Abstract
Glucocorticoid receptor (GR) ontogeny and distribution in postnatal rat brain have been demonstrated, but onset and distribution of GR gene expression during fetal life has not been reported. This study focuses on the distribution of GR-mRNA in the fetal and postnatal rat fore-brain, with emphasis on hypothalamic and limbic structures. Time pregnant rats were decapitated at 8:30–9:30 AM on Gestational Days 14 (F14), F16, F17, F18, and F19. Postnatally, rats were sacrificed on Days 1, 4, 6,10, and 16. Cryostat sections were subjected to in situ hybridization, using a cRNA probe directed to the GR-mRNA. GR-mRNA was detectible in the hippocampo-septal formation as early as F14. By F16, GR gene expression was evident in the hypothalamic paraventricular nucleus (PVN) as well. During late gestation (F17–F19), GR-mRNA was localized also in the thalamus, hippocampus, amygdala, and discrete cortical regions. Postnatally, GR-mRNA abundance was high in the PVN, CA1/CA2 hippocampal field, piriform cortex and dorsal endopiriform nucleus, specific amygdaloid nuclei, and the suprachiasmatic nucleus. In PVN, GR-mRNA was present prior to the onset of CRH gene expression (F17), which may suggest a role for GR in neuronal differentiation.
INTRODUCTION
The ontogeny of glucocorticoid receptor (GR), and of GR-messenger RNA (GR-mRNA) distribution has been studied in postnatal rat brain, using immunohistochemistry and in situ hybridization (ISH), respectively (1, 2). Whole brain GR-mRNA ontogeny has been demonstrated using Northern blot analysis (3). However, the onset of GR gene expression in specific limbic structures and the localization and relative abundance of GR-mRNA in fetal rat brain have not been reported. Such information is of importance in view of the putative role of glucocorticoids (GC) in neuronal development and differentiation (4–6). This study focuses on the distribution of GR-mRNA in the fetal and neonatal rat brain, with emphasis on hypothalamic and limbic structures.
MATERIALS AND METHODS
Animals
Time-pregnant Sprague-Dawley-derived rats (Zivic-Miller, Zelienople, PA; Day 0 defined by sperm-positive smear) were housed under a 12-h light regimen (light on at 7 AM) and given unlimited access to lab chow and water. All animals were decapitated at 8:30–9:30 AM, to avoid diurnal variability in GR-mRNA abundance (7). Pregnant rats were sacrificed on Gestational Days 14 (Fl4), F16, F17, F18, and F19. Fetuses were rapidly harvested and decapitated, and heads transferred onto finely powdered dry ice. For postnatal rats, delivery was assessed every 12 h, and rats were sacrificed on Postnatal Days 1, 4, 6, 10, and 16 (day of birth = 0). Pups were decapitated, brains were removed from skulls and frozen on dry ice. A minimum of three brains, derived from two separate litters, was used per age group.
Tissue Preparation
Brains were stored at −80°C. Coronal sections (20 μm) were mounted on gelatin-coated slides and stored at −80°C. Sections were subjected to ISH. Separate sections were stained with crystal violet to identify brain structures. Prior to ISH, slides were brought to room temperature, air-dried, and fixed in buffered paraformaldehyde. Following a graded ethanol treatment (8, 9), sections were exposed to acetic anhydride-triethanolamine and dehydrated through 100% ethanol (8–10).
Probe Preparation and ISH Procedure
The GR probe was a 456-nucleotide fragment of a cDNA clone directed against the protein coding region and the 3′ untranslated region of the GR-mRNA. The GR-specific cDNA clone (originally from Dr. K. Yamamoto) was obtained from Drs. J. P. Herman and S. J. Watson (11). A sense-strand was used as specificity control. Both sense and antisense S35-cDNA probes were synthesized using S35-UTP (Amersham), and Sp6 and T7 RNA polymerase, respectively. The specific activity of the probes was 1.4 ×109 cpm/μg. The ISH procedure has been described (8–10). Briefly, acetic anhydride-treated, ethanol-dehydrated sections were exposed to 20-μl hybridization solution containing 5 ×107 cpm/ml of labeled probe at 58°C for 15 h. Subsequently, coverslips were removed in 4X SSC, and the slides treated with 20 μg/ml RNAse A for 30 min at 37°C. Serial washes with decreasing concentrations of SSC (containing 1 mMDTT), were followed by a high stringency wash (0.1× SSC) at 75°C for 1 h. The slides were then dehydrated through increasing concentrations of ethanol solutions containing 0.3 M NH4Ac. Dried sections were apposed to film (XAR5, Kodak, Rochester, NY). Several sections were subsequently dipped in emulsion (NTB-2; Kodak) and developed as previously described (8, 9), with the exception that the emulsion was not diluted.
