ABSTRACT
CodY is known to regulate various virulence properties in several Gram-positive bacteria but has not yet been studied in the important histotoxic and intestinal pathogen Clostridium perfringens. The present study prepared an isogenic codY-null mutant in C. perfringens type D strain CN3718 by insertional mutagenesis using the Targetron system. Western blot analysis indicated that, relative to wild-type CN3718 or a complementing strain, this isogenic codY mutant produces reduced levels of epsilon toxin (ETX). Using supernatants from cultures of the wild-type, codY-null mutant, and complementing strains, CodY regulation of ETX production was shown to have cytotoxic consequences for MDCK cells. The CodY regulatory effect on ETX production was specific, since the codY-null mutant still made wild-type levels of alpha-toxin and perfringolysin O. Sialidase activity measurements and sialidase Western blot analysis of supernatants from CN3718 and its isogenic derivatives showed that CodY represses overall exosialidase activity due to a reduced presence of NanH in culture supernatants. Inactivation of the codY gene significantly decreased the adherence of CN3718 vegetative cells or spores to host Caco-2 cells. Finally, the codY mutant showed increased spore formation under vegetative growth conditions, although germination of these spores was impaired. Overall, these results identify CodY as a global regulator of many C. perfringens virulence-associated properties. Furthermore, they establish that, via CodY, CN3718 coordinately regulates many virulence-associated properties likely needed for intestinal infection.
IMPORTANCE
Clostridium perfringens is a major human and livestock pathogen because it produces many potent toxins. C. perfringens type D strains cause intestinal infections by producing toxins, especially epsilon toxin (ETX). Previous studies identified CodY as a regulator of certain virulence properties in other Gram-positive bacteria. Our study now demonstrates that CodY is a global regulator of virulence-associated properties for type D strain CN3718. It promotes production of ETX, attachment of CN3718 vegetative cells or spores to host enterocyte-like Caco-2 cells, and spore germination; the last two effects may assist intestinal colonization. In contrast, CodY represses sporulation. These results provide the first evidence that CodY can function as a global regulator of C. perfringens virulence-associated properties and that this strain coordinately regulates its virulence-associated properties using CodY to increase ETX production, host cell attachment, and spore germination but to repress sporulation, as would be optimal during type D intestinal infection.
Introduction
Clostridium perfringens is a Gram-positive, anaerobic, spore-forming bacterium that ranks among the most common pathogens of humans and livestock (1, 2). Among the diseases caused by C. perfringens are several important human and livestock intestinal diseases, usually manifesting as enteritis or enterotoxemia (2–4). These intestinal illnesses are true infections rather than intoxications, so their pathogenesis involves several distinct properties of C. perfringens, including toxin production, the ability to form spores for environmental persistence and disease transmission, and attachment of vegetative cells or spores to the intestinal epithelium to promote colonization, thus allowing C. perfringens to reach sufficient bacterial numbers in vivo to produce enough toxins for inducing host damage (1, 2, 5).
This bacterium can produce more than 16 different toxins, but strain-to-strain variations occur in expression of this toxin arsenal (2, 4, 6, 7). These toxin production differences allow C. perfringens isolates to be classified into five types (A to E), based upon production of four (α, β, ε, and ι) typing toxins (1, 6). Type D strains cause C. perfringens intestinal infections (2, 3). These strains cause enterotoxemias in livestock when they grow in the intestines and produce toxins, particularly epsilon toxin (ETX), that are then absorbed into the circulation to damage target organs, like the brain, kidneys, and lungs (8). Type D strains also cause acute or chronic enteritis in goats (2, 3). CN3718 is a type D animal disease isolate that produces ETX, alpha-toxin (PLC), and perfringolysin O (PFO) (9). In addition to toxin production, CN3718 produces three sialidases (5). NanI, the major secreted sialidase of this strain, was shown to increase ETX binding and cytotoxicity to cultured host cells (5). NanI also mediates the in vitro adherence of CN3718 vegetative cells to Caco-2 cells (5). These NanI-enhanced effects may be important for type D intestinal infections by promoting ETX action and bacterial colonization (5).
Our understanding of how C. perfringens regulates production of its virulence-associated properties remains rudimentary. There is still relatively little knowledge of the regulatory processes initiating or repressing the sporulation or germination of this bacterium (10). Furthermore, there is limited information on how C. perfringens controls production of sialidases (11). Whether this bacterium regulates its ability to adhere to host cells has not been examined. Even the control of toxin production by C. perfringens has received relatively minimal attention. This is particularly true for ETX, where knowledge is limited to expression of this toxin by CN3718 being independent of the VirS/VirR two-component regulatory system but controlled by the Agr-like quorum-sensing (QS) system (9). However, in two type B strains, ETX production levels are not affected by inactivation of this QS system (12).
The global gene regulator CodY was first identified in Bacillus subtilis in 1993 (13) but has since been studied in several other Gram-positive pathogenic bacteria, including Bacillus anthracis, Bacillus cereus, Clostridium difficile, Listeria monocytogenes, Streptococcus pneumoniae, and Staphylococcus aureus (14–16). CodY represses or, less commonly, activates the transcription of target genes when bound to promoter regions during log-phase growth, where these bacteria are in a nutrient-rich environment that increases cytoplasmic GTP and amino acid pools (14, 15, 17). CodY is known to regulate the virulence properties of several low-G+C, Gram-positive bacteria (14). For example, in C. difficile, CodY represses toxin A and toxin B production during both the stationary and exponential growth phases (18). CodY was also shown to control production of B. anthracis toxins (19), B. cereus toxins (16), and the hemolysins of S. aureus (20). Similarly, CodY regulates the host cell adherence of several Gram-positive pathogens, including S. pneumoniae (21).
To date, there has been no investigation of whether CodY controls C. perfringens virulence-associated properties. Therefore, the current study explored this issue by phenotyping a codY-null mutant and a complementing strain of type D strain CN3718.
RESULTS
Construction and characterization of a CN3718 codY-null mutant.
Bioinformatic analyses indicated that the C. perfringens CodY protein has a very conserved, 258-amino-acid (aa) sequence among all sequenced C. perfringens genomes deposited in GenBank. Comparison of those GenBank data further showed the C. perfringens CodY protein shares about 60 to 70% identity and 75 to 90% similarity with the CodY protein in other Clostridium species, such as C. difficile, C. botulinum, C. novyi, and C. butyricum. Particularly conserved are the sequences in the C-terminal region containing a helix-turn-helix motif that mediates CodY binding to DNA (22).
