Abstract
Tibial muscular dystrophy (TMD) is an autosomal dominant late-onset distal myopathy linked to chromosome 2q31. The linked region includes the giant TTN gene, which encodes the central sarcomeric protein, titin. We have previously shown a secondary calpain-3 defect to be associated with TMD, which further underscored that titin is the candidate. We now report the first mutations in TTN to cause a human skeletal-muscle disease, TMD. In Mex6, the last exon of TTN, a unique 11-bp deletion/insertion mutation, changing four amino acid residues, completely cosegregated with all tested 81 Finnish patients with TMD in 12 unrelated families. The mutation was not found in 216 Finnish control samples. In a French family with TMD, a Leu→Pro mutation at position 293,357 in Mex6 was discovered. Mex6 is adjacent to the known calpain-3 binding site Mex5 of M-line titin. Immunohistochemical analysis using two exon-specific antibodies directed to the M-line region of titin demonstrated the specific loss of carboxy-terminal titin epitopes in the TMD muscle samples that we studied, thus implicating a functional defect of the M-line titin in the genesis of the TMD disease phenotype.
Introduction
Tibial muscular dystrophy (TMD [MIM 600334]) is an autosomal dominant late-onset distal myopathy that was first described, in Finnish patients, by Udd et al. (1991, 1993). In TMD, weakness and atrophy are usually confined to the anterior compartment of the lower leg—in particular, the tibialis anterior muscle. Clinical symptoms usually occur at age 35–45 years—or much later. Cardiomyopathy has not been diagnosed in patients with TMD, and there is no facial-muscle involvement (Udd et al. 1992, 1993).
We have assigned linkage of TMD to a 1-cM region on chromosome 2q31, between markers D2S2173 and D2S2310, in the Finnish families with TMD (Haravuori et al. 1998). All Finnish patients with TMD carry a common core haplotype for the linked markers, which indicates that there is a single ancestral founder mutation responsible for TMD in the Finnish population. Three patients with a more severe childhood-onset limb-girdle muscular dystrophy (LGMD) phenotype were found to be homozygous for the TMD haplotype (Haravuori et al. 1998). The prevalence of TMD is >6/100,000, which makes it one of the most common myopathies in Finland. Linkage to this same chromosome 2q31 locus has also been confirmed in a French family with TMD that showed another haplotype for the linked markers (de Seze et al. 1998).
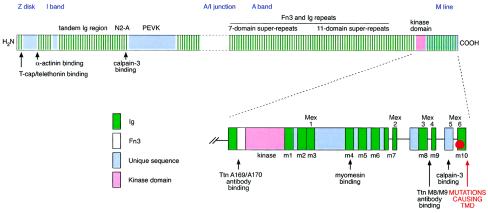
The major positional and functional candidate gene at the TMD locus is the TTN gene, encoding the giant muscle protein titin (formerly named “connectin”) (Haravuori et al. 2001). The entire coding region of titin consists of 363 exons, which encodes 38,138 amino acid residues (4,200 kD). The size of skeletal-muscle titin is 3,700 kD, and the physical length in vivo is 2 μm (Bang et al. 2001). Titin has mechanical, developmental, and regulatory roles in striated muscles. It stretches over the length of one-half of the sarcomere, from the Z disk to the M line (Labeit and Kolmerer 1995). Titin has many properties that make it a highly interesting candidate for TMD. One of the main functions of titin is to keep the contractile elements of the sarcomere in place, and it is responsible for muscle elasticity. Titin also provides multiple ligand binding sites for a large number of other muscle proteins, including the muscle-specific protease calpain-3, α-actinin, myosin, myomesin, and telethonin (Labeit et al. 1992; Trinick 1994; Freiburg and Gautel 1996; Young et al. 1998). Z-disk titin contains telethonin/T-cap binding sites and multiple α-actinin binding sites (Sorimachi et al. 1997; Gregorio et al. 1998; Mues et al. 1998), and it interacts indirectly with myotilin via α-actinin (Salmikangas et al. 1999). Mutations in calpain-3, myosin, telethonin, and myotilin are known to cause other muscular dystrophies. Close to its C-terminus, titin has a Ser/Thr–kinase domain, homologous to myosin light-chain kinases (Labeit et al. 1992), which potentially phosphorylates telethonin during early myocyte differentiation (Mayans et al. 1998) (see fig. 1).
Figure 1.
Structure of skeletal-muscle titin. The location of calpain-3 binding sites and the site of Mex6 mutations are indicated. Lower schematic picture shows the 3′/C-terminal modular structure of titin, providing a more detailed illustration of ligand and antibody binding sites.
