Abstract
The BCR gene is implicated in the development of Ph-positive leukemia through its fusion with the nonreceptor tyrosine kinase gene ABL. The normal 160 kDa Bcr protein has several functional domains, and recently one specific role for Bcr was established in the regulation of respiratory burst activity in white blood cells. Bcr expression levels are relatively constant throughout mouse development until adulthood in brain and in hematopoietic tissues, a pattern that is distinctly different from that of the functionally related n-chimerin gene. In the present study, RNA in situ hybridization was used to explore the normal cellular function of Bcr in rodent brain and hematopoietic organs. The data pinpoint the high bcr expression in the brain to the hippocampal pyramidal cell layer and the dentate gyrus, and to the piriform cortex and the olfactory nuclei, reflecting a potentially interesting function for Bcr in these highly specialized brain regions.
Keywords: bcr, Brain, Expression, Mouse
INTRODUCTION
The human BCR locus was first identified in a break-point cluster region on chromosome 22 in Philadelphia (Ph) positive leukemia (Groffen et al., 1985; Heisterkamp et al., 1983): through a t(9;22)(q34;ql 1) translocation, the BCR gene becomes fused with the ABL gene, a nonreceptor tyrosine kinase normally located on chromosome 9 (Heisterkamp and Groffen, 1991; Rowley, 1990). The leukemogenic role of BCR/ABL has been demonstrated experimentally in transgenic mice carrying BCR/ABL fusion-transgenes and by retroviral transduction experiments in which mice receive BCR/ABL-positive bone marrow transplants (Daley et al., 1990; Elefanty et al., 1990; Heisterkamp et al., 1990; Voncken et al., 1992).
The BCR gene encodes a 160 kDa cytoplasmic protein with several functional domains and is a protein likely to participate in signal transduction. Its N-terminal domain encoded by exon 1 has a serine/threonine kinase activity, can oligomerize, and is capable of binding other protein factors, including p145Abl (McWhirter et al., 1993; Sawyers et al., 1991). The central part of Bcr has homology to the G-nucleotide exchange factor (GEF) Dbl, and the carboxy terminal part is homologous to the n-chimerin protein, which is selectively expressed in neuronal tissue (Diekman et al., 1991; Ron et al., 1991). As is the case for n-chimerin, this domain of Bcr has GTP-ase activating protein (GAP) activity toward a subfamily of p21Ras-related small GTP-binding proteins, p21Rho/Rac (Diekman et al., 1991). Recently, we have established at least one function for Bcr as a regulator of p21Rac-mediated superoxide production by the NADPH-oxidase in leukocytes (Voncken et al., 1995).
In the mouse, bcr expression is detectable as early as the zygote stage, continues throughout embryogenesis (unpublished observations), and is found in most adult tissues examined (Heisterkamp et al., 1993). In man, BCR has also been shown to be ubiquitously expressed. Notwithstanding its role in normal and leukemic blood cells (Campbell and Arlinghaus, 1991; Collins et al., 1987; Voncken et al., 1995), the Bcr protein is found in high levels in the brain. This observation has intrigued us as it suggests a possible different function for p160Bcr. This notion is corroborated by the seemingly highly significant function of G-proteins in the brain (Hopkin, 1994). In an attempt to further delineate a putative cellular function of Bcr in the brain, we have studied the regional expression of bcr in the rodent brain by means of RNA in situ hybridization. We also report data on the expression kinetics of bcr during mouse development in hematopoietic organs and in the brain.
MATERIALS AND METHODS
Tissue preparation
Paraffin embedded adult mouse brain sections were obtained from Novagen Inc. (Madison, WI). Adult Sprague–Dawley rats were decapitated rapidly and the brains were removed onto dry ice and stored at −80°C. Coronal sections (20 µm) were mounted on gelatine-coated slides and stored at −80°C.