Given those observations, the Clostridium-modified Targetron-mediated insertional mutagenesis system (23) was used to inactivate the codY gene in CN3718. The identity of a putative CN3718 codY-null mutant (CN3718::codY) was demonstrated by PCR using primers specific for internal codY open reading frame (ORF) sequences (see Fig. S1A and B in the supplemental material). These internal PCR primers specifically amplified a PCR product of 340 bp from wild-type CN3718 DNA. However, due to the insertion of a 900-bp intron into the codY ORF, the same pair of primers amplified a 1,240-bp PCR product from the DNA of the putative codY-null mutant. Southern blot analyses using an intron-specific probe were then performed to confirm that CN3718::codY carried only a single intron insertion. The Southern blot analysis detected no hybridization of the intron-specific probe to CN3718 DNA. In contrast, using DNA from the codY-null mutant strain, a single band was observed, indicating that only one intron had inserted into CN3718::codY (see Fig. S1C). Once the the genotypic identity of the CN3718::codY-null mutant had been confirmed, a codY-complementing strain (CN3718comp) was then prepared by electroporation of the plasmid pJIR750codYcomp into CN3718::codY. PCR confirmed the presence of the wild-type codY ORF in the complementing strain (see Fig. S1B). To demonstrate the phenotype of the codY-null mutant and CN3718comp, a CodY Western blot assay was performed using pelleted bacteria from overnight culture of CN3718, CN3718::codY, and CN3718comp. The results of those CodY Western blots showed that both wild-type CN3718 and the complementing strain express the 25-kDa CodY, while no CodY production was detected from the mutant strain (see Fig. S1D).
To evaluate whether the codY gene altered vegetative growth properties, the vegetative growth rates of CN3718, CN3718::codY, and CN3718comp strains were compared in SFP medium (Shahidi-Ferguson perfringens medium). Results of this analysis showed (see Fig. S1E in the supplemental material) that, under these culture conditions, all three strains grew at the same rate from the time of inoculation until ~4 to 5 h, a time when the codY-null mutant entered stationary phase, while the wild-type and complementing strains continued growing for slightly longer.
Effects of a codY-null mutation on ETX production.
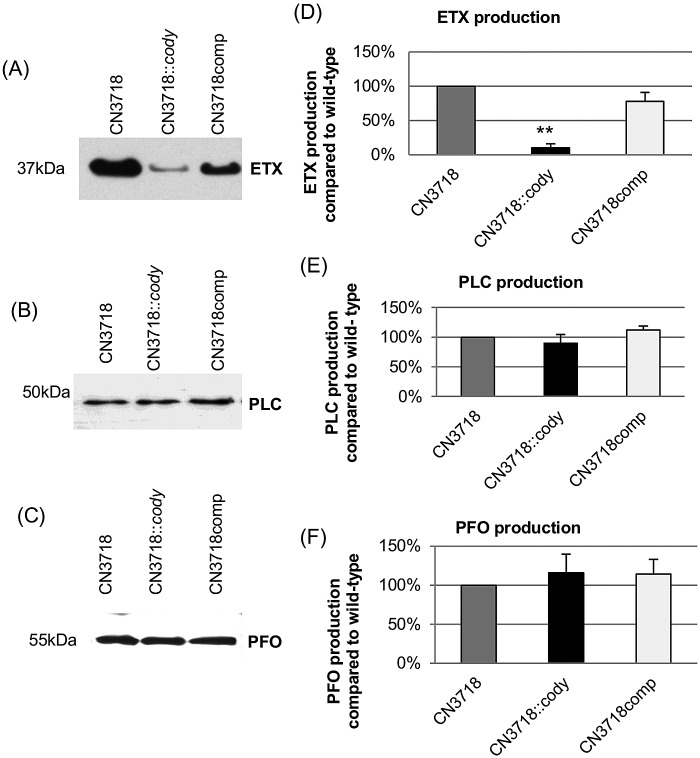
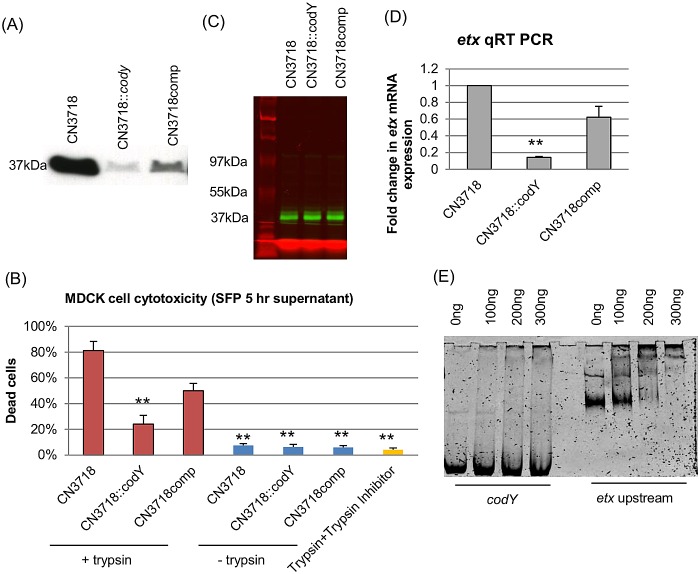
Previous studies have shown that CodY regulates the production of several toxins by other Gram-positive pathogens, e.g., the anthrax toxins of B. anthracis (19), the hemolysins of S. aureus (20) and the TcdA and TcdB toxins of C. difficile (18). However, the involvement (if any) of CodY in regulating C. perfringens toxin production has not yet been evaluated. Therefore, Western blot analyses were performed in the current study, first using overnight culture supernatants of CN3718, CN3718::codY, and CN3718comp. Results from those experiments revealed that a codY-null mutation decreased ETX production compared with that in the parent strain (Fig. 1A and D). This effect was not due to a secondary mutation in the codY mutant, since complementation substantially restored ETX production. Because the codY-null mutant entered the stationary phase slightly earlier than CN3718 and the complementing strain, the decreased ETX production by the codY-null mutant in overnight culture supernatants might arguably have been due to lower numbers of bacteria rather than to CodY regulation of ETX production. However, Western blot analysis of 5-h culture supernatants of CN3718, CN3718::codY, and CN3718comp showed the same pattern of ETX production as for the overnight culture (Fig. 2A). The conclusion that CodY positively regulates ETX production in 5-h cultures was confirmed by an experiment comparing ETX-induced cytotoxicity for MDCK cells. After trypsin treatment to activate ETX (9), supernatants of CN3718 or the complementing strain caused about 4-fold more cytotoxicity than the trypsin-activated supernatant from a culture of the codY-null mutant. Controls causing <10% cytotoxicity in MDCK cells included non-trypsin-activated culture supernatants, mock treatment with trypsin and trypsin inhibitor (no culture supernatants), and preincubation of activated supernatants with an ETX-neutralizing monoclonal antibody (Fig. 2B and data not shown).
FIG 1 .
Western blot analysis of toxin production. Shown are Western blot results for ETX (A), PLC (B), and PFO (C) production in overnight SFP medium culture supernatants of CN3718, CN3718::codY, and CN3718comp. Protein sizes are at left. Quantitative analysis of ETX (D), PLC (E), and PFO (F) production by wild-type CN3718 (value was set as 100%), the codY-null mutant, and the complemented strains. The band intensities of the Western blots were compared by ImageJ analysis. **, P < 0.001 (Student’s t test, compared to the wild type). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
FIG 2 .