All sections were subjected to ISH together with adult brain sections at the level of the hippocampus, as positive controls; all ISH films were analyzed together. Semi-quantitative and statistical analyses have been described (8–10). Briefly, optical density (OD) was determined over discrete brain regions using the MCID software image analysis system (Imaging Research, Ontario, Canada). Each point was derived from 6–12 sections from a minimum of three individual rats. OD values were compared to brain-paste standards (12). Ratios of (region-OD)/ (background-OD) were also determined, thus eliminating background variability (8–10). Fetal and postnatal brain structures were identified according to Paxinos et al. (13) and Sherwood and Timiras (14), respectively.
RESULTS
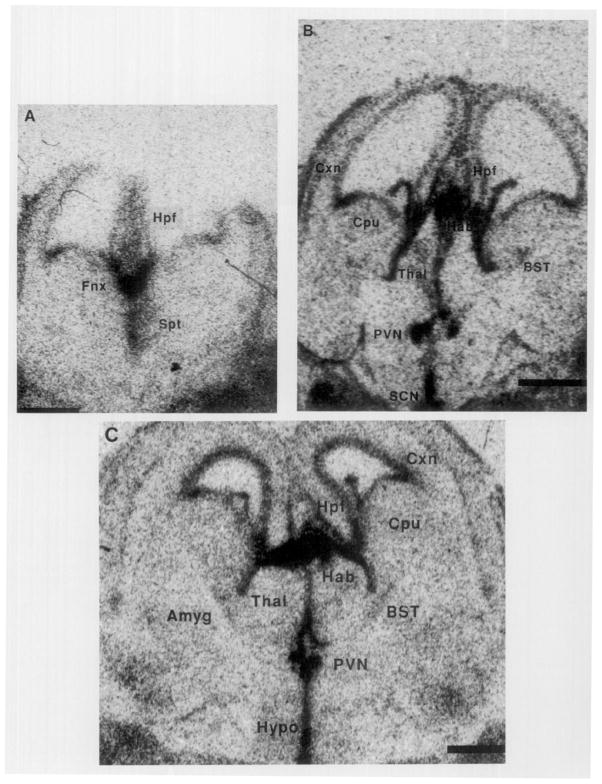
GR-mRNA was detectible in the septohippocampal formation as early as F14 (Fig. 1A). By F16, GR gene expression was evident in the hypothalamic paraventricular nucleus (PVN), hippocampal formation and some cortical areas (Fig. IB, Table 1). By F18, robust GR-mRNA hybridization was present over PVN, hippocampal formation (probably CA3), and superficial cortical layers (Fig. 1C, Table 1).
FIG. 1.
Photomicrographs of the glucocorticoid receptor gene expression in fetal rat forebrain. In situ hybridization was performed using a cDNA probe directed against GR-mRNA. GR gene expression is evident in the septohippocampal region on Fetal Day 14 (A). GR-mRNA is present also in PVN by Fetal Day 16 (B) and Fetal Day 18 (C). Amyg, amygdala; BST, bed nucleus stria terminalis; Cpu, caudate putamen; Cxn. cortical neuroepithelium; Fnx, fornix; Spt, septum; Hab, habenular nucleus; Hpf, hippocampal formation; Hypo, hypothalamus; PVN, paraventricular nucleus (hypothalamus); SCN, suprachiasmatic nucleus; Thal, thalamus. Bar, 1 mm.
TABLE 1.
