Abstract
Most of the approximately 90,000 cases of Breast Cancer (BC) documented annually in Pakistan are not diagnosed properly because of lack of suitable markers. We performed serum proteome expression profiling of BC and benign breast disease (BBD) patients with the aim to identify biomarkers that can be helpful for diagnosis and prognosis of the disease. Sera of patients were analyzed by one-dimensional SDS polyacrylamide gel electrophoresis (PAGE). Differentially expressed proteins were subjected to identification through LC-MS/MS analysis. In majority of the BC cases some acute phase proteins (APP) and some complement system components (C3 and C8) containing fractions were up-regulated with the exception of transthyretin (TTR) which was predominantly (68.75%) down-regulated (n = 33/48) in the sera of these patients. Varying expression patterns were observed in BBD patients and healthy controls. These differentially expressed proteins have the potential to serve as diagnostic biomarkers for BC as well as benign breast diseases.
Keywords: acute phase proteins (APPs), biomarker, breast cancer (BC), haptoglobin (Hp), inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4), serum amyloid A (SAA), transthyretin (TTR), apolipoprotein A1 (apoA-1)
Introduction
Breast Cancer (BC), the third most frequent type of cancer in the world, is known to be the most common type of cancer among females both in developed and developing countries.1,2 This leading cause of death in the world is the second leading cause of death in USA.3,4 Annually, more than one million cases of BC are reported globally and approximately 90,000 cases are documented in Pakistan.2 Based on the BC incidence rate in 1995–2007, American Cancer Society predicted that 230,480 new cases of invasive BC and 57,650 cases of in-situ BC will be expected in 2011.3 In Pakistan, BC accounts for 38.5% of all other types of cancers in the country. The ratio of developing BC is increasing at an alarming rate among Pakistani females and it has been estimated that one out of every nine women in Pakistan is at risk of developing BC.5 Contrary to western females, Pakistani females suffer from the BC at an early age, having a higher risk of metastatic cancers. They also present with large lesions. The infiltrating ductal carcinoma has been identified as the most common type of BC among Pakistani females.6
Unfortunately, Pakistan has the highest prevalence of BC in Asia and, on the average, about 40,000 women die per year due to this disease.7,8 More than 50% of the BC cases in Pakistan have been found in the Punjab province. According to one report, only 10% of the BC cases (ie, 2250 out of 90,000) are diagnosed and treated while 75% of the patients do not go for treatment and die within five years after diagnosis. Survival rate of BC patients is dependent on early diagnosis and treatment, as it has been observed that early diagnosis of the BC can increase the chances of survival up to more than 90%.7,9 Early detection of BC is therefore extremely important, but remains elusive due to the non-availability of reliable biomarkers.10 Overall five-year survival of BC patients is greatly reduced as the stage of BC progresses. The five-year survival rate has been reported to be 85%, 60% and 20% in stages II, III and IV BC patients, respectively.11
Since diagnosis of BC at the earliest possible stage cannot be achieved through mammographic screening,12 blood proteome profiling may help to identify the molecular markers which are usually present at a very low concentration and can be utilized for the early detection and treatment of BC.13–15 High throughput biomolecular profiling techniques such as transcriptomics, proteomics and metabolomics are commonly used for the discovery of biomarkers of various diseases.16 Proteins that are found to be differentially expressed can be used as biomarkers for the diagnosis and prognosis of the disease and may be used as therapeutic targets for future treatment and management of the disease.17
Although estrogen, progesterone and Human Epidermal Growth Factor Receptor 2 (HER2) receptors represent a group of biomarkers that are used as molecular tools for the prognosis of BC, at present there is no plasma or serum protein that can be assigned the status of validated biomarker for the early diagnosis of BC.18 Certain proteins including autoantibodies (RS/DJ-1, p53, HSP60, HSP90, mucin-related), serum proteins (CA-15-3, RS/DJ-1, HER-2/neu) and ductal proteins (α-2-HS-glycoprotein, lipophilin B, beta-globin, hemopexin, vitamin–D-binding protein) have been identified as potential biomarkers of BC by studying humoral response, serum and nipple aspirate fluid profiling.18 Proliferation marker Ki67, estrogen receptor β (ERβ) and cell cycle regulatory proteins (cyclin D1and cyclin E) have also been projected as emerging biomarkers of BC.19–25
Considering the importance of the issue, we initiated the current study with the aim to identify potential biomarkers that can help in BC diagnosis. Polyacrylamide gel electrophoresis (PAGE), 1 dimensional (1D) or 2 dimensional (2D), in combination with mass spectrometry is widely used for the identification of tissue-based or circulating diagnostic and therapeutic biomarkers for breast cancer. 2D-gel electrophoresis is an expensive tool and requires technical skill.26–30 We therefore started our search to recognize the differentially expressed proteins in the sera of breast cancer patients using 1D sodium dodecyl sulfate (SDS)-PAGE. When we compared with the electrophoretic profile of normal sera we were able to see some differentially expressed bands in the diseased cases. Although a protein band in 1D SDS PAGE may contain many different proteins, the absence of one band would indicate that multiple proteins are involved in the pathogenesis of breast cancer simultaneously. For multiple proteins containing fractions, we planned to perform 2D-gel electrophoresis as part of the second phase of study.