CodY upregulates etx transcription and ETX production. (A) Western blot analysis for ETX production by 5-h SFP medium culture supernatants of CN3718, CN3718::codY, and CN3718comp. The protein size is at left. (B) MDCK cell cytotoxicity after treatment with trypsin-activated supernatant from 5-h SFP broth cultures of CN3718, CN3718::codY, and CN3718comp. Cell cytotoxicity was measured by an LDH release assay. **, P < 0.001 (Student’s t test, compared to wild type). Results are averages from three repetitions; the error bars indicate standard errors of the means. Controls included untreated supernatants of CN3718, CN3718::codY, and CN3718comp, trypsin, and trypsin inhibitor alone (no culture supernatant) or supernatant treated with an ETX-neutralizing monoclonal antibody (data not shown). (C) Proteolytic degradation of ETX prototoxin was assessed using 5 µg of pETX (in green) incubated for 2 h at 37°C with overnight culture supernatant from CN3718, CN3718::codY, and CN3718comp. The left lane shows protein marker (in red) sizes. (D) Quantitative PT-PCR analyses of etx transcription was performed with 20 ng of the RNA isolated from 5-h SFP medium cultures of CN3718, CN3718::codY, and CN3718comp. Average CT values were normalized to the housekeeping 16S RNA gene, and the fold differences were calculated using the comparative CT method (2−ΔΔCT). The values are the calculated fold change relative to wild-type CN3718. **, P < 0.001 (Student’s t test, compared to the wild type). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations. (E) Gel mobility shift assay for the binding of rCodY to the 650-bp sequences upstream of the etx ORF. The negative DNA fragment that corresponded to 350 bp of internal codY gene sequences was used. Each DNA fragment was incubated with increasing concentrations of rCodY.
Since protease production significantly increases after inactivation of the codY gene in Lactococcus lactis (15), we assessed whether the decreased ETX levels detected in codY-null mutant cultures might involve C. perfringens protease degradation of ETX. For this purpose, fluorescently labeled ETX prototoxin (pETX) was used, since the prototoxin is the ETX form initially secreted from C. perfringens prior to its processing and activation by intestinal proteases (24). When the same amount of pETX was added to overnight culture supernatants from CN3718, CN3718::codY, and CN3718comp for 2 h at 37°C, no ETX prototoxin degradation was detected in any supernatants by SDS-PAGE analysis (Fig. 2C).
Since the decreased ETX production of the codY-null mutant was not due to increased proteolytic degradation, we next explored whether the lower ETX production of the codY-null mutant might involve reduced etx transcription. When quantitative reverse transcription-PCR (qRT-PCR) for the etx gene was performed, the results demonstrated that etx mRNA levels in 5-h cultures of the codY-null mutant strain were reduced compared with CN3718 or the complementing strain CN3718comp (Fig. 2D).
Studies with other Gram-positive bacteria have shown that CodY regulates gene expression by DNA binding; this can involve either a direct effect where CodY binds upstream of the controlled gene’s ORF or by an indirect effect where CodY binds to a regulatory gene that then controls expression of the controlled gene (14, 15). To begin exploring whether CodY might directly regulate ETX expression by binding upstream of the etx ORF, a gel mobility shift assay was performed that assessed the ability of recombinant CodY (rCodY) to bind to a DNA fragment containing sequences present immediately upstream of the etx ORF. The results showed that, even at relatively low levels (200 ng), rCodY binds to sequences upstream of the etx ORF (Fig. 2E). This binding was specific, since no shift was observed in the mobility of a control DNA fragment even when the rCodY concentration was increased to 500 ng (Fig. 2E and data not shown).
Effects of a codY-null mutation on PLC and PFO production.
Besides ETX, CN3718 also produces PLC and PFO (9). Therefore, Western blot studies were performed to start assessing whether CodY is a general regulator of C. perfringens toxin production or a more specific regulator that controls production of only certain toxins. These studies found that, in contrast to the CodY regulation of ETX production shown in Fig. 1A and D, there were no significant differences in PLC production among overnight cultures of CN3718, CN3718::codY, and CN3718comp (Fig. 1B and E). Similarly, no significant differences were observed in PFO production levels between overnight cultures of these strains (Fig. 1C and F).
CodY regulation of sialidase production.
Since previous studies have shown that NanI enhances the binding and cytotoxic activities of ETX for MDCK cells (5), the current study examined whether the exosialidase activity of CN3718 is controlled by the codY gene. The results demonstrated that for both 5-h and overnight culture supernatants, CN3718::codY showed significantly stronger sialidase activity than the parent or complementing strain (Fig. 3A).
FIG 3 .
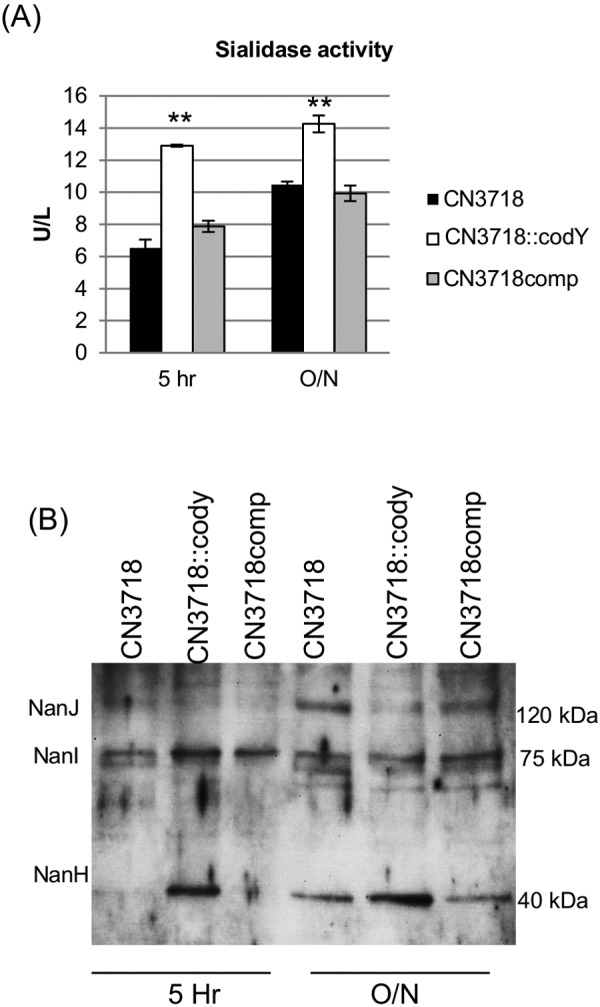
Involvement of CodY in controlling culture supernatant sialidase activity and sialidase content. (A) Sialidase activity in 5-h or overnight SFP medium culture supernatants of CN3718, CN3718::codY, and CN3718comp. **, P < 0.001 (Student’s t test, compared to the wild type). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations. (B) Western blot analyses for the presence of three sialidases in 5-h or overnight SFP medium supernatants from cultures of CN3718, CN3718::codY, and CN3718comp. Protein sizes are at right.
CN3718 produces three sialidases, named NanJ (129 kDa), NanI (77 kDa), and NanH (43 kDa) (5), so a sialidase Western blot analysis (Fig. 3B) was performed to determine which sialidase(s) was responsible for the increased supernatant sialidase activity resulting from inactivation of the codY gene. Results of this analysis indicated that more NanH is present in the supernatants of the codY-null mutant. In contrast, less NanJ is present in the supernatants of the codY mutant, while the presence of NanI in culture supernatants was relatively unaffected by inactivation of the codY gene.