Probes
A 400-bp fragment of mouse bcr was amplified by RT/PCR with the primers ALLE (Kawasaki et al., 1988) and BEX3 (5' AACCCACTTTCTCATCTCCAG 3') as previously described.(Heisterkamp et al., 1990). The RT/PCR product was gel purified and digested with EcoRI and Clal yielding a 251 -bp fragment that was subcloned directionally into pSK (Stratagene). The identity of the cloned fragment was confirmed by sequencing both strands using the Sequenase 2.0 system (USB). To obtain antisense and sense RNA probes, the construct was linearized with BamHI (antisense) or Xhol (sense) and transcribed in vitro with α-35SUTP using T7 and T3 RNA polymerase, respectively, essentially as described by the manufacturer (Novagen Inc., Madison WI). A second bcr antisense RNA probe was produced by digesting a 1 8-kb HindIII/EcoRI genomic fragment (Zhu et al., 1990) cloned in pSK with Narl and transcription with T7 RNA polymerase. Probes were labeled to high specific activity, typically 2.0 × 108 cpm/µg. The oligonucleotide probe BEX2 (5' GAGGAAGAAGGTGAATCATCG 3') was labeled on the 3' end with 35SdATP using terminal deoxy-nucleotidyl transferase (Bethesda Research Lab.). An oligonucleotide probe for an unrelated gene, CRH, was similarly generated and labeled as specificity control (Baram and Schultz, 1991; Yi et al., 1993).
In situ hybridization procedures
In situ hybridization on paraffin-embedded mouse tissues was performed using the SureSiteII System from Novagen. Briefly, paraffin was removed by three washes in xylene and rehydrated through a graded ethanol series. Tissue pretreatment consisted of ribosome disruption in 0.2 N HC1 for 5 min, deproteinization in 1 µg/ml proteinase K for 5 min, followed by acetylation in 0.1 M triethanolamine-HCl, pH 8.0, containing 0.25% (v/v) acetic anhydride for 5 min. Prehybridization, hybridization with antisense and sense probes on parallel sections, and subsequent washings were performed according to the manufacturers instructions. Slides were apposed to XAR2 films for 1–2 d, then dipped into Kodak NTB-2 emulsion, exposed in a sealed container for 2−3 weeks at +4°C, and finally developed and fixed using Kodak developer D-19 and Kodak fixer, respectively. Slides were stained in 5% aqueous Giemsa, examined and photographed with bright and darkfield optics using an Olympus BH-2 microscope equipped with an Olympus C-35AD-2 camera. In situ hybridization on rat brain sections using the oligonucleotide probes described above was performed essentially as previously described (Baram and Schultz, 1991, 1992; Yi et al., 1993). After prehybridization for 1 h and hybridization for 20 h at 40°C in a humidity chamber, serial washes were performed (4 washes at 2×SSC for 15 min at 40°C, followed by one wash in 1×SSC and one in 1×SSC at room temperature), followed by dehydration through ethanol-0.3M NH4Ac and apposition to X-ray film (Hyperfilm B-max, Amersham) for 24–48 h.
Northern blot analysis
Tissues from C57B1/6 × B6CBA F1 mice were minced in a polytron, and total RNA was extracted using the acid guanidinium thiocyanate phenol-chloroform method (Chomczynski and Sacchi, 1987). At embryonic day E15, E17, and E20 the head region was separated from the remainder of the embryo. At parturition (P0) and postnatal day 1, 5, 10, 15, 20, 25, 30, and 35, the entire brain including cerebellum was dissected. To compare the developmental expression of bcr in the brain with bcr expression in hematopoietic tissue, RNA was prepared from the liver at E15, E17, E20, at parturition, and from the spleen at postnatal day 1, 5, 10, 15, 20, 25, 30, and 35. Samples (10 µg) were run on formaldehyde/formamide gels, blotted as described (Sambrook et al., 1989), and subsequently hybridized to the 1.8-kb HindIII/EcoRI probe containing coding sequences from mouse bcr exon 1. Filters were washed at low stringency (2.5×SSC, 65°C for 20 min). At this stringency the probe also cross-hybridizes to the 28S and 18S ribosomal bands, making it possible to check for equal loading of the lanes. Densitometric scanning was performed using an in-house manufactured densitometric device.
