionate according to quality and to edit the called genotypes (Palsson et al. 1999). In regions that demonstrated linkage by use of the framework marker set, marker density was further increased (i.e., fine mapping of locus) by additional microsatellite markers, to obtain an average coverage of 0.2 cM in these regions.
Statistical Analyses
A genomewide linkage scan was performed using a framework map of 976 microsatellite markers. We analyzed the data by use of the Allegro program (Gudbjartsson et al. 2000) and determined statistical significance by applying affecteds-only allele-sharing methods (not specifying any particular inheritance model). The Allegro program, a linkage program developed at deCODE Genetics, calculates LOD scores on the basis of multipoint calculations (Kruglyak et al. 1996; Kong and Cox 1997; Gudbjartsson et al. 2000) and is available at no cost for noncommercial use by sending e-mail to allegro@decode.is. Our standard linkage analysis approach uses the Spairs scoring function (Whittemore and Halpern 1994; Kruglyak et al. 1996), the exponential allele-sharing model (Kong and Cox 1997), and a family weighting scheme that is halfway, on the log scale, between weighting each affected pair equally and weighting each family equally. All genotyped individuals who are unaffected are treated as “unknown.” P values are calculated on the basis of the large-sample theory; Zlr = √[2 loge (10) LOD] is approximately distributed as a standard normal distribution under the null hypothesis of no linkage (Kong and Cox 1997), and the observed LOD score is compared with its complete data sampling distribution under the null hypothesis (Gudbjartsson et al. 2000). The information measure we use is part of the Allegro program output (Nicolae 1999) and is closely related to a classical measure (Dempster et al. 1977). The information value equals zero if the marker genotypes are completely uninformative and equals one if the genotypes determine the exact amount of allele sharing by descent among the affected relatives. The marker order and positions for the framework mapping set were obtained using a high-density genetic map developed at DeCODE (Kong et al. 2002). Data from 146 Icelandic nuclear families (sibships with genotypes for two to seven siblings and for both parents), providing 1,257 meioses, were analyzed to estimate the genetic distances. By comparison, distances in the Marshfield genetic map were estimated on the basis of 188 meioses. Intermarker distances in the peak region, after enrichment with four markers, were estimated using an adaptation of the expectation-maximization algorithm (Dempster 1977) within Allegro.
Results
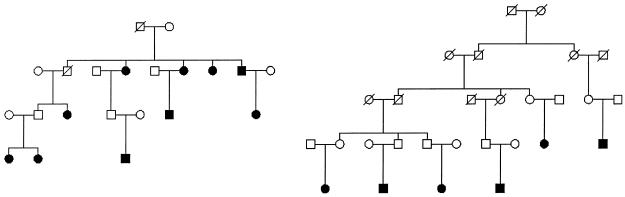
We cross-matched a list of >7,000 patients from the National University Hospital of Iceland with the genealogy database. In the present study, we included patients with physician-diagnosed asthma who were related by six or fewer meiotic events to other patients with asthma. There were 596 patients in 175 families with asthma. In the present study >30 families had at least 6 affected members each. Two of these families are displayed in figure 1. The patients’ demographic data, geometric mean IgE values, and results of lung function tests (including percent predicted FEV1, and FEV1:FVC ratio), MCh-challenge test, and skin tests for the most common aeroallergens in Iceland are shown in table 1. Seventy-three percent of the patients were atopic, as defined by a positive skin-test reaction.
Figure 1.
Examples of asthma pedigrees used in the linkage analysis. Unaffected siblings of patients are not shown, and sex indicators have been shuffled for some individuals in the top two generations, to protect privacy. The darkened squares and circles represent affected men and women, respectively. The slashed symbols represent deceased individuals.
Table 1.
Demographic Data, Atopic Status, and Lung Function Values for the Study Population
| Variable | Valuea |
| Mean age | 39.3 years |
| Sex (M/F) | 38/62 |
| Smoking history (in pack-years): | |
| >1 | 33 |
| >10 | 17 |
| >20 and >55 years of age | .5 |
| Atopy: | |
| Positive skin test | 73 |
| Allergic to: | |
| Animals | 43 |
| Pollen | 40 |
| Dust mites | 12 |
| Mold | 4 |
| IgE | 137 ± 242b |
| Lung function: | |
| FEV1 | 98.4 ± 17.3b |
| FEV1/FVC | 82.2 ± 9.5b |
| MCh challenge:c | |
| ⩽8 mg/ml | 90 |
| ⩽2 mg/ml | 67 |
| Asthma severity: | |
| Mild to moderate | 90 |
| Severe | 10 |
Data are percentages, except as otherwise noted.