CodY regulates adherence of CN3718 vegetative cells and spores to Caco-2 cells.
A previous study (5) had shown that CN3718 adheres specifically to enterocyte-like cultured cells, including Caco-2 cells. Since CodY has been implicated in the host cell adherence of some other Gram-positive pathogens (14), the current study assessed whether inactivating codY affects CN3718 vegetative cell or spore adherence to Caco-2 cells. The vegetative cells of the isogenic codY-null mutant showed significantly lower levels of attachment to Caco-2 cells than the wild type or complementing strains (Fig. 4). Interestingly, we also found that washed CN3718 spores adhere well to Caco-2 cells and determined that this effect is CodY dependent (Fig. 4).
FIG 4 .
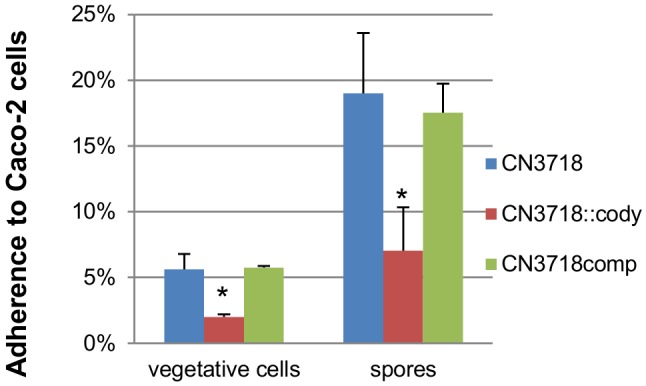
Vegetative cell and spore adherence to Caco-2 cells. Caco-2 cell monolayers were incubated for 2 h with washed vegetative cells or spores of CN3718, CN3718::codY, and CN3718comp at 37°C under anaerobic conditions. Monolayers were then washed three times with HBSS buffer and lysed in HBSS buffer, and total bacteria or spores were plated onto BHI agar plates for counting. For spore counting, the collected spores were heat shocked for 20 min at 70°C before plating. Attachment was then expressed as the percentage of attached bacteria or spores relative to the total number of input bacteria or spores. *, P < 0.05 (Student’s t test, compared to wild type). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
CodY regulates C. perfringens sporulation and germination.
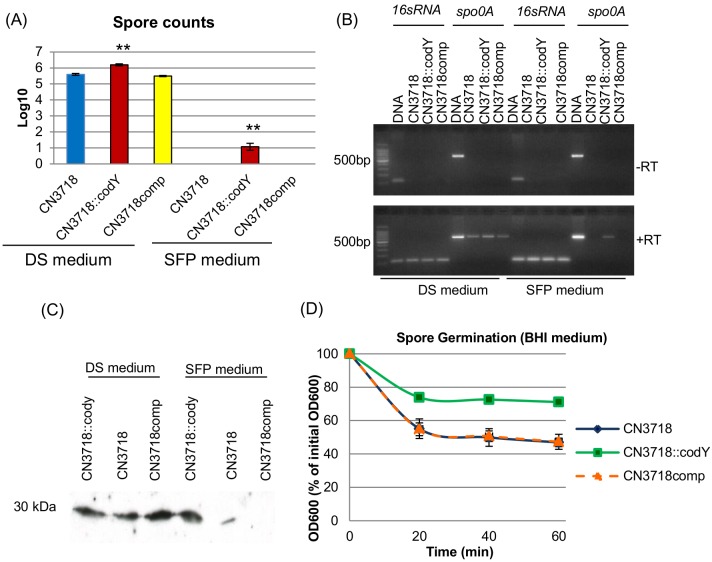
CodY is known to repress sporulation in B. subtilis (15), so we next examined the involvement of this regulatory protein in controlling C. perfringens sporulation. When the sporulating abilities of CN3718, CN3718::codY, and CN3718comp were quantitatively compared by measuring their formation of heat-resistant spores in overnight Duncan-Strong (DS) sporulation medium cultures, the isogenic codY-null mutant made ~10-fold more spores than wild-type CN3718 (Fig. 5A). This increase in sporulation involved the inactivation of the codY locus, since the complementing strain formed approximately the same number of heat-resistant spores/ml as did wild-type CN3718. Interestingly, in SFP vegetative culture medium, the wild-type and complementing strains did not make heat-resistant spores, but the codY-null mutant strain produced ~10 heat-resistant spores/ml (Fig. 5A).
FIG 5 .
Spore formation, germination, spo0A gene transcription, and Spo0A production by CN3718, CN3718::codY, and CN3718comp. (A) Heat-resistant spore formation by CN3718, CN3718::codY, and CN3718comp. The bacteria were grown in DS or SFP medium overnight at 37°C; the overnight cultures were then heat shocked for 20 min at 70°C. After a 10-fold serial dilution with distilled water, the heat-shocked cultures were then plated onto BHI agar plates and grown overnight at 37°C for colony counting. **, P < 0.001 (Student’s t test, compared to the wild type). All experiments were repeated three times, and mean values (log10 scale) are shown. The error bars indicate standard deviations. (B) RT-PCR analyses for spo0A transcription by CN3718, CN3718::codY, and CN3718comp grown for 5 h in DS or SFP medium. The leftmost lane shows a 100-bp DNA ladder. (Top) Samples lacking reverse transcriptase to demonstrate the absence of DNA contamination. (Bottom) Samples receiving reverse transcriptase. (C) Western blot analyses for Spo0A production by CN3718, CN3718::codY, and CN3718comp using a 5-h DS medium culture or SFP medium culture cell pellet. The protein size is at left. (D) Germination of spores of CN3718, CN3718::codY, and CN3718comp in BHI broth. Heat-activated spores were incubated at 40°C in BHI broth, and each OD600 was measured as described in Materials and Methods. All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
Previous studies showed that, in cultures of B. subtilis growing in rich media, CodY inhibits the initiation of sporulation by repressing the expression of the early sporulation gene spo0A (25). Therefore, the current study used RT-PCR to investigate whether CodY represses expression of spo0A by CN3718 during growth under vegetative, but not sporulation-inducing, conditions. Results of this analysis showed that, under the relatively rich growth conditions of SFP medium, only the codY-null mutant strain expressed spo0A mRNA. However, in DS sporulation medium, CN3718, CN3718::codY, and CN3718comp strains expressed spo0A mRNA (Fig. 5B). A Spo0A Western blot experiment then confirmed those RT-PCR results, i.e., CN3718, CN3718::codY, and CN3718comp each produced Spo0A when grown in DS sporulating medium. In contrast, when these strains were grown in SFP vegetative medium, only the codY-null mutant strain expressed spo0A or produced Spo0A (Fig. 5C), consistent with this being the only strain that produced spores under these normally vegetative culture conditions.
While the plate count results indicated that the codY-null mutant strain produced more heat-resistant spores than the wild-type and complementing strains, we noted by microscopy that the codY-null mutant appeared to produce even more spores than had been indicated by the plate counting results, suggesting that the spores of the codY mutant might have a germination defect. Therefore, an experiment was performed to evaluate whether codY gene expression affects C. perfringens spore germination. Results from that experiment, indicated that spores of the codY-null mutant possess lower germination ability than the wild-type strain spores. Complementation of the codY mutant restored wild-type germination ability to spores (Fig. 5D).