Using the relatively simple and cost-effective technique of 1D SDS PAGE in combination with Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS), we have identified apolipoprotein A1 (apoA-1), the Ig kappa (κ) chain C region, and transthyretin (TTR), along with some other acute phase proteins (eg, haptoglobin (Hp), serum amyloid A (SAA), inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4), serum albumin) and some components of a complement system (i.e, C8, C3 and C4) as potential biomarkers for breast carcinoma.
Materials and Methods
Sample collection
After approval from the Institutional Ethical Committee, blood samples (n = 61) were collected from patients presenting at Bahawal Victoria (BV) Hospital, Bahawalpur, Pakistan, following standard procedures and written consent taken from each patient on a prescribed form. Complete demographic, disease and medication history of each patient was recorded in the questionnaire. Collected sera were first screened for HBV/HCV/HIV infection. No sample was found to be HIV positive. However, few samples were positive for HBV (n = 1) and HCV (n = 7). A fraction of BC patients (n = 8) had undergone chemotherapeutic treatment. The patients found positive for HBV/HCV infection and those who had been treated chemotherapeutically were excluded from the main cohort of samples. Excluded samples were treated as a separate group and analyzed to get an idea about combinatorial effect of BC and either HBV/HCV infection or chemotherapy. Female patients with BC (n = 48) and non-malignant pre-cancerous benign breast lesions (n = 13) were included in the study. The mean age of the patients was 45.54 ± 13.15 years.
Sample processing and protein gel electrophoresis
Sera obtained using standard methodology were stored at −70 °C in small aliquots until further analysis took place. Proteins were quantified using the Bradford method.31 Following the optimized gel conditions, 60 μg of serum proteins were resolved on 10% SDS PAGE.32 Gels were run for 2 hours (h) at 125 volts and stained using 0.25% solution of Coomassie brilliant blue (G-250) for 2 h. Sera of normal healthy female subjects were used as a normal control comparison. Differentially expressed protein bands, compared with the sera of normal individuals, were cut from the gel and identified using LC-MS/MS.33 Scores of peptide match and protein sequence coverage% (Table 1) provided an account of the degree of confidence associated with the identification of proteins by mass spectrometry. A higher peptide match and protein sequence coverage score is the indicator of higher confidence with which the protein is identified.34
Table 1.
Differentially expressed proteins identified by LC-MS/MS analysis in the sera of BC patients.