Mean ± SD.
Patients are grouped according to amounts of MCh required to produce a 20% decrease in FEV1.
More than 400 of the patients were tested for airway reactivity by use of MCh challenge. As shown in table 1, although 67% of the patients tested had a ⩾20% decrease in FEV1 at an MCh concentration of ⩽2 mg/liter, >90% of the patients tested positive at an MCh concentration of ⩽8 mg/liter. This would be consistent with moderate-to-severe airway hyperresponsiveness in two thirds of our study population. The spirometric values reported in table 1 are those obtained during the study, at which time the majority of the patients had stable asthma and were receiving full therapy; however, the medical charts of all these patients show previous spirometric values with FEV1 <80% predicted, at one or more earlier time points (data not shown). Bronchodilator reversibility was tested in selected patients, including those who had negative results of an MCh challenge and patients for whom the clinician determined the test to be necessary to support the asthma phenotype.
Thirty-three percent of the patients gave a history of having smoked for >1 pack-year. Of those, 47% had smoked for <10 pack-years. We cannot exclude the possibility that some of the study participants who are smokers had mild coexisting chronic obstructive pulmonary disease (COPD); however, only 0.5% of the study patients who were ⩾55 years had smoked for ⩾20 pack-years (table 1). In contrast, all of our study subjects have asthma—as defined by the ATS criteria (American Thoracic Society 1995), which is the phenotype used for this study—thereby minimizing the potential of confounding effects from COPD.
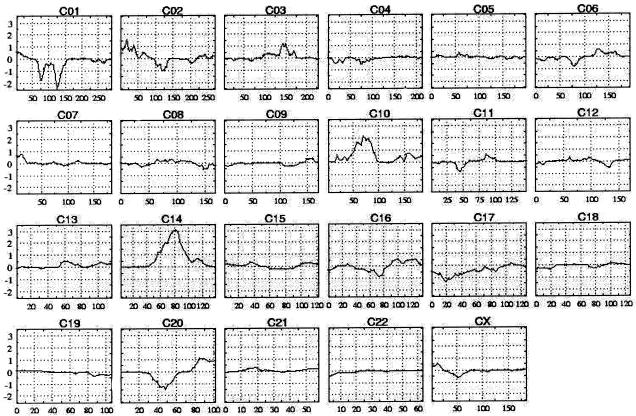
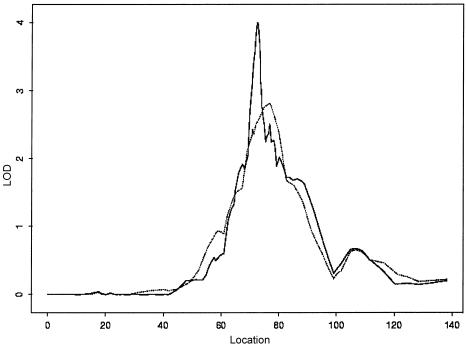
Five hundred ninety-six patients and 538 of their unaffected (i.e., nonasthmatic) relatives were genotyped using 976 microsatellite markers in a genomewide linkage scan. The disease status of relatives was obtained by use of a questionnaire. We analyzed the data and determined statistical significance by applying affecteds-only, allele-sharing methods (which do not specify any particular inheritance model) (Gulcher et al. 2001). Since our linkage scan is performed on affected status only, there is no possible confounding effect from the relatives, even if we failed to identify an affected relative in the group. Figure 2 shows the LOD score–tracing genome scan using the framework markers. The genomic region that showed the most evidence of linkage to the asthma phenotype was chromosome 14q24, with a LOD score of 2.66 (single test P=2.16×10-4). Given that the information content on identity-by-descent sharing in the region was <85% (i.e., 0.77), which is less than we prefer, we added another 34 microsatellite markers under this peak to ensure that the results are a true reflection of the information contained in the material. This increased the information content at the peak to >95% and produced a LOD score of 4.00 (single test P=8.70×10-6) (fig. 3). This locus is significant on a genomewide basis, even allowing for the multiple-marker testing, and it corresponds to a genomewide adjusted P value of <.05 (Lander and Kruglyak 1995). We designate this locus as “asthma locus one” (AS1). The locus peak is centered on markers D14S588 and D14S603, which are spaced 84 kb apart. The locus, defined by a decrease of ∼1.0 in the LOD score, is between markers D14S1069 and D14S289, centromeric and telomeric, respectively. The segment with a 1-LOD decrease is ∼3.9 cM, and we estimate that it corresponds to ∼3.0 million bases.