DISCUSSION
CodY regulates virulence genes in many Gram-positive bacteria, e.g., B. anthracis, Streptococcus pyogenes, S. pneumoniae, L. monocytogenes, Enterococcus faecalis, and B. cereus (14, 16). However, CodY regulation of virulence genes had been studied in only one clostridial pathogen, i.e., C. difficile, where CodY was shown to control TcdA and TcdB production (18). CodY involvement in regulating clostridial virulence-associated properties beyond toxin production had not yet been studied. We now report that CodY is a global regulator of C. perfringens virulence-associated properties.
The current study first found that CodY regulates production of some but not all toxins in C. perfringens strain CN3718. Specifically, inactivation of the codY gene in CN3718 significantly decreased ETX production but did not affect production levels of PFO or PLC. Determining that CodY increases ETX production provides the first evidence for positive regulation of toxin gene expression by CodY in a pathogenic Clostridium spp. This result contrasts with C. difficile, where a codY-null mutant still made wild-type levels of binary toxin but expressed more TcdA and TcdB than its wild-type parent (18). CodY typically represses toxin production in most Gram-positive bacteria examined to date (14), but there is a precedent for CodY-mediated positive regulation of toxin production; i.e., a B. cereus codY-null mutant produced less toxins than its wild-type parent (16). Determining that CodY is a positive regulator of ETX production in CN3718 is important because so little is known regarding how C. perfringens regulates production of this toxin, which is critical for the virulence of type D strains (8). Since ETX production by CN3718 is independent of VirS/VirR but activated by the Agr-like QS system (9), future studies will examine possible cross talk between the Agr-like QS system and CodY in CN3718. Several other Gram-positive pathogens possess both an Agr-like QS system and CodY; previous studies indicated variability in cross talk between these two regulators (14, 20, 26). In S. aureus, CodY represses the Agr QS (20); however, in L. monocytogenes, CodY activates the Agr-like QS (26). In addition, ETX production in C. perfringens type B strains CN1793 and CN1795 is Agr independent (12), so it may also be informative for future studies to explore CodY regulation of ETX production in those strains.
In other bacteria, CodY regulates genes directly, by binding to their regulatory regions, or indirectly, by controlling expression of regulatory genes (14). In our study, gel shift analyses showed that CodY binds specifically to sequences immediately upstream of the etx ORF, which is consistent with direct CodY positive regulation of ETX production. While further studies are needed, bioinformatic analyses offer additional support for this possibility by identifying two potential CodY binding boxes, located 21 bp and 354 bp upstream of the etx start codon. There is ample documentation of gene repression by CodY directly binding to CodY boxes in other Gram-positive bacteria (14), but only one example showed that CodY is a positive regulator of the ackA gene of B. subtilis by directly binding to CodY boxes upstream of this gene (17).
Bacterial adherence to host cells is thought to play a significant role in many intestinal infections caused by C. perfringens and other clostridial enteropathogens, including C. difficile (5, 27, 28). To our knowledge, no regulatory genes mediating the host cell adherence properties of any clostridial enteropathogen had been identified prior to the current identification of CodY as a positive regulator of C. perfringens vegetative cell and spore adherence to Caco-2 cells. However, CodY was shown to promote the host cell adherence of some other Gram-positive pathogens, e.g., S. pneumoniae (21). The mechanisms used by C. perfringens vegetative cells to adhere to host cells are just being explored, but previous studies reported that the NanI sialidase is necessary for the specific adherence of CN3718 vegetative cells to the Caco-2 cells (5). Therefore, it is notable that we found that CodY regulates CN3718 vegetative cell adherence without affecting NanI levels in CN3718 culture supernatants. One explanation for this observation could be that one or more surface adhesins of CN3718 are CodY regulated. This hypothesis is consistent with previous findings (5) that NanI pretreatment of Caco-2 cells promotes C. perfringens binding, strongly suggesting that NanI itself is not the adhesin used by CN3718 to attach to Caco-2 cells. Future studies will also evaluate CodY-regulated mechanisms of C. perfringens spore adherence to Caco-2 cells, as demonstrated in this study, which could be relevant for initial colonization of a host during potentially spore-transmitted C. perfringens diseases such as antibiotic-associated diarrhea (2).
While NanI levels in culture supernatants were not changed by inactivation of the codY gene in CN3718, the codY-null mutation did affect supernatants levels of the other two sialidases produced by this strain, i.e., there was less NanJ, but more NanH, present in culture supernatants of the codY mutant than wild-type CN3718. Combined with the observation that the codY mutant produced the same NanI levels as wild-type CN3718, these results suggest that production of these three sialidases involves different regulatory mechanisms. The substantial amounts of NanH detected in 5-h supernatants of the codY mutant is interesting, since this enzyme has a cytoplasmic location in log-phase culture of CN3718 (Fig. 3B and 5). However, NanH appears in the supernatants of overnight cultures of wild-type CN3718 (Fig. 3B), so the presence of NanH in 5-h supernatants of the codY mutant may reflect the slightly earlier entry of this strain, relative to the wild-type parent, into stationary phase. Why the codY mutant enters more rapidly into stationary phase is not clear, but a codY mutant of C. difficile showed a similar growth phenotype (18).
To our knowledge, this study presents the first direct evidence that CodY can repress sporulation in Clostridium spp., although previous microarray analyses had shown that CodY regulates some early sporulation genes in C. difficile (18). Similar to our C. perfringens results, CodY also represses sporulation by B. subtilis in rich medium (25), but it does not universally affect sporulation by Gram-positive sporeformers, since a B. cereus codY-null mutant reportedly does not show increased sporulation in rich media (16). Our study found that, as for B. subtilis, CodY represses spo0A transcription and Spo0A production when C. perfringens grows in rich vegetative media. This effect likely explains the CodY repression of C. perfringens sporulation, since Spo0A is necessary for sporulation initiation in this bacterium (29).
Our study also presents the first clear demonstration that CodY can affect spore germination in any Gram-positive pathogen. Some clues into potential mechanisms for this effect may be gleaned from earlier microarray studies with C. difficile (18), which determined that CodY activates expression of a germination-specific N-acetylmuramoyl-l-alanine amidase in C. difficile. Therefore, CodY could regulate spore germination by activating the C. perfringens cortex-lytic enzyme (CLE) SleC, which was reported to have N-acetylmuramoyl-l-alanine amidase activity (30) and to act as the sole CLE in C. perfringens spore cortex hydrolysis (31).
The current study identifies CodY as a global regulator of C. perfringens virulence-associated properties, including the production of some toxins, host cell adherence properties of vegetative cells and spores, sporulation, and germination. This finding provides the first evidence for coordinate regulation of diverse virulence-associated properties in this bacterium. Given the pathogenesis of type D intestinal infections, it makes good “pathogenic sense” for CN3718 to simultaneously increase production of ETX and host cell adherence. Similarly, since type D diseases are caused by vegetative cells, it would favor pathogenesis if CN3718 used CodY to repress sporulation under rich growth conditions.