| Spot | Acc | Name | Mass | Ip | Score | Peptide matched | Coverage % |
|---|---|---|---|---|---|---|---|
| SEB1a | P00738 | HPT_HUMAN haptoglobin | 45177 | 6.13 | 303 | 18 (7) | 10 |
| SEB1b | P07360 | CO8G_HUMAN complement component C8 gamma chain | 22264 | 8.49 | 119 | 2 (1) | 12 |
| SEB2 | P02766 | TTHY_HUMAN transthyretin | 15877 | 5.52 | 261 | 10 (6) | 24 |
| SEB3 | P02766 | TTHY_HUMAN transthyretin | 15877 | 5.52 | 140 | 5 (3) | 17 |
| SEB4 | Q549N7 | HBB_HUMAN hemoglobin subunit beta | 15988 | 6.75 | 289 | 18 (8) | 46 |
| SEB5a | Q6FG67 | SAA_HUMAN serum amyloid A protein | 13524 | 6.28 | 241 | 10 (3) | 24 |
| SEB5b | Q9P157 | ALBU_HUMAN | 69321 | 5.92 | 189 | 5 (2) | 7 |
| SEB5c | P18985 | HBB_CALAR hemoglobin subunit beta | 15942 | 6.85 | 135 | 6 (1) | 21 |
| SEB6a | P01024 | CO3_HUMAN complement C3 | 187030 | 6.02 | 625 | 21 (10) | 9 |
| SEB6b | Q14624 | ITIH4_HUMAN inter-alpha-trypsin inhibitor heavy chain H4 | 103293 | 6.51 | 259 | 9 (2) | 6 |
| SEB7a | Q9P157 | ALBU_HUMAN serum albumin | 69321 | 5.92 | 236 | 7 (2) | 10 |
| SEB7b | Q5JQM8 | Complement C4-A | 192650 | 6.65 | 102 | 6 (1) | 2 |
| SEB8a | P01834 | Ig kappa chain c-region | 11602 | 5.58 | 429 | 10 (9) | 80 |
| SEB8b | P02647 | Apolipoprotein A1 | 30759 | 5.56 | 138 | 5 (3) | 20 |
Abbreviation: Ip, Isoelectric point; Acc, Accession number.
Notes: Peptide matched = Number of (unique) peptides matched out of our MS/MS analysis to a certain protein (identified protein in this case) and number in parenthesis is the count of peptides with significant matches, Protein sequence coverage% = % age of amino acids from identified protein covered in this MS/MS analysis which is bases of the identification, Protein score = It is non-probabilistic method of ranking protein hits, SEB1-8 = protein bands cut from normal samples.
The protein expression pattern was documented using the UV-Pro gel system (USA). Gel images were processed for the densitometric quantification of various protein bands using GelQuantNet software (BiochemLabSolutions, Princeton, NJ, http://biochemlabsolutions.com/GelQuantNET.html). Protein bands were analyzed and densities of the bands observed in test samples (ie, sera of breast cancer and benign breast disease patients) were compared with those of normal controls. On the basis of this comparison test samples were identified to have either more expression (up-regulation), less expression (down-regulation) or the same level of expression (no change). Results were finally reported as the percentage of breast cancer and benign breast disease patients having either up- or down-regulated expression.
Results are presented as percentages because the overall detection rate of the bands is simply based upon recording the presence or the absence of a particular protein band on gel, and is therefore less informative. Overall detection rate of the bands is not as informative as the comparison of densities, which reveals the nature and extent of change in the expression level of different proteins (Table 2).
Table 2.
Expression pattern of various proteins in the sera of BC and benign breast disease patients.
| Sr. no. | Protein/s | Expression level w.r.t. normal | Total patients (n = 61) % age | BC Sera (n = 48) % age | BBDs Sera (n = 13) % age | P-value (X2-test) |
|---|---|---|---|---|---|---|
| 1. | SEB1a = Haptoglobin, SEB1b = Complement component C8 gamma chain | Down-regulation | 47.54 | 50.00 | 38.46 | 0.670a |
| Up-regulation | 52.46 | 50.00 | 61.54 | |||
| 2. | SEB2 = Transthyretin | Down-regulation | 63.93 | 68.75 | 46.15 | 0.238a |
| Up-regulation | 30.07 | 31.25 | 53.85 | |||
| 3. | SEB3 = Transthyretin (modified) | Only expressed in diseased subjects | 27.87 | 25.00 | 38.46 | 0.541a |
| 4. | SEB4 = Hemoglobin subunit β | Up-regulation | 4.92 | 2.08 | 15.39 | 0.213a |
| No change | 95.08 | 97.92 | 84.62 | |||
| 5. | SEB5a = Serum amyloid A protein, SEB5b = Albumin, SEB5c = Hemoglobin subunit β | Down-regulation | 37.71 | 39.58 | 30.77 | 0.796a |
| Up-regulation | 62.30 | 60.42 | 69.23 | |||
| 6. | SEB6a = Complement C3 SEB6b = Inter-alpha-trypsin inhibitor heavy chain H4 |
Down-regulation | 44.26 | 37.50 | 69.23 | 0.119a |
| Up-regulation | 54.10 | 60.42 | 30.77 | |||
| No change | 1.64 | 2.08 | – | |||
| 7. | SEB7a = Serum albumin SEB7b = Complement C4-A |
Down-regulation | 60.66 | 58.33 | 69.23 | 0.694a |
| Up-regulation | 39.34 | 41.67 | 30.77 | |||
| 8. | SEB8a = Ig Kappa chain c-region SEB8b = Apolipoprotein A1 |
Down-regulation | 40.98 | 43.75 | 30.77 | 0.099a |
| Up-regulation | 54.10 | 50.00 | 69.23 | |||
| No change | 4.92 | 6.25 | – |
Notes:
Significance level for variation amongst the BC and BBDs, n = no of subjects/individuals. X2 analysis was performed on actual outcome/numerical values not on percentages. P-value is the probability that gives the measure of deviation between observed and expected outcome.