Figure 2.
Genealogy-based genomewide linkage scan of all 596 affected family members, using 976 microsatellite markers. The multipoint LOD score is shown on the vertical axis, and the distance (in cM) from the p terminus of the chromosome is on the horizontal axis.
Figure 3.
Multipoint allele-sharing LOD score of chromosome 14. A framework genome scan is indicated by a dotted line. A fine-mapping LOD score of 4.00 was detected within the peak region after 34 microsatellite markers were added to obtain a marker density of ⩽0.2 cM, using deCODE's high-density genetic map to determine genetic distances. The multipoint LOD score is shown on the vertical axis, and distance (in cM) from the p terminus of the chromosome is shown on the horizontal axis.
Discussion
Our genomewide scan revealed significant linkage of asthma to chromosome 14q24, indicating that this region harbors a gene that contributes to the development of asthma. We included individuals who had a physician-confirmed diagnosis of asthma and who were taking asthma medications. The majority of the patients with asthma were monitored on a regular basis in private clinics by three allergists/pulmonologists. Accurate information about medication history and current use of asthma medication was recorded for all participants. The extensive computerized genealogy database on Icelanders enabled us to identify extended families of patients with asthma, wherein the initial genomewide linkage scan (using microsatellite markers with the average genomewide density of 3–4 cM) demonstrated significant linkage to chromosome 14q24 (fig. 2). Several genomewide linkage scans in asthma and atopy have been published; however, most of them have not reported results that meet criteria for significance at a genomewide level (reviewed by Wjst and Immervoll [1998], Hakonarson and Wjst [2001], and Hakonarson et al. [2001]). A notable exception is the European linkage study of 199 families with atopic dermatitis, which demonstrated significant linkage to chromosome 3q21 (Lee et al. 2000). Three other studies have reported linkage results of genomewide significance (Lander and Kruglyak 1995) to the following asthma-associated phenotypes: bronchial hyperresponsiveness (Xu et al. 2001a), elevated IgE (Xu et al. 2001b), and mite-sensitive asthma (Yokoucki et al. 2000). A recent study of asthma in the Kainuu population of Finland showed linkage on 7q14-15 to high levels of IgE (nonparametric linkage score 3.9, P=.0001); however, the P value did not reach the threshold for genomewide significance (Laitinen et al. 2001).
Genomewide searches for genes that contribute to asthma and asthma-associated phenotypes (atopy, high IgE levels, and bronchial hyperresponsiveness) have been completed by several investigators, each studying independent sets of families (Marsh et al. 1994; Daniels et al. 1996; The Collaborative Study of the Genetics of Asthma 1997; Hizawa et al. 1998; Ober et al. 1998; Dizier et al. 2000; Wjst et al. 1999; Collings et al. 2000; Cho et al. 2001). However, it is noteworthy that, apart from the locus reported by Lee et al. (2000), in their study of 199 families with atopic dermatitis (eczema), none of the linkages reported reach the level of genomewide statistical significance. The first genomewide scan for linkage to quantitative traits underlying asthma identified evidence of linkage on chromosomes 4q, 6p (near the locus for the major histocompatibility complex), 7, 11q (containing FcεRI-β), 13q, and 16 (Daniels 1996). A replication sample of families in the same study demonstrated suggestive linkage to chromosomes 4, 11, 13, and 16 (Daniels 1996). A genome scan for responsiveness to house-dust–mite allergen found suggestive linkage to chromosomes 2q, 6p (near the MHC), 13q, and 8p (Hizawa et al. 1998). A genome scan in American families from three racial groups revealed weak linkage to broad regions on chromosomes 2q, 5q, 6p, 12q, and 13q that may constitute replications of the results of some previous studies (The Collaborative Study of the Genetics of Asthma 1997) and to 14q. A two-stage scan in Hutterite families from the United States found suggestive evidence of linkage and replication for loci on chromosomes 5q, 12q, 19q, and 21q (Ober et al. 1998). A screen in German families identified suggestive linkage to asthma on chromosomes 2q (near the IL-1 family cluster), 6p, 9, and 12q (The Collaborative Study of the Genetics of Asthma 1997). A two-stage genome scan in French families found replicated linkage on chromosomes 1p, 12q, and 17q (Dizier et al. 1999). A recent genomic screen for QTL that underly asthma in 533 Chinese families encompassing 2,551 individuals reported significant linkage of airway hyperresponsiveness to the D2S1780 marker on chromosome 2 (Xu et al. 2001a). A study of 47 Japanese families with mite-sensitive atopic asthma reported significant linkage to the IL-12B gene locus on 5q31 (Yokouchi et al. 2000). Genome screens in a Dutch population (Xu et al. 2001b) showed genomewide significant results, when IgE was used as a QTL. Thus, the loci most consistently identified by these screens are on chromosomes 5, 6, 11, 12, and 13.