RESULTS
In situ hybridization patterns
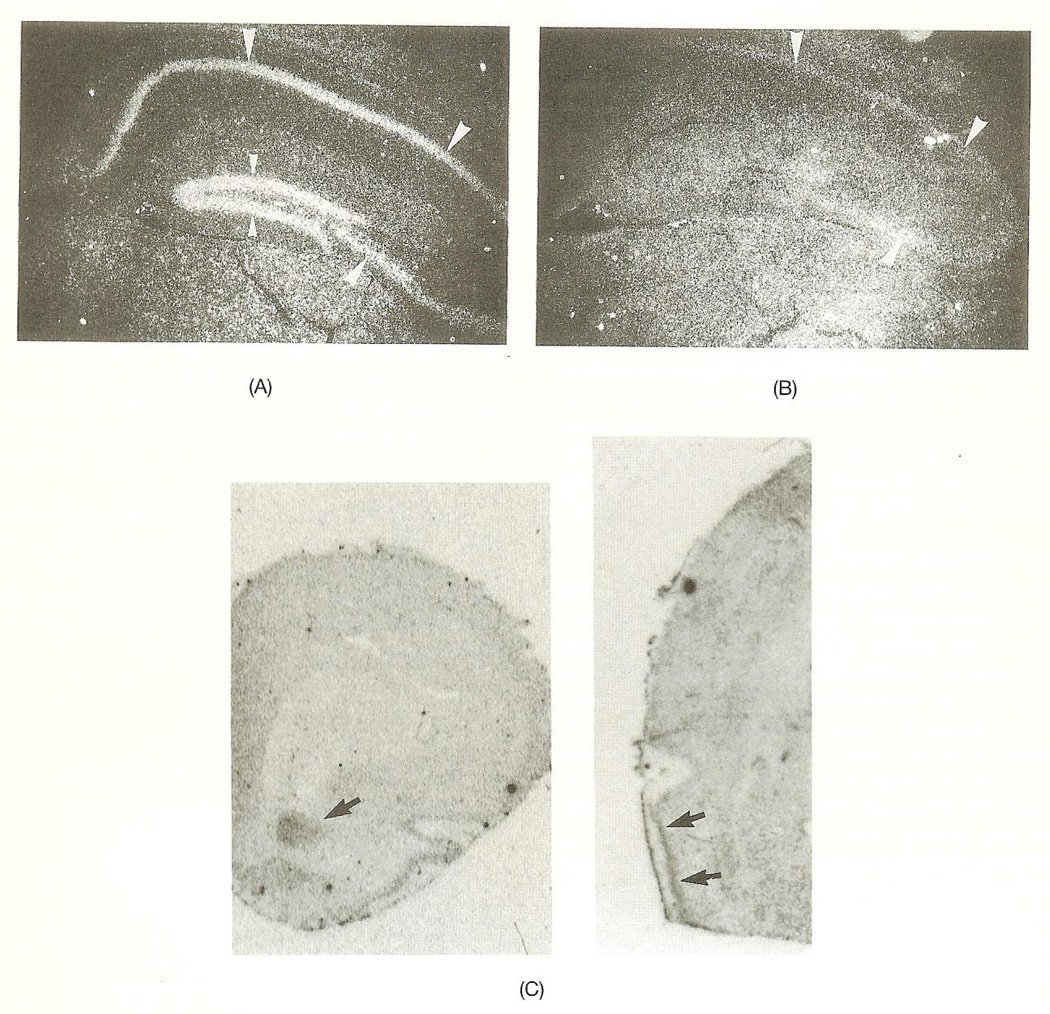
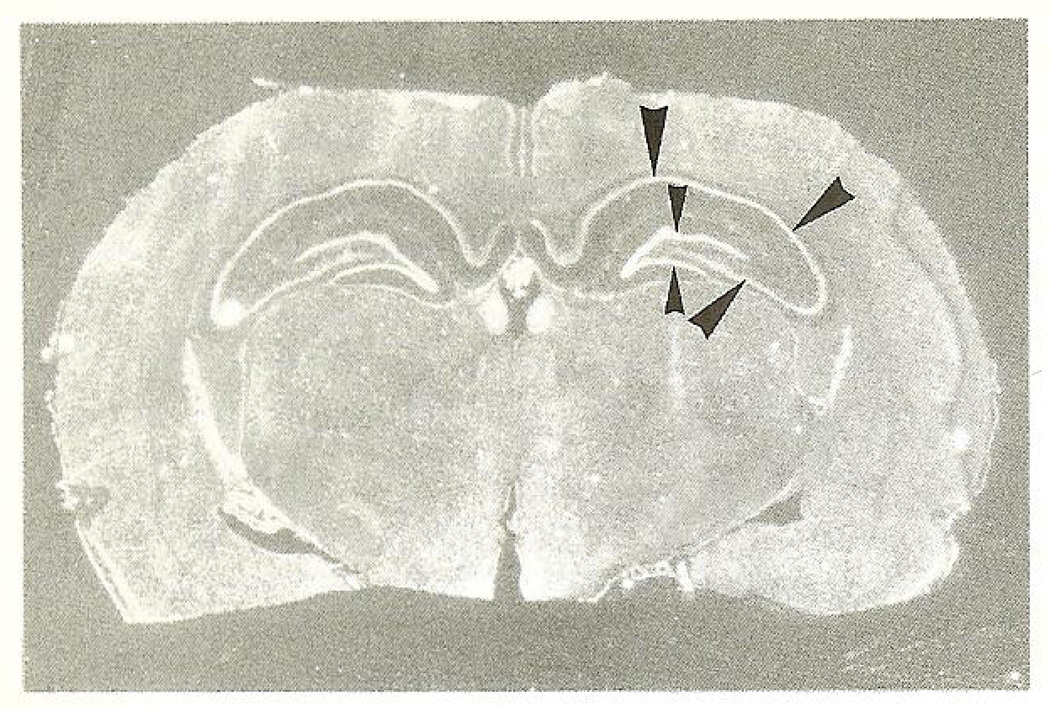
Coronal sections of adult mouse brain revealed specific hybridization mainly localized to cells of neuronal morphology when hybridized with either of the two (nonoverlapping) antisense bcr probes described above. The sense probe did not produce any specific signal in any of the experiments performed. High concentration of bcr mRNA was present in hippocampus, and to a somewhat lower degree in the cerebral cortex. In hippocampus and dentate gyrus, region CA1–CA3, the pyramidal cell and granular cell layers were strongly labeled (Figs. 1A and 1B). In the cortex, neurons in layers II–VI were moderately labeled (not shown), whereas a stronger labeling was noted in the piriform cortex (Fig. 1C). Some thalamic nuclei showed a weak hybridization (not shown). Neurons in amygdala and hypothalamus were not labeled above background hybridization. The hybridization pattern in the cerebellum was not investigated. In the rat, anterior olfactory nuclei displayed a specific hybridization pattern (Fig. 1C). In situ hybridization with labeled oligo-nucleotides further revealed similar expression patterns to that of the mouse: dentate gyrus granular cells, the hippocampal pyramidal layer (CA1–CA3 region), and the piriform cortex clearly displayed a specific hybridization pattern, diminishing in intensity in the order indicated above (Fig. 2).
Fig. 1.
(A) Darkfield photograph of a coronal section of a mouse brain showing strong hybridization to the hippocampus (large arrow-heads) and dentate gyrus (small arrow-heads) when hybridized wilh the antisense her probe. (B) Daikfield photograph of a parallel section to the one presented in Fig. IA showing only weak background hybridization to the hippocampus (large arrowheads) and no hybridization signal to the dentate gyrus when hybridized with the sense bcr probe. (C) Magnifications of rat olfactory bulb (to the left) and whole brain sections (to the right) showing specific hybridization of a bcr-specific oligonucleotide to anterior olfactory nucleus and piriform cortex (arrows). Autoradiographic image.
Fig. 2.
Coronal section of the rat brain showing strong hybridization to the hippocampus (large arrow-heads) and dentate gyms (small arrow-heads) when hybridized to the bcr oligonucleotide (see the Material and Methods section). Reversed autoradiographie image.