Ontogeny of GR-mRNA in the Prenatal Rat Brain
| F14 | F16 | F18 | |
|---|---|---|---|
| Cortex | |||
| Cortical neuroepithelium | + | + | ++ |
| Cortical plate | ++ | ++ | ++ |
| Hippocampal formation | + | ++ | ++ |
| Fornix | +++ | ++ | ++ |
| Septum | ++ | ++ | + |
| Habenular nucleus | ++ | ++ | |
| Bed nucleus stria teminalis/caudate putamen | + | + | |
| Thalamus | + | + | |
| Amygdaloid area | + | ||
| Hypothalamus | |||
| Paraventricular nucleus | +++ | +++ | |
| Ventromedial nucleus | + | ||
| Suprachiasmatic nucleus | +++ | ||
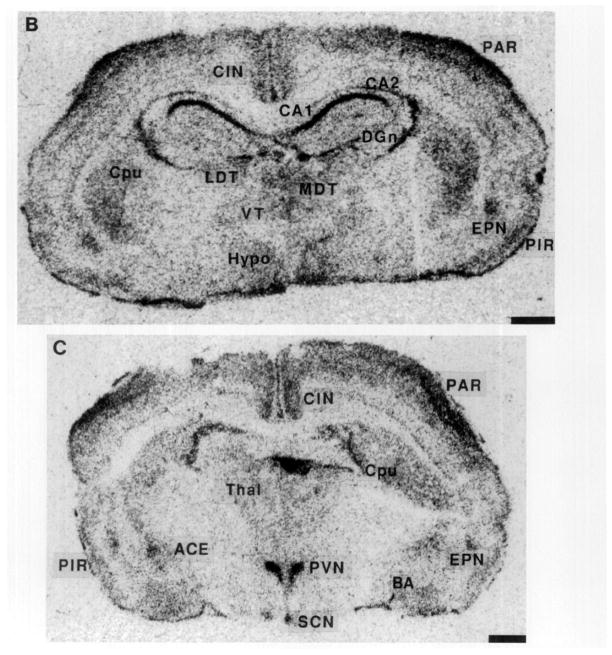
On Postnatal Day 1, GR-mRNA was clearly localized to the CA1 hippocampal area (Fig. 2A), with little message in CA2, CA3, or dentate gyrus (Table 2). The cingulate and frontoparietal cortical areas, thalamus and amygdaloid complex contained GR-mRNA as well: semiquantitative analysis revealed higher GR-mRNA abundance in the PVN than in any other forebrain structure (Table 2). GR-mRNA signal was much stronger over the CA1 region in Day 4 than in Day 1 (Fig. 2B, Table 2).
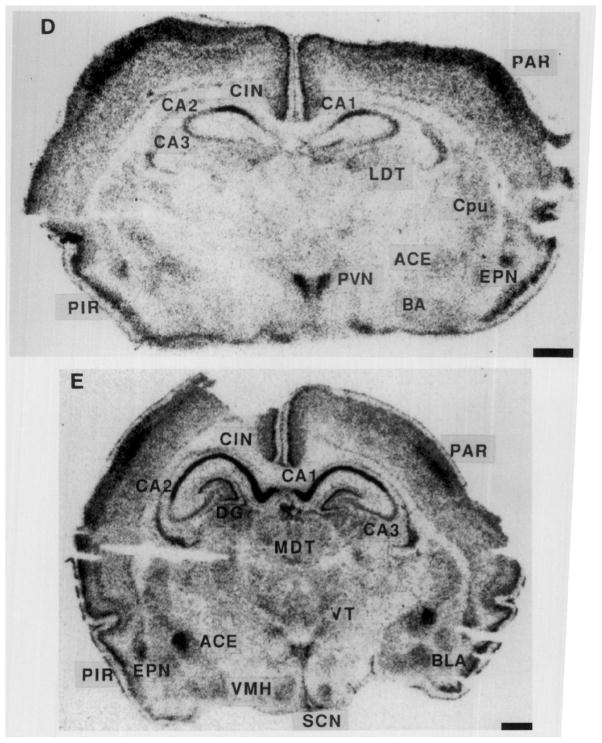
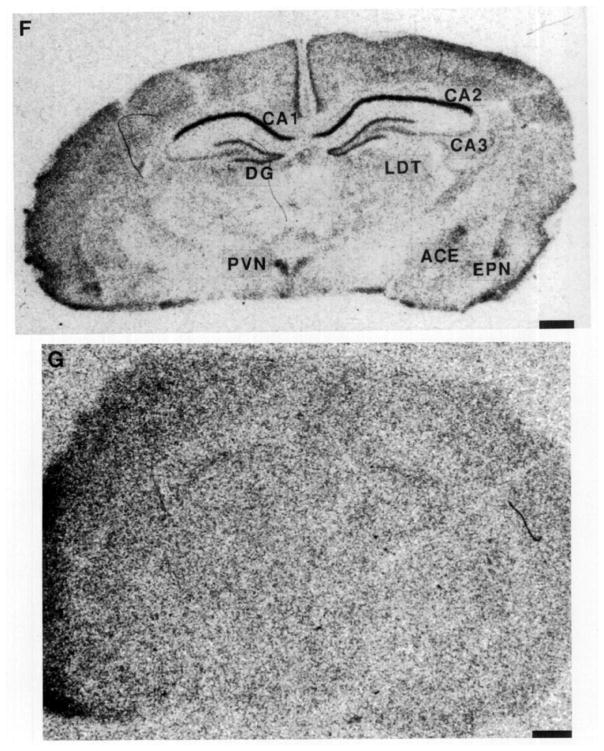
FIG. 2.