TTN has several sites for alternative splicing, causing isoforms of different lengths to appear in different muscles (Bang et al. 2001). I-band isoforms have been observed to be longer in skeletal (3,700 kD) than in cardiac (2,970–3,300 kD) muscle, whereas, in the Z disk, cardiac-muscle titins contain more repeat motifs than do skeletal-muscle titins (Gautel et al. 1996; Sorimachi et al. 1997). Interacting with obscurin, a C-terminally truncated 700-kD isoform of titin is expressed in cardiac muscle (Bang et al. 2001). The M-line region of titin is also differentially expressed, and two different splice isoforms have been identified: Mex5+ and Mex5− (Labeit and Kolmerer 1995; Kolmerer et al. 1996; Sorimachi et al. 1997). On the ultrastructural level, both the carboxy-terminal Mex6 titin epitope and the catalytic titin kinase domain have been localized within the periphery of the M-line lattice (Obermann et al. 1997). Therefore, the Mex5/Mex6 titin epitopes, the catalytic titin kinase domain, and the p94 and muscle-specific ring finger–1 (MURF-1) proteins that are bound to this region of titin may form a signaling complex (Sorimachi et al. 1997; Centner et al. 2001). The occurrence of several different titin isoforms may be relevant for the selective involvement of muscles in anatomically restricted myopathies (Sorimachi et al. 1997). Recently, autosomal dominant dilated cardiomyopathy was shown to be associated with A-band TTN mutations in exon 326, which cause a frameshift, and with a mutation in the Z-disk TTN (Siu et al. 1999; Gerull et al. 2002).
Autosomal recessive LGMD2A is caused by mutations in the gene, on chromosome 15, that encodes muscle-specific calpain-3 (Richard et al. 1995). Myonuclear apoptosis and perturbation of the IκBα/nuclear factor–κB pathway have been associated with absence of functional calpain-3 in LGMD2A muscle (Baghdiguian et al. 1999). Because of the similarities between LGMD2A and the LGMD phenotype that is associated with homozygous state of TMD, we previously screened for—and found—secondary calpain-3 deficiency in TMD (Haravuori et al. 2001). Myonuclear apoptosis and altered IκBα/nuclear factor–κB pathway were also observed (Haravuori et al. 2001).
In mice, a spontaneous mutation in the proximal part of chromosome 2, a region syntenic to human chromosome 2q31, causes muscular dystrophy with myositis (MDM) (Siracusa et al. 1990; Müller-Seitz et al. 1993). The mouse titin gene (Ttn) colocalizes with this disease locus, and a deletion of the N2-A–line calpain-3 binding site of mouse titin was recently reported to cause MDM (Garvey et al. 2002), a result that is consistent with our previous demonstration of secondary calpain-3 deficiency in the MDM mouse (Haravuori et al. 2001).
Titin has at least two binding sites for calpain-3, and it has been suggested that it stabilizes calpain-3 from autolytic degradation (see fig. 1). The three immunoglobulin (Ig) repeats (exons I81–I83) on the N2-A line of I-band titin constitute one binding site, and the other is in M-line titin (Sorimachi et al. 1995).
The complete 294-kb sequence of TTN is organized into 363 exons (GenBank accession number AJ277892). All exons originally known to be expressed in human soleus skeletal muscle (348 of a total of 363) of TTN were sequenced. Samples sequenced were from two Finnish heterozygous patients with TMD, three unrelated French heterozygous patients with TMD, one Finnish homozygous patient with TMD, and unaffected control individuals.
Material and Methods
Patient Material
Patient material was obtained from the four Finnish families (with 68 affected and 70 healthy members total) and one French family (with 3 affected and 4 healthy members) that were included in the previous linkage studies (de Seze et al. 1998; Haravuori et al. 1998). In the present study, we included 13 patients, from eight unrelated families, who have not been described elsewhere, and their 6 healthy relatives. DNA extracted from blood lymphocytes and the muscle biopsy samples for immunohistochemical analyses were available from previous studies (Haravuori et al. 1998, 2001). All the samples were obtained from informed and consenting individuals. The study protocol was approved by the ethical board of Vasa Central Hospital.