Gene expression throughout development
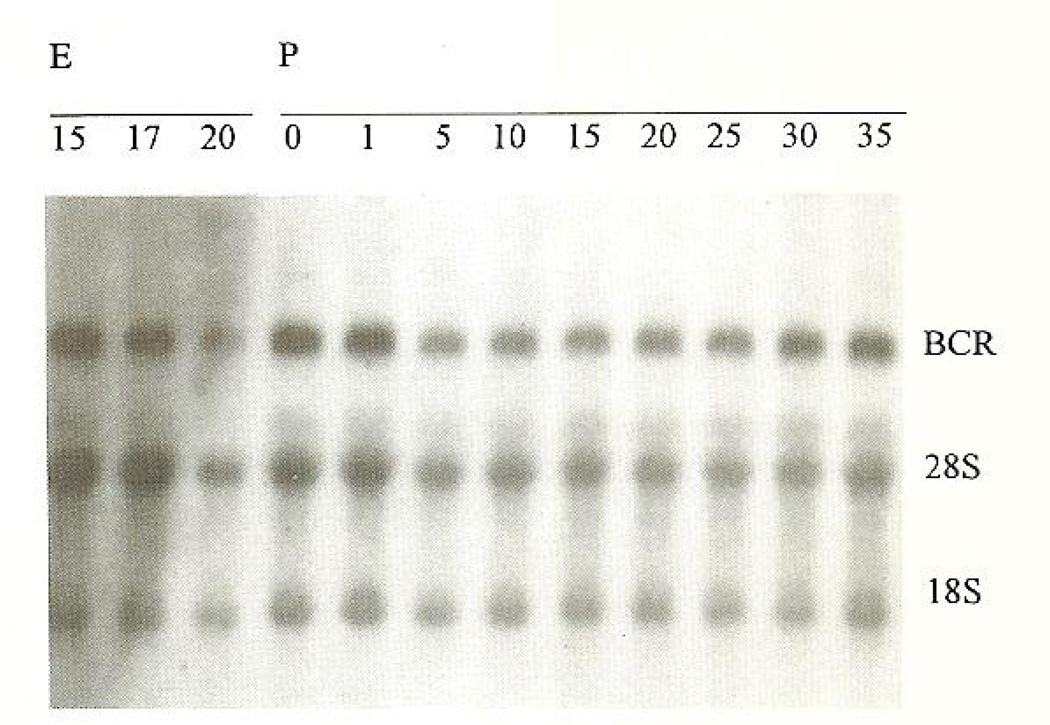
Northern blot analysis of bcr expression in the mouse brain showed a transcript of approximately 7 kb that was present at E15 and remained constant throughout the embryonal period of development, as well as through postnatal days 1–35 (Fig. 3). Densitometric scanning revealed no clear changes in her expression level over time, when corrected for unequal loading on the basis of background hybridization to 18 and 28S ribosomal RNA. The role of Bcr in blood cells has recently been validated (Voncken et al., 1995). Northern blot analysis of liver and spleen samples, organs that have a distinct hematopoietic function during embryogenesis and for some period after birth, showed a lower level of bcr expression throughout the same developmental stages, but, similar to bcr expression in the brain, the level remained constant throughout pre- and postnatal development (data not shown).
Fig. 3.
Autoradiogram showing a constant expression of bcr in the mouse brain from embryonal day 15 (E15) to postnatal day 35 (P35). The bcr transcript of approximately 7 kb and the 28 and 18S ribosomal bands are indicated.
DISCUSSION
The Bcr protein is a multidomain protein and is involved in Ph-positivc leukemogenesis through its fusion to the nonreceptor tyrosine kinase Abl (Heisterkamp and Groffen, 1991; Van Etten, 1993). Although we have recently established a function for Bcr in neutrophils, the reason why bcr is highly expressed in the brain remains elusive. We have used in situ hybridization to study bcr expression patterns in the rodent brain. Our data show that bcr is highly expressed in the hippocampus and dentate gyrus, and to a lower level in the piriform cortex and anterior olfactory nuclei.
Granule cells of the dentate gyrus are primarily generated postnatally. In rodents, granule cells proliferate and differentiate almost exclusively in the first 2 postnatal weeks. In the mouse and the rat, a small percentage of these cells is constantly generated in the adult (Gage, 1994; Gould et al., 1994; and references therein). The hippocampus and olfactory bulb are sites of postnatal neurogenesis and intense synaptic plasticity. Small GTP-binding proteins as well as their regulatory molecules are frequently implicated in processes that control cellular growth in tissues, including the brain. The constitutive activation of p21RAS in neurofibromatosis by deactivation of neurofibromin (Basu et al., 1992; Bollag and McCormick, 1992; DeClue et al., 1992), a GTP-ase activating protein (GAP) for RAS, is a good example. The BCR gene itself has been implicated in human leukemia, and the BCR-related ABR gene appears to be deleted in particular cancers (Isomura et al., 1994; Morris et al., 1995; Saxena et al., 1992).