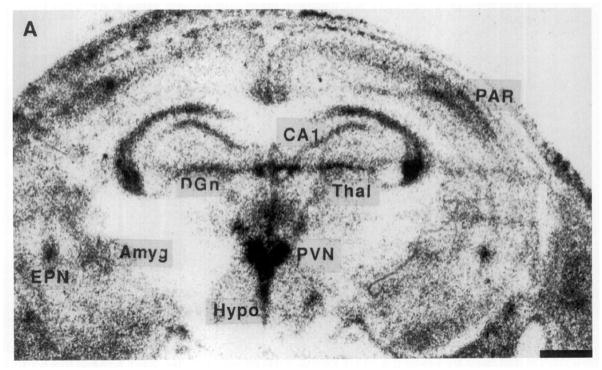
Photomicrographs of the GR gene expression in postnatal rat forebrain, demonstrating the distribution of GR-mRNA on Postnatal Days 1, 4, 6, 10, and 16. On Day 1 (A), GR-mRNA is evident in the hippocampal CA1 as well as in the PVN. A similar distribution is seen on Day 4 (B), with increased signal over the CA1/CA2 region and parietal cortex. GR-mRNA is evident in the amygdala and endopiriform nucleus by Day 6 (C). The distribution of GR message on Postnatal Day 10 is seen in two coronal levels (D, E). By Day 16 (F), GR-mRNA is abundant in dentate gyrus. Hybridization with a sense probe (G) was presented to show the specificity of the signal. ACE, central nucleus (amygdala); BA, basal nucleus (amygdala); BLA, basolateral nucleus (amygdala); CIN, cingulate cortex; DGn, dentate gyrus neuroepithelium; EPN, endopiriform nucleus; LDT, laterodorsal nucleus (thalamus); MDT, mediodorsal nucleus (thalamus); PAR. parietal cortex; PIR, piriform cortex; VMH, ventromedial nucleus (hypothalamus); VT, ventral nucleus (thalamus). Bar, 1 mm.
TABLE 2.
Ontogeny of GR-mRNA in the Postnatal Rat Brain
| P1 | P4 | P6 | P10 | P16 | |
|---|---|---|---|---|---|
| Cortex | |||||
| Cingulate cortex | + | + | ++ | ++ | + |
| Frontoparietal | ++ | +++ | +++ | +++ | ++ |
| Piriform | + | + | + | +++ | ++ |
| Hippocampus | |||||
| CA1 | ++ | +++ | +++ | ++++ | ++++ |
| CA2 | +++ | +++ | +++ | ++++ | |
| CA3 | + | ++ | + | ||
| Dentate gyrus | ++a | ++a | +++ | +++ | |
| Habenular nucleus | + | + | + | ++ | + |
| Caudate putamen | + | ++ | ++ | + | + |
| Thalamus | |||||
| Laterodorsal | ++ | + | ++ | ++ | + |
| Mediodorsal | + | + | + | ||
| Ventral | ++ | + | + | + | + |
| Amygdala | |||||
| Central nucleus | ++b | + | ++ | +++ | +++ |
| Basal | + | ++ | + | ||
| Endopiriform nucleus | ++ | ++ | ++ | +++ | ++ |
| Hypothalamus | |||||
| Paraventricular | +++ | +++ | +++ | +++ | +++ |
| Suprachiasmatic | ++ | +++ | ++ | ||
| Other hypoth nuclei | + | + | + | ++ | + |
The signal represented the dentate gyrus neuroepithelium, rather than the dentate gyrus.