Sequencing
The entire TTN coding region as known at the time, including 348 of 363 exons, was sequenced using patient and control DNA. Ninety-three unaffected population control individuals from France were screened, for exon Mex6 mutations, by sequencing. The titin sequences were amplified by PCR and were run in agarose gels to confirm fragment size and specificity of the PCR. PCR products were purified by Qiagen columns before sequencing. The sense and antisense strands were sequenced using a BigDye Terminator Cycle sequencing Ready Reaction DNA kit (Applied Biosystems), according to the manufacturer's instructions (Chadwick et al. 1996). Extension products were purified, and the samples were run on an ABI 377 sequencer (Perkin Elmer). Sequencher 3.1.1 software was used for analysis of the sequences.
SSCP Analysis
SSCP analysis, as described by Vidal-Puig and Moller (1994), was used to screen patients and control individuals for the putative TMD-causing mutation in exon Mex6 of TTN. PCRs were performed using a primer pair designed for a 184-bp fragment, including the 11-bp mutated area, of the Mex6 exon. Primers used were as follows: A (CAGCATTGATGAAGGCAAAG) and B (TCCACCATCTTGTTTCTGTACG). DNA samples from 78 heterozygous and 3 homozygous patients with TMD, as well as from 76 healthy relatives, were tested. Two hundred sixteen unaffected population control individuals from Finland (56 from the National Public Health Institute and 160 from the Finnish Red Cross) were screened for the mutation. The PCR products were mixed with loading buffer, were denatured at 95°C for 5 min, and were placed on ice. Gel electrophoresis was performed at 3 W for 15 h in a 0.5 × MDE polyacrylamide gel (BioWhittaker Molecular Applications) in 0.6 × Tris-borate–EDTA buffer. The gels were stained using the silver staining method and were fixed and dried (Vidal-Puig and Moller 1994; Donner et al. 2002). The gels were scanned at 300 dpi with an Agfa Snapscan 1236s image scanner. Adobe Photoshop, version 5.5, was used for image enhancement and detection of the bands. The sequences containing the mutation showed aberrant migration and, by this method, proved to be very clearly distinguishable from the wild-type sequence.
Primers
More than 500 primers for PCR amplification and sequencing were designed using the Primer3 software (developed, at the Whitehead Institute for Biomedical Research, by Steve Rozen and Helen J. Skaletsky; available at the Primer3 Software Distribution Web site). Primer sequences are available on request.
RNA Extractions and RT-PCRs
Total RNA was extracted with Purescript RNA Isolation Kit (Gentra) from muscle biopsy samples that were surgically obtained, were immediately frozen in liquid nitrogen, and were stored at −80°C. cDNA was synthesized by use of 200 ng total RNA as template, with 200 U M-MLV reverse transcriptase (Promega), 20 U RNAse inhibitor (Rnasin; Promega), 0.8 μM specific primer, 0.6 mM deoxyribonucleoside triphosphate, and buffer in a total volume of 25 μl.
Immunohistochemical Methods
Muscle biopsy samples from one homozygous patient with TMD, from three heterozygous patients with TMD, and from an unaffected control individual were analyzed. Standard immunohistochemistry and immunofluorescence protocols were used on unfixed frozen 6-μm longitudinal muscle sections. For obtainment of exon-specific titin antibodies, the Ig/Fn3 domains A169/A170 (corresponding to the Mex1 exon and containing the MURF-1 binding site) and the Ig domains M8/M9 (corresponding to Mex3/Mex4) were expressed in Escherichia coli essentially as described elsewhere (Gregorio et al. 1998). Rabbit polyclonal antibodies were raised to the purified antigens, and obtained antisera were affinity purified (Biogenes). Both affinity-purified polyclonal antibodies were used at 1:200 dilutions. The monoclonal anti-myomesin B4 (Grove et al. 1984) was used at dilution 1:10. A Ventana Nexes automated immunostainer (Ventana Medical Systems) was employed for immunohistochemistry, by the avidin-biotin–complex method followed by 3,3′-diaminobenzidine (DAB) detection. Immunofluorescent detection was manually performed using fluorescein isothiocyanate (FITC)–conjugated secondary antibody (DAKO 0205; DAKO A/S). A control section from healthy muscle was placed on every slide.