Although we cannot entirely exclude an involvement of Bcr in neuronal proliferation, the developmental profile of Bcr expression and the presence of Bcr in postmitotic neurons of the hippocampus suggests that Bcr functions also in the absence of neuronal proliferation.
Postmitotic neurons in bcr expressing regions, the olfactory bulb, cortex, and hippocampal formation undergo intensive activity-related alteration of synaptic efficacy. It is intriguing to consider a role for Bcr in energy-consuming processes underlying cellular mechanisms of synaptic plasticity. We are currently examining this hypothesis by measuring neuronal Bcr at baseline conditions and subsequent to physiological stress.
The C-terminal part of Bcr has a high degree of homology with that of n-chimerin, which, like Bcr, possesses GTP-ase activity toward p21rac, a member of the RAS superfamily. The regional expression pattern of bcr in the brain overlaps in part that of n-chimerin. N-chimerin expression is restricted to neurons, with the highest levels of expression in hippocampal pyramidal cells, granulae cells of the dentate gyrus, in cortical neurons, and in the Purkinje cells of the cerebellum (Lim et al., 1992). The developmental expression of bcr, however, displays different dynamics than that of n-chimerin: whereas n-chimerin expression rises sharply after birth to maximal levels at 20 d postpartum (Lim et al., 1992), a period coinciding with cellular differentiation and synaptogenesis, bcr expression remained constant from E15 through day 35 after birth. Northern blot analysis of hematopoietic organs (liver and spleen) also revealed a constant level of bcr expression from embryonal stage E15 throughout development until adulthood.
It is difficult to assess the significance of the relatively high level of bcr expression in the hippocampal region of the brain. Recently, n-chimerin expression in the canary forebrain was connected with seasonal changes in song learning and behavior; the conserved structure and pattern of n-chimerin expression in the song circuit suggested a fundamental function for this gene in the vertebrate forebrain and a role in the regulation of neural plasticity (George and Clayton, 1992). In the young mouse brain, the developmental expression pattern of n-chimerin seems to disclose a potential role in the development of particular cognitive functions (Lim et al., 1992). The finding that bcr expression is more or less constant throughout brain maturation would seem to point to a regulatory role of a more fundamental character in cellular processes.
In the rodent brain, bcr mRNA in the hippocampus is most abundant in dentate gyrus cells, lower in CA3, and lowest in CA1. The functional organization of the hippocampus is such that input through the perforant path reaches granule cell layers. The granular cells, through axons (mossy fibers), synapse on CA3 neurons, which then impinge on CA1. The gradient of bcr expression levels along this functional neuronal path may reflect a potentially interesting yet presently poorly understood function of Bcr in these highly specialized structures.
A growing number of GTPases and GAPs has been implicated in brain biology over the most recent years. Often these proteins (i.e. n-chimerin, α2-chimerin, Bcr, Abr) display a brain-specific expression pattern (Hall et al., 1993; Leung et al., 1994; Lim et al., 1992; Tan et al., 1993) or are, as described above, implicated in brain physiology (n-chimerin) or neoplasia (neurofibromin). Members of the extended family of GTPases have also been implicated in synaptic vesicle transport in nerve terminals (Liu et al., 1994; Von Gersdorff and Matthews, 1994), further indicating the importance of such protein factors in fundamental brain-related processes. This study, concurrent with the recently demonstrated function of Bcr as a regulator of p21Rac-mediated superoxide production by the respiratory burst oxidase in neutrophils (Voncken et al., 1995), suggests differences in tissue-specific functioning of p160Bcr. Increased knowledge of these cellular function(s) of Bcr should strengthen our understanding of the role of Bcr in leukocytes, its contribution to Ph-positive leukemogenesis, and its biological relevance in the brain.
Acknowledgements
This work was supported by grants from the Swedish Cancer Society (T.F.), NIH grant NS 28912 (T.B.), and grant DB-70 from the American Cancer Society (N.H.).