Many studies are needed to explore CodY regulation of C. perfringens virulence-associated properties. These include molecular biology studies to identity CodY boxes in this bacterium, analysis of CodY regulation of other C. perfringens toxins, and examination of the cross talk between CodY and other regulators of C. perfringens virulence-associated properties. It would also be of interest to examine whether CodY affects C. difficile adherence to host cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, medium, and culture conditions.
CN3718 is an ETX-positive C. perfringens type D animal disease strain (9). Escherichia coli Top10 cells (Invitrogen) were used as the cloning host. Plasmids used in this study included the Targetron vector pJIR750ai (Sigma) and C. perfringens/E. coli shuttle plasmid pJIR750 (32). Media used for culturing C. perfringens included FTG medium (fluid thioglycolate medium; Becton Dickinson), SFP medium (0.5% yeast extract, 0.75% proteose peptone no. 3, 0.75% pancreatic digest of casein, 0.5% Soytone [Becton Dickinson] supplemented with 0.1% sodium thioglycolate [Sigma-Aldrich]), Duncan-Strong sporulation medium (33), and BHI (brain heart infusion) and SFP agar plates (Becton Dickinson). For culturing E. coli, Luria-Bertani (LB) broth and agar (1.5% agar [Becton Dickinson]) were used. All antibiotics used were purchased from Fisher Scientific.
Construction of a codY-null mutant and complementing strain of CN3718.
The codY gene of CN3718 was inactivated by the insertion of a targeted group II intron using the Clostridium-modified Targetron system (23). Based upon the reported codY sequence of type D strain (GenBank accession number CJD_2278) and the Sigma Targetron website, an intron was targeted to insert, in the sense orientation, into the codY ORF between nucleotides 645 and 646. The primers used for PCR targeting of the intron were 645|646s-IBS (5′ AAAAAAGCTTATAATTATCCTTAGGAATCACTAGAGTGCGCCCAGATAGGGTG 3′), 645|646s-EBS1d (5′ CAGATTGTACAAATGTGGTGATAACAGATAAGTCACTAGA TCTAACTTACCTTTCTTTGT 3′), and 645|646s-EBS2 (5′ TGAACGCAAGTTTCTAATTT CGATTATTCCTCGATAGAGGAAAGTGTCT 3′). The 350-bp PCR product was inserted into pJIR750ai to construct a codY-specific Targetron plasmid (pJIR750codYi). The mutant preparation and selection were performed as described previously (5). Bacterial cells carrying an intron insertion were screened using PCR primers codYKOF (5′ AATGATGATGAC CTAGTTTTAGCAG 3′) and codYKOR (5′-CCCAAGACTTCTTGATTCAATA-3).
The CN3718 codY-null mutant was complemented by cloning the codY ORF, flanked by 600 bp of upstream sequence and 300 bp of downstream sequence, into pJIR750 and then transforming this new plasmid into CN3718::codY. Briefly, DNA was isolated from wild-type CN3718 using the MasterPure Gram-positive DNA purification kit (Epicentre, Madison, WI). PCR was then performed using the long-range Taq DNA polymerase (New England Biolabs) and primers codYF (5′ TTACgaattcGAATTCTTTACTCCATTAAAGGAAGACATAGAAAAG 3′ with an added EcoRI site [lowercase]) and codYR (5′ TAGAggatccCCGGCTTATAGATTCGA TAATGTGTGTATT 3′ with an added BamHI site [lowercase]). The resultant 1.69-kb PCR product was cloned into pJIR750, producing plasmid pJIR750codycomp. CN3718::codY was transformed by electroporation with pJIR750codYcomp; the complementing strain, named CN3718comp, was then selected on BHI agar plates containing 15 µg/ml of chloramphenicol.
Southern blot analysis of the codY-null mutant.
Aliquots (3 µg each) of CN3718 and CN3718::codY DNA samples were digested overnight with EcoRI at 37°C, according to the manufacturer’s instructions (New England Biolabs). The intron-specific Southern blot was performed as described previously (5).
RNA extraction, RT-PCR, and qRT-PCR.
Total C. perfringens RNA was extracted from pelleted cells of 5-h cultures grown in SFP medium or DS medium by using saturated phenol (Fisher Scientific), as described in a previous study (5). RT-PCR analysis of spo0A gene transcription was then performed using the AccessQuick RT-PCR kit from Promega Company. The PCR primers used to amplify the 16S RNA gene sequences (as a control housekeeping gene) were 16sF (5′ CCTTACCTACACTTGACATCCC 3′) and 16sR (5′ GGACTTAACCCAACATCTCACG 3′), and the spo0A primers were Spo0A-F (5′ AACAACCAGATTTAGTTGTATTAG 3′) and Spo0A-R (5′-TCCCCAAGC-3′). The RT-PCR conditions used were as follows: 95°C for 2 min; 45°C for 1 h; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 72°C for 40 s; and finally a single extension at 72°C for 5 min.
Quantitative RT-PCR (qRT-PCR) was performed using the iScript one-step RT-PCR kit with SYBR green (Bio-Rad) and Applied Biosystems real-time PCR instruments with a 96-well reaction module. qRT-PCRs were performed in triplicate with 20 ng of total RNA and a 500 nM concentration of each primer. The reaction condition was 1 cycle at 50°C for 10 min, 1 cycle at 95°C for 10 s, and 40 cycles of 95°C for 15 s and 55°C for 30 s. Melting curves were generated by a PCR cycle of 95°C for 15 s and 55°C for 15 s and 80 cycles of 55°C with 0.5°C increments. All qRT-PCR primers were designed from the IDT website. The 16S RNA gene served as a housekeeping gene and the primers were described above for the RT-PCR. The primers used for amplifying etx sequences were qetxF (5′ AAAGGAAATGTAAAGTTAGTAGGACAAG 3′) and qetxR (5′ CCATCCCTAGGAAAAGCTAAATAAC 3′). After qRT-PCR, the relative quantitation of mRNA expression was normalized to the constitutive expression of the housekeeping 16S RNA gene and calculated by the comparative threshold cycle (CT) (2−ΔΔCT) method (34).
Western blot analyses of CodY, ETX, PLC, PFO, sialidase, and Spo0A production.
For Western blot analyses of CodY, ETX, PLC, PFO, and sialidases, 0.2-ml aliquots of overnight FTG cultures of CN3718, CN3718::codY, and CN3718comp were inoculated into 10 ml of SFP medium, which was then cultured at 37°C for 5 h or overnight, as specified. After this incubation, the SFP medium culture was centrifuged, and the pellet was analyzed for CodY by Western blotting using a rabbit polyclonal antiserum against B. subtilis CodY (35), kindly provided by Abraham Sonenshein. The supernatant was examined for the presence of ETX, PLC, PFO, and sialidases. ETX and PLC were detected on the blots using ETX and PLC mouse monoclonal antibodies, kindly provided by Paul Hauer. PFO and sialidases were detected on the blots using PFO (33) and sialidase (LifeSpin BioSciences, Inc.) rabbit polyclonal antibodies. Western blotting was then completed using previously described procedures (5, 9).