Results
More than 20 SNPs were detected in TTN in patients with TMD (for previously reported polymorphisms, see Haravuori et al. 2001). None of these SNPs were frameshift mutations, and only five of them either were close to intron/exon borders or, in exons, changed an amino acid residue; the amino acid charge was not changed in these cases (see table 1). In the Finnish TMD samples, a unique mutation was identified in Mex6, the 363rd and last exon of TTN. Mex6 of TTN encodes an Ig domain that, in situ, is localized at the periphery of the M-line lattice. Mex6, together with Mex5, is in the region determining the calpain-3 binding site of M-line titin. The mutation consists of an 11-bp change, AAGTAACATGG→TGAAAGAAAAA, at position 293,269–293,279 in the TTN sequence. The mutation changes four amino acids—GAA→GTG (Glu→Val), GTA→AAA (Val→Lys), ACA→GAA (Thr→Glu), TGG→AAA (Trp→Lys) (see fig. 2)—close to the C-terminal end of the titin protein but does not cause a frameshift or a stop codon. Each of the four amino acids is changed to an amino acid of another charge, and the overall charge is changed from acidic (−1) to basic (+1). Neither the particular Mex6 DNA sequence variant nor the resultant amino acid sequence of this variant part (underlined) of titin, DEGKVLTVACAFTGEPTPVKEKCGGR, has been reported elsewhere, as tested by performance of a nucleotide sequence and a protein search (see the National Center for Biotechnology's BLAST Web site) (Altschul et al. 1997). The affected titin protein segment is relatively well conserved in humans, pigs, mice, and chickens. There are some very small differences in mouse and chicken protein sequences, compared to human titin.
Table 1.
New TTN Sequence Differences between Finnish Patients with TMD and Control Individuals[Note]
| Exon (Position) in TTN | Mutation | Effect |
| −4 of PEVK66 (162,097) | T→C | Close to splice site |
| A45 (233,784) | ACT→ATT | Thr→Ile |
| A149 (274,193) | AGT→GGT | Ser→Gly |
| A166 (283,464) | GTT→ATT | Val→Ile |
| Mex6 (293,268–293,280) | 11-bp change | 4-amino-acid change |
| Mex6 UTR (293,564) | A insertion | Ins in 8 A stretch |
Note.— Only changes that cosegregated with TMD, either in exons causing an amino acid change or at exon/intron borders, are listed. The changes in A45 and A149 cause the exchange from neutral polar amino acid to neutral nonpolar amino acid. The insertion at position 293,564 is outside the translated region, in the last exon. (For previously reported polymorphisms, see Haravuori et al. 2001.)
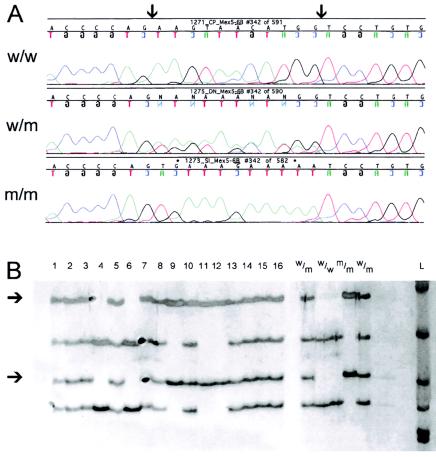
Figure 2.
The TMD-associated 11-bp mutation at positions 293,268–293,280 in exon Mex6 of TTN. A, Nucleotide sequence with borders of mutated area indicated by arrows. B, SSCP analysis of exon Mex6 of TTN, including the associated mutation, from patients with TMD and unaffected control individuals. The migration of PCR products matches the haplotypes and phenotypes of the patients. Bands indicating the mutation are marked by arrows. Samples are denoted “1”–“16.” w/w = Wild-type sequence from an unaffected control individual; w/m = TMD heterozygote; m/m = TMD homozygote; L = 100-bp DNA ladder.
The 11-bp mutation was easily detected by SSCP analysis (Vidal-Puig and Moller 1994) (see fig. 2). SSCP studies showed that this mutation did not occur in the 216 Finnish unaffected population control individuals who were tested. When a total of 78 TMD heterozygotes, 3 TMD homozygotes, and 76 healthy first-degree relatives were tested, the mutation showed cosegregation with TMD in 12 unrelated pedigrees, in agreement with full penetrance. One family was an exception: three offspring aged 50–55 years did show the mutation without clinically manifesting the disease. However, in this family, the transmitting affected parent in the older generation had very late onset of symptoms, at 65 years. These three individuals were followed up for detection of exceptional late manifestation. All four families in the previous linkage studies were included, as well as eight new unrelated families.