Given that asthma genes have been difficult to map, various attempts have been made to perform meta-analysis, combining the evidence from several studies on multiple markers in a particular candidate region, and to summarize the multipoint LOD values evaluated at the location of each marker. However, this is not always practical, because of the variability in methods and models among different studies. Lonjou et al. (1999) presented a meta-analysis of a retrospective collaboration for positional cloning of asthma susceptibility by the Consortium on Asthma Genetics. Combination of evidence over multiple samples with 1,037 families supported loci contributing to asthma susceptibility in the cytokine region on 5q, with a maximum LOD score of 2.61 near the locus of IL-4, but no evidence of linkage to atopy. Comparable results were obtained from the analysis of linkage data from five independent studies, including 3,175 subjects in 569 families from five countries, who were sampled for the 5q31-33 region (Palmer et al. 2001). Our genome scan shows, for the first time, significant linkage of asthma to markers on chromosome 14q24. It should be noted that suggestive linkage to this locus has been reported elsewhere, by investigators in the United Kingdom (Mansur 1999) and the United States (The Collaborative Study of the Genetics of Asthma 1997). Chromosome 14q24 contains many genes that could contribute to the susceptibility to asthma, including genes that encode for the epidermal growth factor–response factor 1, phosphatidylinositol glycan class H, secreted modular calcium-binding protein 1, a disintegrin, metalloproteinase domain–20 and –21, and RNA polymerase II transcriptional regulation mediator, to name a few. However, given that the size of the region marked by a decrease of 1 in LOD score is currently ∼3.9 cM, further fine mapping by use of marker spacing of 0.03–0.05 cM is needed and is under way. This approach allows for single-point and haplotype-association analysis and is aimed at narrowing the region of 1-LOD decrease to the most likely candidate. We conclude that we have mapped a high-risk asthma gene on chromosome 14q24.
Acknowledgments
We would like to thank the patients and their families, whose contributions have made this study possible. We thank the nurses and study assistants at the National University Hospital of Iceland (Vifilstadir) and at the Phlebotomy Station in Nóatún, for the excellent assistance, and the deCODE genotyping staff, for their valuable contribution.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- deCODE Genetics Inc., http://www.decode.com/ for deCODE GT program [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for asthma [MIM 600807])
References
- American Thoracic Society (1995) Standardization of spirometry, 1994 Update. Am J Respir Crit Care Med 152:1107–1136 [DOI] [PubMed] [Google Scholar]
- Bleecker ER, Postma DS, Meyers DA (1997) Evidence for multiple genetic susceptibility loci for asthma. Am J Respir Crit Care Med 156:S113–S116 [DOI] [PubMed] [Google Scholar]
- Cho SH, Son JW, Koh YY, Min KU, Kim YY, Kim YK (2001) Linkage between bronchial responsiveness to methacholine and gene markers of IL-4 cytokine gene cluster and T-cell receptor α/δ gene complex in Korean nuclear families. Clin Exp Allergy 31:103–109 [PubMed] [Google Scholar]
- Cockcroft DW, Killian DN, Mellon JJ, Hargreave FE (1977) Bronchial reactivity to inhaled histamine: a method and clinical survey. Clin Allergy 7:235–243 [DOI] [PubMed] [Google Scholar]
- Collaborative Study on the Genetics of Asthma, the (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–392 [DOI] [PubMed] [Google Scholar]
- Collings A, Ennis S, Tapper W, Morton NE (2000) Mapping of oligogenes for atopy and asthma by meta-analysis. Am J Hum Genet 66:A1366 [Google Scholar]
- Cookson W (1999) The alliance of genes and environment in asthma and allergy. Nature 25:B5–B11 [DOI] [PubMed] [Google Scholar]