The signal appeared over the amygdaloid complex, which contains the central nucleus.
By Postnatal Day 6 (Fig. 2C), GR-mRNA was abundant in parietal cortex, piriform cortex, endopiriform nucleus, and in specific amygdaloid nuclei, particularly the central nucleus (Table 2). GR-mRNA was also present in the hypothalamic suprachiasmatic nucleus (SCN) (Fig. 2C, Table 2). GR-mRNA abundance in PVN remained high throughout the first postnatal week.
By the 10th postnatal day, (Figs. 2D and 2E) GR-mRNA was maximal in the hippocampal CA1/CA2 region, PVN, and central amygdaloid nucleus, followed by frontoparietal and piriform cortex, dentate gyrus, endopiriform nucleus, and cingulate cortex in that order (Fig. 3, Table 2). GR-mRNA was expressed also in hypothalamic ventromedial and amygdaloid basolateral and basomedial nuclei (Table 2, Fig. 3).
FIG. 3.
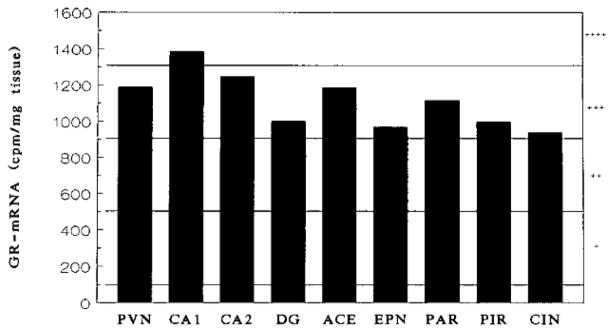
A representative graph for the distribution of GR-mRNA in 10-day-old rat brain. This graph was generated by analyzing the signal shown in Fig. 2E, using a MCID image analysis system. In order to illustrate at a glance the developmental changes of GR-mRNA abundance in various brain areas, the intensity of the signal was simplified from actual numbers to degrees: a range of intensity between 100 and 500 cpm/mg tissue was given a rate of +. (background signal <100). Similarly, a rate of ++ was given to the intensity between 500 and 900, +++ to 900 and 1300, and ++++ to 1300 and 1700 cpm/mg tissue. That information is provided in Tables 1 and 2.
In the 16-day-old rat (Fig. 2F), GR-mRNA abundance was maximal in the hippocampal CA1/CA2 and PVN (Table 2). Intermediate GR-mRNA signal was observed over the dentate gyrus, central nucleus of amygdala, endopiriform nucleus, and cortical areas (Table 2). Of interest, GR-mRNA abundance in dentate gyrus increased between Day 10 and 16, in agreement with Vazquez et al. (15).
DISCUSSION
This study demonstrates the presence of GR-mRNA in rat telencephalon as early as the 14th fetal day (F14). We further find that hippocampal/septal structures express GR-mRNA prior to hypothalamic nuclei.
In hippocampus, the fimbria region is the first to contain GR-mRNA, followed by hippocampal formation by F16–17. By the first postnatal day, robust GR-gene expression is visible in CA1, with little message in CA3 or dentate gyrus. The differential distribution of GR-mRNA in hippocampus (CA1/CA2 > CA3) persists throughout the first postnatal week to Postnatal Day 16. These results are in overall agreement with those of Van Eekelen (2) and Vazquez et al. (15), though these authors found low amounts of GR-mRNA in CA3-CA4 as well.
We found GR-mRNA in the hypothalamic SCN on Postnatal Day 6 (Fig. 2C). Van Eekelen et al. were unable to demonstrate GR-mRNA presence in this region, though the same group had previously demonstrated GR-containing cells in the SCN. The reason for the discrepancy is not clear, but may relate to diurnal variability in GR gene expression in cells of this “biological clock” structure. Van Eekelen et al. (2) did not indicate the time of sacrifice.