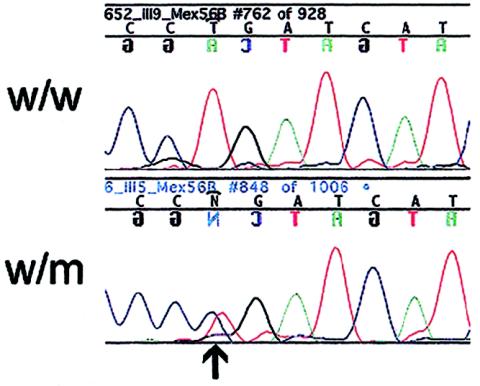
SSCP analysis for this Mex6 region on DNA samples from the chromosome 2q31–linked French family did not show any aberrant bands. However, sequencing samples from the affected persons in this family revealed another potential mutation in Mex6 at position 293,357. This mutation changed CTG to CCG (Leu→Pro) (see fig. 3). Both amino acids are neutral hydrophobic. The mutation was detected in three affected individuals and was not present in four unaffected individuals from the same family. The mutation was not present in the 93 French unaffected population control samples (186 alleles), as analyzed by sequencing. In addition, the mutation was not found in major SNP databases (see the Web sites dbSNP Home Page, SNP Consortium Ltd., and JNSPs Home).
Figure 3.
The French TMD-associated base substitution CTG→CCG at position 293,357, causing a Leu→Pro change in exon Mex6 of TTN. Nucleotide sequence with the mutation indicated by an arrow. w/w = Wild-type sequence from healthy relative; w/m = heterozygous affected relative.
We performed RT-PCR and sequenced the Mex5/Mex6 RNA from biopsy samples of affected tibialis anterior muscle from a Finnish heterozygous patient with TMD. The results showed that the Mex5/Mex6 cDNA sequence is indeed heterologous and that both wild-type and mutant RNAs are expressed in the affected muscle. The sequences were in frame, indicating that the mutation does not affect the splicing of these exons.
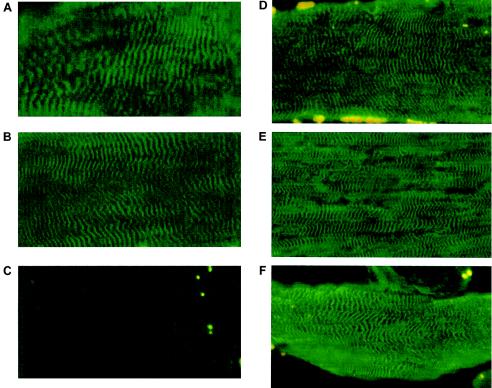
Immunohistochemical analyses of muscle biopsy samples that were homozygous for the Finnish mutation indicated loss of the M8/M9 titin epitopes, which are encoded by the Mex3/Mex4 exons, when either DAB or FITC detection methods were used. The loss of C-terminal titin epitopes appeared to be specific, since TMD heterozygotes and an unaffected control individual showed normal cross-striated sarcomeric M-line–specific labeling when this antibody was used (see fig. 4). Interestingly, the nearby A169/A170 titin epitopes could be detected in biopsy samples from both homozygous and heterozygous patients with the Finnish mutation, as well as in unaffected control individuals (see fig. 4).
Figure 4.
Muscle sections from an unaffected control individual (A), from a heterozygous patient with TMD (B), and from a homozygous patient with TMD and LGMD (C), immunostained with titin M8/M9 antibody, and sections from an unaffected control individual (D), from a heterozygous patient with TMD (E), and from a homozygous patient with TMD and LGMD (F), immunostained with titin A169/A170 antibody and detected by FITC.
Myomesin is a titin ligand relevant to the structure of the M line in the sarcomere (Grove et al. 1984). Immunohistochemistry showed normal expression of myomesin in muscle from homozygous and heterozygous patients with TMD (data not shown), consistent with unaltered titin binding with myomesin in the M4 domain upstream from the M8/M9 titin epitopes (Obermann et al. 1997). In conclusion, our immunohistochemical data suggest the specific loss of titin epitopes C terminally of M4. This localized titin defect appears to be compatible with the assembly of normal sarcomeric ultrastructures in TMD (Udd et al. 1993).
Discussion
Our recent linkage studies have suggested that TTN is a promising candidate gene for TMD—although, because of the giant coding size of titin, no mutations could be identified so far. Here, we describe two sequence alterations in patients with TMD. We identified a complex 11-bp mutation in the large Finnish population with TMD and a 1-bp exchange in a French family with TMD, and both of these mutations were localized in Mex6 (exon 363 in TTN [GenBank accession number AJ277892]).