Normalization of proteins loaded on gel
The differentially expressed proteins were identified by comparing gel profiles of patients with those of normal controls. Equal quantities of total proteins were loaded in each lane. A protein band on the gels exhibiting minimum variations across the samples was taken as the internal control for densitometric analysis. For further normalization of the loaded proteins and better assessment of differentially expressed bands, serum albumin quantification data was considered (data not shown). The main aim of all the efforts was to document the true picture of differential expression and to minimize the chances of erroneous documentation of various proteins in sera of BC and benign breast disease patients.
In-gel tryptic digestion and peptide sequence analysis
Standard methodology was used for in-gel tryptic digestion and peptide sequence analysis.33 Specific bands excised from the Coomassie brilliant blue (G-250) stained gel were subjected to destaining and in-gel digestion using optimized protocol for ESI-QTQF MS/MS as described elsewhere.35 Chromatographically-separated peptides were ultimately analyzed using Q-TOF Ultima Global (Micromass, Manchester, UK) mass spectrometer provided with a nanoflow ESI Z-spray source in positive ion mode. MassLynx (v 4.0) software on Windows NT PC was used for data acquisition. Further processing of the data was performed on Protein-Lynx- Global-Server (v 2.1; Micromass, Manchester, UK). To identify the proteins present in the analyzed sample, processed data was searched in MSDB and Swiss-Prot databases through a Mascot search engine with a peptide mass tolerance of ±0.5 Da and fragment mass tolerance of ±0.5 Da. Search criteria were set to the maximum and only one missed cleavage was allowed by trypsin. Protein modifications were set to methionine oxidation and carbamidomethyl cysteine, whatever was appropriate.33
Statistical analysis
To know the significance level of the variations among different groups of BC and benign breast disease patients, a Chi-square test (X2) was performed, using SPSS 18.0, on the actual numerical values/outcomes but not on percentage data. Results of the X2 test guided us to document the expression trends of various proteins in the pathogenesis of BC and benign breast diseases. Further, the statistical analysis helped to explore whether the differential expression of various proteins can be potentially used to discriminate between BC and benign breast disease patients. To document the effect of chemotherapeutic treatment and HCV infection on expression of different proteins, we grouped BC patients into various groups based on the status of chemotherapeutic treatment and HCV infection. Patients suffering from different types of BC were placed into separate groups to explore the type of existing relationship, if any, between the differential expression of studied proteins and the type of breast cancer. Patient groups containing either one or two representative samples were not subjected to X2 analysis.