- Cookson W, Moffatt MF (2000) Genetics of asthma and allergic disease. Hum Mol Genet 9:2359–2364 [DOI] [PubMed] [Google Scholar]
- Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, le Souef PN, Lathrop GM, Musk AW, Cookson WO (1996) A genome wide search for quantitative trait underlying asthma. Nature 383:247–250 [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc B 39:1–38 [Google Scholar]
- Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, Charpin D, et al (2000) Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med 162:1812–1818 [DOI] [PubMed] [Google Scholar]
- European Community Respiratory Health Survey Group (1997) Genes for asthma? An analysis of the European community respiratory health survey. Am J Respir Crit Care Med 156:1773–1780 [DOI] [PubMed] [Google Scholar]
- Fjalldal JB, Benediktsson K, Sigurdsson J, Ellingssen LM (2001) Automated genotyping: combining neural networks and decision trees to perform robust allele calling. Proc Int Joint Conf Neural Networks A1–A6 [Google Scholar]
- Green SL, Gaillard MC, Song E, Dewar JB, Halkas A (1998) Polymorphisms of the beta chain of the high-affinity immunoglobulin E receptor (FcεRI-β) in South African black and white asthmatic and nonasthmatic individuals. Am J Respir Crit Care Med 158:1487–1492 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Kong A, Stefansson K (2001) The role of linkage studies for common disease. Curr Opin Genet Dev 11:264–267 [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Kristjansson K, Gudbjartsson H, Stefansson K (2000) Protection of privacy by third-party encryption in genetic research in Iceland. Eur J Hum Genet 8:739–742 [DOI] [PubMed] [Google Scholar]
- Gulcher JR, Stefansson K (1998) Population genomics: laying the groundwork for genetic disease modeling and targeting. Clin Chem Lab Med 36:523–527 [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Bjornsdottir US, Ostermann E, Arnason T, Adalsteinsdottir AE, Halapi E, Shkolny D, Kristjansson K, Gudnadottir SA, Frigge ML, Gislason D, Gislason T, Kong A, Gulcher J, Stefansson K (2001) Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med 164:2036–2044 [DOI] [PubMed] [Google Scholar]
- Hakonarson H, Wjst M (2001) Current concepts on the genetics of asthma. Curr Opin Pediatr 13:267–277 [DOI] [PubMed] [Google Scholar]
- Helgason A, Sigurðardóttir S, Gulcher JR, Ward R, Stefánsson K (2000) mtDNA and the origin of the Icelanders: deciphering signals of recent population. Am J Hum Genet 66:999–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizawa N, Freidhoff LR, Chiu YF, Ehrlich E, Luehr CA, Anderson JL, Duffy DL, Dunston GM, Weber JL, Huang SK, Barnes KC, Marsh DG, Beaty TH (1998) Genetic regulation of Dermatophagoides pteronyssinus–specific IgE responsiveness: a genome-wide multipoint linkage analysis in families recruited through 2 asthmatic sibs. Collaborative Study on the Genetics of Asthma. J Allergy Clin Immunol 102:436–442 [DOI] [PubMed] [Google Scholar]
- Holloway JW, Beghé B, Holgate ST (1999) The genetic basis of atopic asthma. Clin Exp Allergy 29:1023–1032 [DOI] [PubMed] [Google Scholar]
- Kauffmann F, Dizier MH, Oryszczyn MP, Le Moual N, Siroux V, Annesi-Maesano I, Bousquet J, Charpin D, Feingold J, Gormand F, Grimfeld A, Hochez J, Lathrop M, Matran R, Neukirch F, Paty E, Pin I, Demenais F (2002) Epidemiologic study of the genetics and environment of asthma, bronchial hyperresponsiveness, and atopy. Chest Suppl 121:27S [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson RE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Koppelman GH, Los H, Postma DS (1999) Genetics and environment in asthma: the answer of twin studies. Eur Respir J 13:2–4 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Laitinen T, Daly MJ, Rioux JD, Kauppi P, Laprise C, Petays T, Green T, Cargill M, Haahtela T, Lander ES, Laitinen LA, Hudson TJ, Kere J (2001) A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population. Nat Genet 28:87–91 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lee YA, Wahn U, Kehrt R, Tarani L, Businco L, Gustafsson D, Andersson F, Oranje AP, Wolkertstorfer A, van Berg A, Hoffmann U, Kuster W, Wienker T, Ruschendorf F, Reis A (2000) A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat Genet 26:470–473 [DOI] [PubMed] [Google Scholar]