The 11-bp Mex6 mutation represents a very unusual type of mutation and causes, on the protein level, four amino acid exchanges within the conserved hydrophobic core of the M10 domain of M-line titin. It is complex but not an inversion, and it does not seem to have originated from a secondary loop in the area. The mutation could have originated from a recombination. The same sequence, among three other locations, was found within an intron that is located (using the “old numbering”) between the A-band exons 246 (Fn3) and 247 (Ig) in the wild-type TTN gene, 87,204 bp upstream from the mutated site in Mex6. One of the four changed amino acids, the tryptophan, is mutated into a charged lysine. The tryptophan residue is generally conserved in all Ig repeats of titin and is known to be crucially important for stabilization of the hydrophobic core of the Ig β-barrel fold (Improta et al. 1996). In the French family, the mutation at position 293,357 in Mex6 changed CTG to CCG (Leu→Pro). Both amino acids are neutral hydrophobic. Since proline differs from leucine because of its circular structure, it may introduce a kink and cause significant conformational change of M-line titin structure. The mutation cosegregated with the disease in the family and was not seen in 93 French unaffected population control samples (186 alleles), suggesting that this base exchange is not merely a polymorphism but could indeed be related to the disease. Similarly, the 11-bp exchange cosegregated with the disease in the Finnish families and was absent from 200 Finnish control chromosomes. The finding of two independent mutations in the same exon, Mex6, in different families with TMD further strengthens the argument that these mutations really are the cause of TMD.
Both mutant and wild-type titin RNAs are transcribed in muscle from heterozygous patients with TMD, as shown by RT-PCR and sequencing in the affected muscles. The sequences of the heterologous cDNA were in frame, indicating that the mutation does not affect the splicing of the exons Mex5 and Mex6. It is in accordance with our immunofluorescence data that the mutant titin protein is present in the muscle from the homozygous patient with TMD, suggesting that the mutant titin is normally translated and incorporated into the myofibril.
The lack of epitope recognition by the M-line M8/M9 antibody and the normal labeling of TMD homozygote muscle with the antibody A169/A170, together with the normal myomesin expression, indicate that the titin structure may be intact up to the level of Mex2 and may be conformationally disrupted in the Mex3–Mex6 region. This part of titin is located within the periphery of the M-line lattice. Interestingly, several protein-protein–interaction sites are localized to this region, which could be involved in myofibrillar signaling—namely, the titin kinase domain (Labeit et al. 1992; Mayans et al. 1998), p94/CAPN3 (Sorimachi et al. 1995), and MURF-1 (Bodine et al. 2001; Centner et al. 2001). Therefore, it remains to be seen if the disruption of the M-line portion of titin could be involved in the perturbation of titin-based myofibrillar signaling. Defective calpain-3 regulation caused by M-line titin mutations could be one part of the mechanism. The facts that Mex6 is expressed in heart-muscle titin and that TMD shows no cardiomyopathy are compatible with defective titin–calpain-3 interaction. Calpain-3 is not expressed in mature heart muscle (Fougerousse et al. 2000). Variable proportions of Mex5+ and Mex5− isoforms in different muscles may be responsible for selective muscle involvement. In addition, still-unknown interactions disturbed by the mutations in M-line titin are indicated.
Defective titin–calpain-3 interaction cannot explain all aspects of the disease mechanism. In primary calpainopathy, LGMD2A, in which there is no functional calpain-3 in the muscle, all muscles are not equally affected, which indicates that other molecules interact and modify the phenotype of individual muscles (Fardeau et al. 1996). Our previous studies showed severe reduction of calpain-3 in homozygous TMD muscle and in the MDM mouse (Haravuori et al. 2001). In the recent report of the MDM mutation, Garvey et al. (2002) confirmed a decrease of calpain-3 in muscle from a 4-wk-old MDM mouse. The phenotype in mice with targeted calpain-3 mutation is less severe than in the MDM mouse (Richard et al. 2000), and the phenotype in homozygous TMD, with small amounts of calpain-3 left in the muscle, is as severe as in LGMD2A. This indicates that other unknown molecular mechanisms are also disrupted by the TMD and MDM mutations in the calpain-3 binding regions.
Functional genomics and the use of the natural animal model, the MDM mouse, will be needed for a better understanding of the titin–calpain-3 interaction, both at the N2-A line and at the M line, and for the detection of the missing parts and functions of the pathomechanism. The selective involvement of skeletal muscles in TMD heterozygotes remains to be explained, and clarification of the sparing of other muscles may also reveal clues for therapeutic hypotheses.