Results
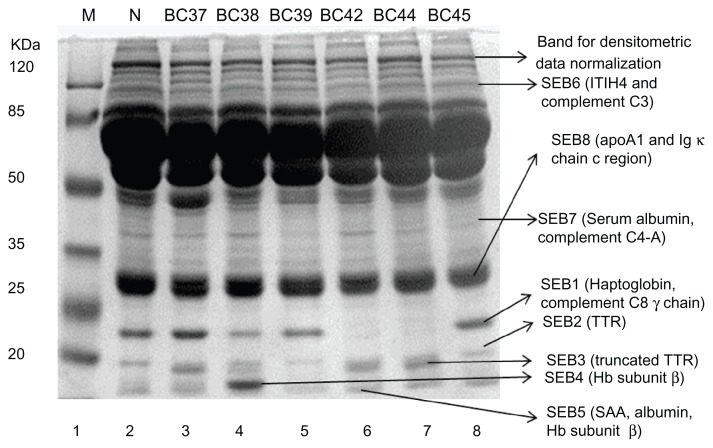
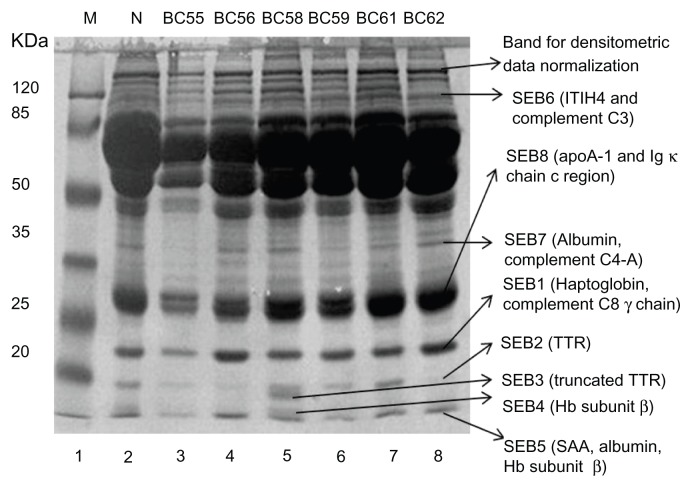
Altered expression levels of eight different protein bands were visually noticed in the sera of patients with BC, inflammation and benign or complications like mammary dysplasia and chronic mastitis as compared to normal female controls. Differentially expressed protein bands were labeled as SEB1-8, sometimes with additional letters to indicate closely spaced bands. Intriguing was the observation that certain protein bands were either completely missing (Fig. 1, lane 6–7, SEB1; lane 5, SEB3, SEB4; lane 5 and 8, SEB7; Fig. 2, lane 3, SEB7, SEB3) or expressed at very low levels (Fig. 1, lane 4, SEB1; lane 4–5 and 8, SEB2; Fig. 2, lane 6 and 8, SEB2). Protein component of these bands were identified through LC-MS/MS and the data is summarized in Table 1. Details can be seen in the supplementary data. The differentially expressed proteins associated with BC and benign or non-malignant breast lesions/inflammations were identified as a number of acute phase proteins (haptoglobin, SAA, serum albumin, TTR, truncated TTR and ITIH4), apolipoprotein A1, Ig kappa chain C region, and some complement system components (including C3, C4-A and C8, Table 1).
Figure 1.
Differential expression profiles of serum proteins in breast carcinoma and other breast complications.
Notes: Sera of patients and normal individuals were subjected to SDS PAGE as described in materials and methods. Variations in protein bands were examined after Coomassie brilliant blue staining.
Abbreviations: M, Molecular weight markers; N, Normal individual; BC37, BC38, Mammary dysplasia; BC39, Infiltrating ductal carcinoma grade II; BC42, Lobular Carcinoma; BC44, Severe, acute, chronic and non-specific inflammation; BC45, Infiltrating ductal carcinoma grade I; BC37, BC38 and BC44, Benign breast disease patients; BC39, BC42 and BC45, Breast cancer patients.
Figure 2.
Comparison of protein expression profiles in the sera of different types of breast carcinoma and fibroadenoma.
Notes: Sera of patients and normal individuals were subjected to SDS PAGE as described in materials and methods. Variations in protein bands were examined after Coomassie brilliant blue staining.
Abbreviations: M, Molecular weight markers; N, Normal individual; BC55, Phylloid tumor; BC56, Infiltrating ductal carcinoma; BC58, Fibroadenoma; BC59, Duct carcinoma; BC61, Lobular carcinoma; BC62, Infiltrating ductal carcinoma; BC55, BC58, Benign breast diseases patients; BC56, BC59, BC61, and BC62, Breast cancer patients.