REFERENCES
- Baram TZ, Schultz L. Ontogeny of somatostatin gene expression in rat diencephalon. Dev. Neurosci. 1991;13:176–180. doi: 10.1159/000112155. [DOI] [PubMed] [Google Scholar]
- Baram TZ, Schultz L. CRH gene expression in the fetal rat is not increased after pharmacological adrenalectomy. Neurosci. Lett. 1992;142:215–218. doi: 10.1016/0304-3940(92)90376-i. [DOI] [PubMed] [Google Scholar]
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation ofras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- Bollag G, McCormick F. NF is enough of GAP. Nature. 1992;356:663–664. doi: 10.1038/356663a0. [DOI] [PubMed] [Google Scholar]
- Campbell ML, Arlinghaus RB. Current status of the BCR gene and its involvement with human leukemia. Adv. Cancer Res. 1991;57:227–256. doi: 10.1016/s0065-230x(08)61000-3. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;163:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collins S, Coleman H, Groudine M. Expression of bcr and bcr—abl fusion transcripts in normal and leukemic cells. Mol. Cell. Biol. 1987;7:2870–2876. doi: 10.1128/mcb.7.8.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210 bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Reckling-hauscn (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- Diekman D, Brill S, Garrctt MD, Totty N, Hsuan J, Monfries C, Hall C, Lim L, Hall A. Bcr encodes a GTPase-activating protein for p21rac. Nature. 1991;351:400–402. doi: 10.1038/351400a0. [DOI] [PubMed] [Google Scholar]
- Elefanty AG, Haribaran IK, Cory S. Bcr–Abl, the hallmark of chronic myeloid leukemia in man, induces multiple hematopoietic neoplasms in mice. EMBO J. 1990;9:1069–1078. doi: 10.1002/j.1460-2075.1990.tb08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Challenging an old dogma: ncurogenesis in the adult hippocampus. J. NIH Res. 1994;6:53–55. [Google Scholar]
- George JM, Clayton DF. Differential regulation in the avian song control circuit of an mRNA predicting a highly conserved protein related to protein kinase C and the bcr oncogene. Mol. Brain Res. 1992;12:323–329. doi: 10.1016/0169-328x(92)90134-w. [DOI] [PubMed] [Google Scholar]
- Gould E, Cameron HA, McEwen BS. Blockade of NMDA receptors increases cell death and birth in the developing rat dentate gyms. J. Comp. Neural. 1994;340:551–565. doi: 10.1002/cne.903400408. [DOI] [PubMed] [Google Scholar]
- Groffcn J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region bcr, on chromosome 22. Cell. 1985;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Hall C, Sin WC, Teo M, et al. Alpha 2-chimerin, an SH2-containing GTPase-activating protein for the ras-related protein p21rac derived by alternate splicing of the human n-chimerin gene, is selectively expressed in brain regions and testes. Mol. Cell. Biol. 1993;13:4986–4998. doi: 10.1128/mcb.13.8.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heislerkamp N, Groffen J. Molecular insights into the Phiiadelphia transposition. Hematol. Pathol. 1991;5:1–10. [PubMed] [Google Scholar]
- Heisterkamp N, Jenster G, ten Hoeve J, Pattengale P, Groffen J. Acute leukemia in BCR/ABL transgenic mice. Nature. 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N, Kaartinen V, van Soest S, Bokoch GM, Groffen J. Human ABR encodes a protein with GAPrac activity and homology to the DBJL nucleotide exchange factor domain. J. Biol. Chem. 1993;268:16903–16906. [PubMed] [Google Scholar]
- Heisterkamp N, Stephenson JR, Groffen J, Hansen PF, de Klein A, Bartram CR, Grosveld G. Localization of the c-abl oncogene adjacent to a translocation break point in chronic myelocytic leukemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- Hopkin K. Synaptie vesicle recycling: a search for the switch. J. NIH Res. 1994;6:34–36. [Google Scholar]
- Isomura M, Tanigami A, Saito H, Harada Y, Katagiri T, lnazawa J, Ledbetter DH, Nakamura Y. Detailed analysis of loss of heterozygosity on chromosome band 17p13 in breast cancer carcinoma on the basis of a high-resolution physical map with 29 markers. Genes Chromosom Cancer. 1994;9:173–179. doi: 10.1002/gcc.2870090305. [DOI] [PubMed] [Google Scholar]