Acknowledgments
We acknowledge the dedicated cooperation of the families with TMD and Drs. K. Pelin, C. Wallgren-Pettersson, and H. Somer, who provided assistance with this study, and we thank Mrs. Merja Soininen and Mr. Philippe Hajdari, for technical assistance, and Dr. J.-C. Perriard, for the generous gift of the monoclonal anti-myomesin B4. Grants were from Association Française contre les Myopathies, Muscular Dystrophy Association, the Sigrid Juselius Foundation, the Vasa Central Hospital Research Funds, Finska Läkaresällskapet, the Finnish Cultural Foundation and the Maud Kuistila Memorial Foundation, and the Deutsche Forschungsgesellschaft (La668/6-2).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for TTN sequence [accession number AJ277892])
- JNSPs Home, http://snp.ims.u-tokyo.ac.jp/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for TMD [MIM 600334])
- Primer3 Software Distribution, http://www-genome.wi.mit.edu/genome_software/other/primer3.html
- SNP Consortium Ltd., The, http://snp.cshl.org/
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdiguian S, Martin M, Richard I, Pons F, Astier C, Bourg N, Hay RT, Chemaly R, Halaby G, Loiselet J, Anderson LVB, Munain ALD, Fardeau M, Mangeat P, Beckmann JS, Lefranc G (1999) Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IκBα/NF-κB pathway in limb-girdle muscular dystrophy type 2A. Nat Med 5:503–511 [DOI] [PubMed] [Google Scholar]
- Bang M-L, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S (2001) The complete gene sequence of titin, expression of an unusual 700-kDa titin isoform and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res 89:1065–1072 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704–1708 [DOI] [PubMed] [Google Scholar]
- Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang M-L, Tombitás K, Granzier H, Gregorio CC, Sorimachi H, Labeit S (2001) Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol 306:717–726 [DOI] [PubMed] [Google Scholar]
- Chadwick RB, Conrad MP, McGinnis MD, Johnston-Dow L, Spurgeon SL, Kronick MN (1996) Heterozygote and mutation detection by direct automated fluorescent DNA sequencing using a mutant Taq DNA polymerase. Biotechniques 20:676–683 [DOI] [PubMed] [Google Scholar]
- de Seze J, Udd B, Haravuori H, Sablonniere B, Maurage CA, Hurtevent JF, Boutry N, Stojkovic T, Schraen S, Petit H, Vermersch P (1998) The first European family with tibial muscular dystrophy outside the Finnish population. Neurology 51:1746–1748 [DOI] [PubMed] [Google Scholar]
- Donner K, Ollikainen M, Ridanpää M, Christen H-J, Goebel HH, de Visser M, Pelin K, Wallgren-Petterson C (2002) Mutations in the β-tropomyosin (TPM2) gene—a rare cause of nemaline myopathy. Neuromuscul Disord 12:151–158 [DOI] [PubMed] [Google Scholar]
- Fardeau M, Eymard B, Mignard C, Tomé FMS, Richard I, Beckmann J (1996) Chromosome 15-linked limb-girdle muscular dystrophy: clinical phenotypes in Reunion Island and French metropolitan communities. Neuromuscul Disord 6:447–453 [DOI] [PubMed] [Google Scholar]
- Fougerousse F, Anderson LVB, Delezoide A-L, Suel L, Durand M, Beckmann JS (2000) Calpain3 expression during human cardiogenesis. Neuromuscul Disord 10:251–256 [DOI] [PubMed] [Google Scholar]
- Freiburg A, Gautel M (1996) A molecular map of the interactions between titin and myosin-binding protein C: implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem 235:317–323 [DOI] [PubMed] [Google Scholar]
- Garvey SM, Chandrika R, Lerner AP, Frankel WN, Cox GA (2002) The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics 79:146–149 [DOI] [PubMed] [Google Scholar]
- Gautel M, Goulding D, Bullard B, Weber K, Fürst DO (1996) The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J Cell Sci 109:2747–2754 [DOI] [PubMed] [Google Scholar]
- Gerull B, Gramlich M, Atherton J, McNabb M, Trombitás K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L (2002) Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet 30:201–204 [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, Sorimachi H, Labeit S (1998) The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J Cell Biol 143:1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove BK, Kurer V, Lehner C, Doetschman TC, Perriard J-C, Eppenberger HM (1984) A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol 98:518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haravuori H, Mäkelä-Bengs P, Udd B, Partanen J, Pulkkinen L, Somer H, Peltonen L (1998) Assignment of the tibial muscular dystrophy locus to chromosome 2q31. Am J Hum Genet 62:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haravuori H, Vihola A, Straub V, Auranen M, Richard I, Marchand S, Voit T, Labeit S, Somer H, Peltonen L, Beckmann JS, Udd B (2001) Secondary calpain3 deficiency in 2q-linked muscular dystrophy: titin is the candidate gene. Neurology 56:869–877 [DOI] [PubMed] [Google Scholar]