In the hypothalamic PVN, GR-mRNA is clearly detectible on F16. The distribution of GR-expressing cells overlaps the parvicellular PVN. In postnatal and adult rats, GR-mRNA has been localized to cells expressing corticotropin releasing hormone (CRH, 16). CRH-mRNA is first detectible using ISH on the morning of F17 (8, 17). In the mature brain, GR participate in mediating GC regulation of the CRH gene promoter (12,18). This effect is at least partially mediated by GR receptors in the PVN (19, 20). In the fetal rat, alteration of circulating GCs does not affect the time of onset or the magnitude of CRH gene expression, at least as assessed by CRH-mRNA abundance (8, 9, and unpublished results). Thus, GR-mRNA presence in the PVN may not correlate with the presence of functional GR, as has been demonstrated in the fetal sheep (21). Alternatively, second messenger cascades transducing GC effects on the CRH gene promoter (18, 22, 23) may not be functional in the fetal rat. GR in the fetus may play other roles in neuronal development and function, possibly through trophic or direct membrane effects (24).
In conclusion, discrete limbic and hypothalamic structures express GR-mRNA during the last third of rat fetal life. GR-mRNA distribution is similar to, but not identical to that in postnatal and adult rats. GR may mediate different functions in developing versus mature neurons.
Acknowledgments
This work was supported by NS01307 and NS28912 (T.Z.B.). J.N.M. was supported by a grant from the American Federation for Aging Research.
References
- 1.Rosenfeld P, Van Eekelen JA, Levine S, de Kloet ER. Ontogeny of the type 2 glucocorticoid receptor in discrete rat brain regions: An immunocytochemical study. Brain Res. 1988;470:119–127. doi: 10.1016/0165-3806(88)90207-6. [DOI] [PubMed] [Google Scholar]
- 2.Van Eekelen JAM, Bohn MC, de Kloet ER. Postnatal ontogeny of mineralocorticoid and glucocorticoid receptor gene expression in regions of the rat tel- and diencephalon. Dev brain Res. 1991;61:33–43. doi: 10.1016/0165-3806(91)90111-u. [DOI] [PubMed] [Google Scholar]
- 3.Kalinyak JE, Griffin CA, Hamilton RW, Bradshaw JG, Perlman AJ, Hoffman AR. Developmental and hormonal regulation of glucocorticoid receptor messenger RNA in the rat. J Clin Invest. 1989;84:1843–1848. doi: 10.1172/JCI114370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwen BS, Angulo J, Cameron H, Chao HM, Daniels D, Gannon MN, Gould E, Mendelson S, Sakai R, Spencer R, Woolley C. Paradoxical effects of adrenal steroids on the brain: Protection versus degeneration. Biol Psych. 1992;31:177–199. doi: 10.1016/0006-3223(92)90204-d. [DOI] [PubMed] [Google Scholar]
- 5.Yehuda R, Fairman KR, Meyer JS. Enhanced brain cell proliferation following early adrenalectomy in rats. J Neurochem. 1989;53:241–248. doi: 10.1111/j.1471-4159.1989.tb07320.x. [DOI] [PubMed] [Google Scholar]
- 6.de Kloet ER, Reul JMHM, Sutanto W. Corticosteroids and the brain. J Steroid Biochem Mol Biol. 1990;37:387–394. doi: 10.1016/0960-0760(90)90489-8. [DOI] [PubMed] [Google Scholar]
- 7.Meaney MJ, Aitken DH, Sharma S, Viau V. Basal ACTH, corticosterone and corticosterone binding globulin levels over the diurnal cycle, and age-related changes in hippocampal type I and type II corticosteroid receptor binding capacity in young and aged, handled and nonhandled rats. Neuroendocrinology. 1992;55:204–213. doi: 10.1159/000126116. [DOI] [PubMed] [Google Scholar]
- 8.Baram TZ, Lerner SP. Corticotropin releasing hormone—Ontogeny of gene expression in rat hypothalamus. Int J Dev Neurosci. 1991;9:473–478. doi: 10.1016/0736-5748(91)90033-i. [DOI] [PubMed] [Google Scholar]
- 9.Baram TZ, Schultz L. CRF gene expression in the fetal rat is not increased after pharmacological adrenalectomy. Neurosci lett. 1992;142:215–218. doi: 10.1016/0304-3940(92)90376-i. [DOI] [PubMed] [Google Scholar]