- Lonjou C, Collins A, Ennis S, Tapper W, Morton NE (1999) Meta-analysis and retrospective collaboration: two methods to map oligogenes for atopy and asthma. Clin Exp Allergy 29:S57–S59 [PubMed] [Google Scholar]
- Mansur AH, Bishop DT, Markham AF, Morton NE, Holgate ST, Morrison JF (1999) Suggestive evidence for genetic linkage between IgE phenotypes and chromosome 14q markers. Am J Respir Crit Care Med 159:1796–1802 [DOI] [PubMed] [Google Scholar]
- Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH (1994) Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science 264:1152–1155 [DOI] [PubMed] [Google Scholar]
- National Institutes of Health (1997) Guidelines for the diagnosis and management of asthma: expert panel report 2, July 1997. US Government Printing Office, Washington, DC. NIH Publication No 97-4051 [Google Scholar]
- Nicolae DL (1999) Allele sharing models in gene mapping: a likelihood approach. PhD thesis. University of Chicago, Chicago [Google Scholar]
- Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, Pettersson A, Prescott J, Richardson A, Schlenker E, Summerhill E, Willadsen S, Parry R (1998) Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Hum Mol Genet 7:1393–1398 [DOI] [PubMed] [Google Scholar]
- Palmer LJ, Cookson WO, Deichmann KA, Holloway JW, Laitinen T (2001) Single region linkage analyses of asthma: description of data sets. Genet Epidemiol Suppl 21:S9–S15 [DOI] [PubMed] [Google Scholar]
- Palmquist M, Balder B, Lindholm NB (1988) Hyperreactivitet hos 420 patienter med nydebuterad astma. Hygiea 97:108 [Google Scholar]
- Palsson B, Palsson F, Perlin M, Gudbjartsson H, Stefansson K, Gulcher J (1999) Using quality measures to facilitate allele calling in high-throughput genotyping. Genome Res 9:1002–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M, Furuta S, Onouchi Y, Watanabe O, Doi S, Fujiwara H, Miyatake A, Fujita K, Tamari M, Nakamura Y (2000) Association studies of 33 single nucleotide polymorphisms (SNPs) in 29 candidate genes for bronchial asthma: positive association with a T924C polymorphism in the thromboxane A2 receptor gene. Hum Genet 106:440–446 [DOI] [PubMed] [Google Scholar]
- Weiss ST (2001) Epidemiology and heterogeneity of asthma. Ann Allergy Asthma Immunol Suppl 87:5–8 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rueschendorf F, Reis A et al (1999) A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics 58:1–8 [DOI] [PubMed] [Google Scholar]
- Wjst M, Immervoll T (1998) Asthma Gene Database: an Internet linkage and mutation database for the complex phenotype asthma. Bioinformatics 14:827–828 [DOI] [PubMed] [Google Scholar]
- Xu X, Fang Z, Wang B, Chen C, Guang W, Jin Y, Yang J, Lewitzky S, Aelony A, Parker A, Meyer J, Weiss ST, Xu X (2001a) A genomewide search for quantitative-trait loci underlying asthma. Am J Hum Genet 69:1271–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, Langefeld CD, Beaty TH, Rich SS, Bleecker ER, Cox NJ, Collaborative Study on the Genetics of Asthma (2001b) Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three US populations. Am J Hum Genet 68:1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi Y, Nukaga Y, Shibasaki M, Noguchi E, Kimura K, Ito S, Nishihara M, Yamakawa-Kobayashi K, Takeda K, Imoto N, Ichikawa K, Matsui A, Hamaguchi H, Arinami T (2000) Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31-q33 near the interleukin 12 B locus by a genome-wide search in Japanese families. Genomics 66:152–160 [DOI] [PubMed] [Google Scholar]