- Improta S, Politou AS, Pastore A (1996) Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. Structure 4:323–337 [DOI] [PubMed] [Google Scholar]
- Kolmerer B, Olivieri N, Witt CC, Herrmann BG, Labeit S (1996) Genomic organization of M line titin and its tissue-specific expression in two distinct isoforms. J Mol Biol 256:556–563 [DOI] [PubMed] [Google Scholar]
- Labeit S, Gautel M, Lakey A, Trinick J (1992) Towards a molecular understanding of titin. EMBO J 11:1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B (1995) Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 270:293–296 [DOI] [PubMed] [Google Scholar]
- Mayans O, van der Ven PFM, Wilm M, Mues A, Young P, Fürst DO, Wilmanns M, Gautel M (1998) Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature 395:863–869 [DOI] [PubMed] [Google Scholar]
- Mues A, van der Ven PFM, Young P, Fürst DO, Gautel M (1998) Two immunoglobulin-like domains of the Z-disc portion of titin interact in a conformation-dependent way with telethonin. FEBS Lett 428:111–114 [DOI] [PubMed] [Google Scholar]
- Müller-Seitz M, Kaupmann K, Labeit S, Jockush H (1993) Chromosomal localization of the mouse titin gene and its relation to “muscular dystrophy with myositis” and nebulin genes on chromosome 2. Genomics 18:559–561 [DOI] [PubMed] [Google Scholar]
- Obermann W, Gautel M, Weber K, Fürst D (1997) Molecular structure of the sarcomeric M-band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J 16:211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C (1995) Mutations in the proteolytic enzyme calpain cause limb-girdle muscular dystrophy type 2A. Cell 81:27–40 [DOI] [PubMed] [Google Scholar]
- Richard I, Roudaut C, Marchand S, Baghdiguian S, Herasse M, Stockholm D, Ono Y, Suel L, Bourg N, Sorimachi H, Lefranc G, Fardeau M, Sebille A, Beckmann JS (2000) Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated IκBα/nuclear factor κB pathway perturbation in mice. J Cell Biol 151:1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmikangas P, Mykkänen O-M, Grönholm M, Heiska L, Kere J, Carpén O (1999) Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum Mol Genet 8:1329–1336 [DOI] [PubMed] [Google Scholar]
- Siracusa LD, Silan CM, Justice MJ, Mercer JA, Bauskin AR, Ben-Neriah Y, Duboule D, Hastie ND, Copeland NG, Jenkins NA (1990) A molecular genetic linkage map of mouse chromosome 2. Genomics 6:491–504 [DOI] [PubMed] [Google Scholar]
- Siu BL, Niimura H, Osborne JA, Fatkin D, MacRae C, Solomon S, Benson DW, Seidman JG, Seidman CE (1999) Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation 99:1022–1026 [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S (1997) Tissue-specific expression and α-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol 270:688–695 [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Kinbara K, Kimura S, Takahashi M, Ishiura S, Sasagawa N, Sorimachi N, Shimada H, Tagawa K, Maruyama K, Suzuki K (1995) Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J Biol Chem 270:31158–31162 [DOI] [PubMed] [Google Scholar]
- Trinick J (1994) Titin and nebulin: protein rulers in muscle? Trends Biochem Sci 19:405–409 [DOI] [PubMed] [Google Scholar]
- Udd B, Kääriäinen H, Somer H (1991) Muscular dystrophy with separate clinical phenotypes in a large family. Muscle Nerve 14:1050–1058 [DOI] [PubMed] [Google Scholar]
- Udd B, Partanen J, Halonen P, Falck B, Hakamies L, Heikkilä H, Ingo S, Kalimo H, Kääriäinen H, Laulumaa V, Paljärvi L, Rapola J, Reunanen M, Sonninen V, Somer H (1993) Tibial muscular dystrophy—late adult-onset distal myopathy in 66 Finnish patients. Arch Neurol 50:604–608 [DOI] [PubMed] [Google Scholar]
- Udd B, Rapola J, Nokelainen P, Arikawa E, Somer H (1992) Nonvacuolar myopathy in a large family with both late adult onset distal myopathy and severe proximal muscular dystrophy. J Neurol Sci 113:214–221 [DOI] [PubMed] [Google Scholar]
- Vidal-Puig A, Moller DE (1994) Comparative sensitivity of alternative single strand conformation polymorphism (SSCP) methods. Biotechniques 17:490–496 [PubMed] [Google Scholar]
- Young P, Ferguson C, Bañuelos S, Gautel M (1998) Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO J 17:1614–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]