For Western blot analysis of Spo0A production, a 0.2-ml aliquot of an overnight FTG culture of CN3718, CN3718::codY, or CN3718comp was inoculated into 10 ml of DS or SFP medium, as specified in Fig. 5C, and cultured at 37°C for 5 h. After 5 h, the pellets from the DS and SFP medium cultures were analyzed for the presence of Spo0A production by a Western immunoblot procedure. Spo0A was detected on the blot using an anti-rabbit polyclonal antiserum against B. subtilis Spo0A (36), kindly provided by Masaya Fujita.
Analysis of ETX prototoxin proteolysis in culture supernatants.
ETX prototoxin was purified and fluorescently labeled using an Alexa Fluor 488 protein labeling kit (Invitrogen) as described previously (5). A 5-µg aliquot of labeled epsilon prototoxin was then added to 50 µl of an overnight culture supernatant of CN3718, CN3718::codY, or CN3718comp. After this mixture had been incubated for 2 h at 37°C, the samples were electrophoresed and detected as previously described (5).
Growth rate measurement for wild-type, codY mutant, and complementing strains.
For vegetative culture growth rate measurements, 0.2-ml aliquots of FTG overnight cultures of CN3718, CN3718::codY, and CN3718comp were inoculated into 10 ml of SFP medium and cultured at 37°C. At 2-h intervals up to 8 h, a 1-ml aliquot culture was collected, and the optical density at 600 nm (OD600) was recorded using a Bio-Rad Smartspec.
Sialidase enzyme activity measurement.
To assay sialidase enzyme activity, 0.2-ml aliquots of overnight FTG cultures of CN3718, CN3718::codY, and CN3718comp were inoculated into 10 ml of SFP medium and cultured at 37°C overnight. The next day, a 0.2-ml aliquot of each of these SFP medium cultures was transferred into 10 ml of fresh SFP medium. After a 5-h or overnight culture, sialidase activity was determined using the EnzyChrom neuraminidase assay kit (BioAssay Systems), which measures the sialic acid released by neuraminidase activity. To avoid the background caused by sialic acid present in the SFP medium, the 5-h or overnight SFP medium culture supernatant was desalted and buffer exchanged with PBS using a Millipore ultrafiltration centrifuge tube (10,000 nominal molecular weight limit [NMWL]) (Millipore).
MDCK cell cytotoxicity and C. perfringens adherence to Caco-2 cells.
Madin-Darby canine kidney (MDCK) epithelial cells and Caco-2 cells were cultured as described previously (5). Cytotoxicity assays using culture supernatants of CN3718 and derivatives were performed as described previously (9).
C. perfringens adherence to Caco-2 cells.
A 1.0-ml aliquot of an SFP medium (for vegetative cells) or DS medium (for spores) overnight culture of CN3718, CN3718::codY, or CN3718comp was washed with Hanks balanced salt solution (HBSS) three times for vegetative cells or ten times for spores. The numbers of bacteria adhering to host cells were determined as described previously (5). For measuring spore adherence, the spores were heat shocked for 20 min at 70°C before plating. A control plate (without Caco-2 cells), which determined the number of CFU added to the mammalian cell culture, was also heat shocked (for spores only), serially diluted, and plated onto BHI agar plates.
Quantitative heat-resistant spore counts and measurement of spore germination.
Overnight DS medium cultures of CN3718, CN3718::codY, or CN3718comp were examined for spores using a phase-contrast microscope and then heated at 70°C for 20 min to kill any vegetative cells and allow detection of mature spores by plate counting. Spore germination was determined as described previously (37).
DNA gel mobility shift assays.
The codY ORF of CN3718 was cloned into the E. coli expression vector pTrcHisA (Invitrogen) using the primers CodYHisF (5′ tagaGGATCCatgtcgacactacttagtaaaactag 3′; capital letters indicate a BamHI site) and CodYHisR (5′ ttacGAATTCttattttatctttttaagttcatccattaac 3′; capital letters indicate an EcoRI site). The His6-tagged rCodY was purified from lysates of the induced transformants as described previously (38), and the purity of the rCodY preparations was assessed by Coomassie blue G250 staining of an SDS-containing polyacrylamide gel containing this sample and by Western blot analysis using a mouse monoclonal antibody against polyhistidine (Sigma-Aldrich) or CodY antibody, described above.
To assess the ability of CodY to bind to sequences immediately upstream of the etx ORF, a DNA fragment corresponding to the 650-bp sequences present immediately upstream of the etx ORF was PCR amplified from CN3718 using the primers 1016gsF (5′ AATTAGAGCGATTTATGTGC 3′) and 1016gsR (5′ AGATTTTTTTTCATAAAACC 3′). That PCR product was then agarose gel purified using a QIAquick gel extraction kit (Qiagen). A control 340-bp DNA fragment was PCR amplified from internal codY sequences using the primers codYKOF and codYKOR. Both DNA fragments were then mixed with increasing amounts of rCodY protein in 10-µl reaction mixtures that contained 20 mM Tris-Cl (pH 8.0), 50 mM sodium glutamate, 10 mM MgCl2, 5 mM EDTA, 0.05% (vol/vol) Nonidet P-40 (Sigma-Aldrich), 5% (vol/vol) glycerol, and 2 mM GTP (18). After incubation of those mixtures for 30 min at room temperature, the binding reaction mixtures were added to 2 µl of 6× electrophoretic mobility shift assay (EMSA) gel-loading solution (electrophoretic mobility shift assay kit; Invitrogen). Those mixtures were then loaded onto a 6% nondenaturing Tris-borate-EDTA (TBE) polyacrylamide gel. The gel was electrophoresed at 4°C for 90 min at 200 V in 0.5× TBE. The gel was stained for 20 min with SYBR green in an EMSA (electrophoretic mobility shift assay kit; Invitrogen), and the stained gel was then imaged using a Typhoon 9400 variable-mode imager (Amersham Biosciences), with fluorescence emission set to detect the Alexa Fluor 488 label using the green laser with a wavelength of 532 nm.
SUPPLEMENTAL MATERIAL
Characterization of CN3718 codY-null mutant and complementing strains. (A) Diagram showing the wild type CN3718 codY gene (777 bp) and the null mutant intron insertion site (between 645 bp and 646 bp). The PCR product size is 340 bp when codYKOF and codYKOR internal primers and wild-type DNA are used; after a 900-bp intron insertion, using the same primers, the PCR product size is 1,240 bp. (B) PCR confirmation of the construction of an isogenic codY-null mutant and complementing strain. PCR products of CN3718 and CN3718comp are 340 bp; the PCR product of the codY-null mutant is 1,240 bp. The left panel shows a 100-bp DNA ladder (Thermo Scientific). (C) Southern blot hybridization of an intron-specific probe with DNA from wild-type CN3718 or the codY-null mutant. The sizes of DNA fragments (in kb) are shown at left. (D) Western blot analysis for CodY expression in pelleted cells from overnight SFP medium cultures of wild-type strain CN3718, the codY-null mutant CN3718::codY, and the complementing stain CN3718comp. The protein size (in kDa) is shown at left. (E) Postinoculation change in optical density (OD600) for cultures of wild-type CN3718, the codY-null mutant, and the complementing strain growing in SFP medium at 37°C, measured using a Bio-Rad Smartspec spectrophotometer. Shown are representative results from three similar repetitions. Download
ACKNOWLEDGMENTS
This research was generously supported by U.S. Public Health Service grant R01AI056177-09 (B.A.M.), by a project grant to B.A.M. from the Middle Atlantic Regional Center of Excellence, grant 2U54AI57168-09, by grant R03 AI105635-01 (J.L.), and by a Department of Defense Multi-disciplinary University Research Initiative (MURI) award through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286 (M.R.S.).