- Kawasaki ES, Clark SS, Coyne MY, Smith SD, Champlin R, Witte ON, McCormick F. Diagnosis of chronic myeloid and acute lymphocytic leukemias by detection of leukemia-specific mRNA sequences amplified in vitro. Proc. Natl. Acad. Sci. USA. 1988;85:5698–5702. doi: 10.1073/pnas.85.15.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, How B-E, Manser E, Lim L. Cerebellar β2-chimerin, a GTPase-activating protein for p21 Ras regulated Rac is specifically expressed in granule cells and has a unique N-terminal SH2 domain. J. Biol. Chem. 1994;269:12888–12892. [PubMed] [Google Scholar]
- Lim HH, Michael GJ, Smith P, Lim L, Hall C. Developmental regulation and neuronal expression of the mRNA of rat n-chimerin, a p21rac GAP:cDNA sequence. Biochem. J. 1992;287:415–422. doi: 10.1042/bj2870415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Sim AT, Robinson P. Calcineurin inhibition of dynaminI GTPascs activity coupled to nerve terminal depolarization. Science. 1994;265:970–973. doi: 10.1126/science.8052858. [DOI] [PubMed] [Google Scholar]
- McWhirter J, Galasso D, Wang JYJ. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of the Bcr-Abl oncoproteins. Mol. Cell. Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C, Benjes S, Haataja L, Ledbetter DL, Heisterkamp N, Groffen J. Spatial organization of ABR and CRK genes on human chromosome band 17p13.3. Oneogene. 1995;10:1009–1011. [PubMed] [Google Scholar]
- Ron D, Zannini M, Lewis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. N. Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- Rowley JD. The Philadelphia chromosome translocation. A paradigm for understanding leukemia. Cancer. 1990;65:2178–2184. doi: 10.1002/1097-0142(19900515)65:10<2178::aid-cncr2820651004>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 7.3–7.52. [Google Scholar]
- Sawyers CL, Denny CT, Witte ON. Leukemia and the disruption of normal hematopoiesis. Cell. 1991;64:337–350. doi: 10.1016/0092-8674(91)90643-d. [DOI] [PubMed] [Google Scholar]
- Saxena A, Clark WC, Robertson JT, Ikejiri B, Oldfield EH, Ali IU. Evidence for the involvement of a potential second tumor suppressor gene on chromosome 17 distinct form p53 in malignant astrocytomas. Cancer Res. 1992;50:7184–7189. [PubMed] [Google Scholar]
- Tan EC, Leung T, Manser E, Lim L. The human active breakpoint cluster region-related gene encodes a brain protein with homology to guanine nucleotide exchange proteins and GTP-ase activating proteins. J. Biol. Chem. 1993;268:27291–27298. [PubMed] [Google Scholar]
- Van Etten RA. The molecular pathogenesis of the Philadelphia-positive leukemias: implications for diagnosis and therapy. In: Freireich E, Kantarjian H, editors. Leukemia: advances research and treatment. Deventer, Netherlands: Kluwer Academic Publishers; 1993. pp. 295–325. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Morris C, Pattengale P, Dennert G, Kikly C, Groffen J, Heisterkamp N. Clonal development and karyotype evolution during leukemogenesis of BCR/ABL transgenic mice. Blood. 1992;79:1029–1036. [PubMed] [Google Scholar]
- Voncken JW, Van Schaick H, Kaartinen V, Landing B, Pattengale PK, Deemer K, Coates T, Dorseuil O, Bokoch GM, Groffen J, Heisterkamp N. Increased neutrophil respiratory burst in bcr null mutants. Cell. 1995;80:719–728. doi: 10.1016/0092-8674(95)90350-x. [DOI] [PubMed] [Google Scholar]
- Von Gersdorff H, Matthews G. Inhibition of endocytosis by elevated internal calcium in a synaptic terminal. Nature. 1994;370:652–655. doi: 10.1038/370652a0. [DOI] [PubMed] [Google Scholar]
- Yi SJ, Masters JN, Baram TZ. The effect of a specific glucocorticoid receptor antagonist on CRH gene expression in the neonatal rat hypothalamus. Dev Brain Res. 1993;73:253–259. doi: 10.1016/0165-3806(93)90145-z. [DOI] [PubMed] [Google Scholar]
- Zhu QS, Heisterkamp N, Groffen J. Unique organization of the human BCR gene promotor. Nucleic Acids Res. 1990;18:7119–7125. doi: 10.1093/nar/18.23.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]