- 10.Yi SJ, Masters JN, Baram TZ. The effect of RU-38486 on CRH gene expression in the neonatal rat hypothalamus. Dev Brain Res. 1993;73:253–259. doi: 10.1016/0165-3806(93)90145-z. [DOI] [PubMed] [Google Scholar]
- 11.Herman JP, Patel PD, Akil H, Watson SJ. Localization and regulation of glucocorticoid and mineralocorticoid receptor messenger RNAs in the hippocampal formation of the rat. Mol Endocrinol. 1989;3:1886–1894. doi: 10.1210/mend-3-11-1886. [DOI] [PubMed] [Google Scholar]
- 12.Young WS, III, Mezey E, Siegel RE. Quantitative in situ hybridization histochemistry reveals increased levels of corticotropin releasing factor mRNA after adrenalectomy in rats. Neurosci Lett. 1986;70:198–203. doi: 10.1016/0304-3940(86)90463-5. [DOI] [PubMed] [Google Scholar]
- 13.Paxinos G, Tork I, Tecott LH, Valentino KL. Atlas of the Developing Rat Brain. Academic Press; San Diego: 1991. [Google Scholar]
- 14.Sherwood NM, Timiras PS. A Stereotaxic Atlas of the Developing Rat Brain. Univ. Calif. Press; Berkeley: 1970. [Google Scholar]
- 15.Vazquez DM, Morano MI, Lopez JF, Watson SJ, Akil H. Short-term adrenalectomy increases glucocorticoid and mineralocorticoid receptor mRNA in selected areas of the developing hippocampus. Mol Cell Neurosci. 1993;4:455–471. doi: 10.1006/mcne.1993.1057. [DOI] [PubMed] [Google Scholar]
- 16.Swanson LW, Simmons DM. Differential steroid hormone and neuronal influences on peptide mRNA levels in CRH cells of the paraventricular nucleus: A hybridization histochemical study in the rat. J Comp Neurol. 1989;285:413–435. doi: 10.1002/cne.902850402. [DOI] [PubMed] [Google Scholar]
- 17.Grino MW, Young WS, III, Burgunder JM. Ontogeny of expression of the CRF gene in the hypothalamic paraventricular nucleus and of the proopiomelanocortin gene in rat pituitary. Endocrinology. 1989;124:60–68. doi: 10.1210/endo-124-1-60. [DOI] [PubMed] [Google Scholar]
- 18.Majzoub JA, Emanuel R, Adler G, Martinez C, Robinson B, Wittert G. Corticotropin Releasing Factor, CJBA Symposium 172. Wiley; Chichester: 1993. Second messenger regulation of mRNA for CRF; pp. 31–43. [DOI] [PubMed] [Google Scholar]
- 19.Sawchenko PE. Evidence for a local site of action for glucocorticoids in inhibiting CRF and vasopressin expression in the paraventricular nucleus. Brain Res. 1987;403:213–224. doi: 10.1016/0006-8993(87)90058-8. [DOI] [PubMed] [Google Scholar]
- 20.Kovacs KJ, Mezey E. Dexamethasone inhibits corticotropin-releasing factor gene expression in the rat paraventricular nucleus. Neuroendocrinology. 1987;46:365–368. doi: 10.1159/000124846. [DOI] [PubMed] [Google Scholar]
- 21.Yang KG, Hammond L, Challis JR. Characterization of an ovine glucocorticoid receptor cDNA and developmental changes in its mRNA levels in the fetal sheep hypothalamus, pituitary gland and adrenal. J Mol Endocrinol. 1992;8:173–180. doi: 10.1677/jme.0.0080173. [DOI] [PubMed] [Google Scholar]
- 22.Van LP, Spengler DH, Holsboer F. Glucocorticoid repression of 3′5′-cyclic adenosine monophosphate-dependent human corticotropin releasing hormone gene promoter activity in a transfected mouse anterior pituitary cell line. Endocrinology. 1990;127:1412–1418. doi: 10.1210/endo-127-3-1412. [DOI] [PubMed] [Google Scholar]
- 23.Seasholtz AF, Thompson RC, Douglass IO. Identification of a cyclic adenosine monophosphate-responsive element in the rat corticotropin releasing hormone gene. Mol Endocrinol. 1988;2:1311–1318. doi: 10.1210/mend-2-12-1311. [DOI] [PubMed] [Google Scholar]
- 24.Joels M, de Kloet ER. Control of neuronal excitability by corticosteroid hormones. Trends Neurosci. 1992;15:25–29. doi: 10.1016/0166-2236(92)90345-9. [DOI] [PubMed] [Google Scholar]