Footnotes
Citation Li J, Ma M, Sarker MR, McClane BA. 2013. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. mBio 4(5):e00770-13. doi:10.1128/mBio.00770-13.
REFERENCES
- 1. McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489 In Doyle MP, Buchanan RL. (ed), Food microbiology: fundamentals and frontiers, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 2. McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins TD. 2006. The enterotoxic Clostridia, p 688–752 In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E. (ed), The prokaryotes, 3rd ed. Springer Press, New York, NY. [Google Scholar]
- 3. Songer JG. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J, Sayeed S, Robertson S, Chen J, McClane BA. 2011. Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog. 7:e1002429. 10.1371/journal.ppat.1002429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 7. Amimoto K, Noro T, Oishi E, Shimizu M. 2007. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 153:1198–1206 [DOI] [PubMed] [Google Scholar]
- 8. Garcia JP, Adams V, Beingesser J, Hughes ML, Poon R, Lyras D, Hill A, McClane BA, Rood JI, Uzal FA. 2013. Epsilon toxin is essential for the virulence of Clostridium perfringens type D infection in sheep, goats and mice. Infect. Immun. 81:2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J, Rood JI, McClane BA. 2011. Epsilon toxin production by Clostridium perfringens type D strain CN3718 is dependent upon the agr operon but not the VirS/VirR. mBio 2:e00275-11. 10.1128/mBio.00275-11 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Paredes-Sabja D, Sarker MR. 2009. Clostridium perfringens sporulation and its relevance to pathogenesis. Future Microbiol. 4:519–525 [DOI] [PubMed] [Google Scholar]
- 11. Hiscox TJ, Harrison PF, Chakravorty A, Choo JM, Ohtani K, Shimizu T, Cheung JK, Rood JI. 2013. Regulation of sialidase production in Clostridium perfringens by the orphan sensor histidine kinase ReeS. PLoS One 8:e73525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, McClane BA. 2012. Role of the agr-Like quorum-sensing system in regulating toxin production by Clostridium perfringens type B strains CN1793 and CN1795. Infect. Immun. 80:3008–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slack FJ, Mueller JP, Sonenshein AL. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J. Bacteriol. 175:4605–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stenz L, Francois P, Whiteson K, Wolz C, Linder P, Schrenzel J. 2011. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol. Med. Microbiol. 62:123–139 [DOI] [PubMed] [Google Scholar]
- 15. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 16. Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovács ÁT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14:2233–2246 [DOI] [PubMed] [Google Scholar]
- 17. Shivers RP, Dineen SS, Sonenshein AL. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62:811–822 [DOI] [PubMed] [Google Scholar]
- 18. Dineen SS, McBride SM, Sonenshein AL. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192:5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Schaik W, Château A, Dillies MA, Coppée JY, Sonenshein AL, Fouet A. 2009. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect. Immun. 77:4437–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hendriksen WT, Bootsma HJ, Estevão S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PW. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joseph P, Ratnayake-Lecamwasam M, Sonenshein AL. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Popoff MR. 2011. Epsilon toxin: a fascinating pore-forming toxin. FEBS J 278:4602–4615 [DOI] [PubMed] [Google Scholar]
- 25. Mirouze N, Prepiak P, Dubnau D. 2011. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 7:e1002048. 10.1371/journal.pgen.1002048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453–1467 [DOI] [PubMed] [Google Scholar]
- 27. Gonzalez-Valencia G, Munoz O, Torres JF. 1991. Toxigenicity and adherence in Clostridium difficile strains isolated from patients with and without diarrhoea. Arch. Invest. Med. (Mex.) 22:189–196 [PubMed] [Google Scholar]
- 28. Dingle T, Mulvey GL, Humphries RM, Armstrong GD. 2010. A real-time quantitative PCR assay for evaluating Clostridium difficile adherence to differentiated intestinal Caco-2 cells. J. Med. Microbiol. 59:920–924 [DOI] [PubMed] [Google Scholar]
- 29. Huang IH, Waters M, Grau RR, Sarker MR. 2004. Disruption of the gene (spo0A) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233–240 [DOI] [PubMed] [Google Scholar]
- 30. Kumazawa T, Masayama A, Fukuoka S, Makino S, Yoshimura T, Moriyama R. 2007. Mode of action of a germination-specific cortex-lytic enzyme, SleC, of Clostridium perfringens S40. Biosci. Biotechnol. Biochem. 71:884–892 [DOI] [PubMed] [Google Scholar]
- 31. Paredes-Sabja D, Setlow P, Sarker MR. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:2711–2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bannam TL, Rood JI. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:223–235 [DOI] [PubMed] [Google Scholar]
- 33. Li J, Chen J, Vidal JE, McClane BA. 2011. The Agr-like quorum-sensing system regulates sporulation and production of enterotoxin and beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infect. Immun. 79:2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kondo T, Johnson SA, Yoder MC, Romand R, Hashino E. 2005. Sonic hedgehog and retinoic acid synergistically promote sensory fate specification from bone marrow-derived pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 102:4789–4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J, Paredes-Sabja D, Sarker MR, McClane BA. 2009. Further characterization of Clostridium perfringens small acid soluble protein-4 (Ssp4) properties and expression. PLoS One 4:e6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paredes-Sabja D, Torres JA, Setlow P, Sarker MR. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shrestha A, McClane 2013. Human claudin-8 and -14 are receptors capable of conveying the cytotoxic effects of Clostridium perfringens enterotoxin. mBio 4:e00594-12. 10.1128/mBio.00594-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of CN3718 codY-null mutant and complementing strains. (A) Diagram showing the wild type CN3718 codY gene (777 bp) and the null mutant intron insertion site (between 645 bp and 646 bp). The PCR product size is 340 bp when codYKOF and codYKOR internal primers and wild-type DNA are used; after a 900-bp intron insertion, using the same primers, the PCR product size is 1,240 bp. (B) PCR confirmation of the construction of an isogenic codY-null mutant and complementing strain. PCR products of CN3718 and CN3718comp are 340 bp; the PCR product of the codY-null mutant is 1,240 bp. The left panel shows a 100-bp DNA ladder (Thermo Scientific). (C) Southern blot hybridization of an intron-specific probe with DNA from wild-type CN3718 or the codY-null mutant. The sizes of DNA fragments (in kb) are shown at left. (D) Western blot analysis for CodY expression in pelleted cells from overnight SFP medium cultures of wild-type strain CN3718, the codY-null mutant CN3718::codY, and the complementing stain CN3718comp. The protein size (in kDa) is shown at left. (E) Postinoculation change in optical density (OD600) for cultures of wild-type CN3718, the codY-null mutant, and the complementing strain growing in SFP medium at 37°C, measured using a Bio-Rad Smartspec spectrophotometer. Shown are representative results from three similar repetitions. Download