Altered expression level of some acute phase proteins and complement system components
The LC/MS/MS data revealed haptoglobin as the part of SEB1 protein band along with complement component C8 gamma chain (Figs. 1–2) and this band was found to be predominantly up-regulated in benign breast disease patients (Fig. 1, lane 3, sample BC37; Fig. 2, lane 5, sample BC58). Down-regulation was seen in a small proportion of the benign breast disease patients. X2 analysis revealed that SEB1 expression cannot help in differentiation between BC and benign breast disease patients (Table 2). The differential expression of SEB1 protein fraction was also found to be associated with the status of chemotherapeutic treatment and HCV infection (P = 0.0005). Up-regulation was more significant in non-chemotherapeutically treated BC patients and a large proportion of chemotherapeutically treated BC patients showed down-regulation. All HCV positive BC patients exhibited down-regulation of this protein fraction. Differential expression of SEB1 protein fraction was not found to be related with any particular type of BC (P = 0.102) except that duct carcinoma patients had predominantly down-regulated expression of this protein fraction (Table 3).
Table 3.
Expression pattern of various proteins in the sera of various breast cancer groups.
| Sr. no. | Protein/s | Expression level w.r.t. normal | Breast cancer patients groups w.r.t. treatment/HCV infection (% age) (A = Nonchemotherapeutically treated, B = Chemotherapeutically treated, C = HCV positive) | P-value (X2-test) | Breast cancer patients groups w.r.t. type of breast cancer (% age) (A = Infiltrating/invasive duct carcinoma, B = in-situ Duct carcinoma, C = Unknown) | P-value (X2-test) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| A (n = 32) | B (n = 8) | C (n = 7) | A (n = 22) | B (n = 7) | C (n = 14) | |||||
| 1. | SEB1a = Haptoglobin, SEB1b = Complement component C8 gamma chain | Down regulation | 34.37 | 62.500 | 100.00 | 0.005 | 40.91 | 85.71 | 42.86 | 0.102 |
| Up-regulation | 65.63 | 37.50 | – | 59.09 | 14.29 | 57.14 | ||||
| 2. | SEB2 = Transthyretin | Down-regulation | 56.25 | 87.50 | 100.00 | 0.035 | 72.73 | 71.43 | 57.14 | 0.605 |
| Up-regulation | 43.75 | 12.50 | – | 27.27 | 28.57 | 42.86 | ||||
| 3. | SEB3 = Transthyretin (modified) | Only expressed in diseased subjects | 28.13 | 37.50 | – | 0.211 | 27.27 | 14.29 | 7.14 | 0.302 |
| 4. | SEB4 = Hemoglobin subunit β | Up-regulation | 3.13 | – | – | 0.787 | – | – | 7.14 | 0.346 |
| No change | 96.88 | 100.00 | 100.00 | 100.00 | 100.00 | 92.86 | ||||
| 5. | SEB5a = Serum amyloid A protein, SEB5b = Albumin, SEB5c = Hemoglobin subunit β | Down-regulation | 31.25 | 37.50 | 71.43 | 0.140 | 36.36 | 42.86 | 42.86 | 0.190 |
| Up-regulation | 68.75 | 62.50 | 28.57 | 63.64 | 57.14 | 57.14 | ||||
| 6. | SEB6a = Complement C3 SEB6b = Inter-alpha-trypsin inhibitor heavy chain H4 |
Down-regulation | 34.38 | 12.50 | 85.71 | 0.048 | 31.82 | 57.14 | 28.57 | 0.595 |
| Up-regulation | 62.50 | 87.50 | 14.23 | 63.64 | 42.86 | 71.43 | ||||
| No change | 3.13 | – | – | 4.55 | – | – | ||||
| 7. | SEB7a = Serum albumin SEB7b = Complement C4-A |
Down-regulation | 46.88 | 75.00 | 85.71 | 0.093 | 54.55 | 71.43 | 50.00 | 0.470 |
| Up-regulation | 53.13 | 25.00 | 14.29 | 45.45 | 28.57 | 50.00 | ||||
| 8. | SEB8a = Ig Kappa chain c-region SEB8b= Apolipoprotein A1 |
Down-regulation | 28.12 | 62.50 | 85.71 | 0.032 | 40.91 | 71.42 | 35.71 | 0.239 |
| Up-regulation | 65.63 | 25.00 | 14.29 | 50.00 | 14.29 | 64.29 | ||||
| No change | 6.25 | 12.50 | – | 9.09 | 14.29 | – | ||||
Another acute phase protein SAA protein, as well as human serum albumin and hemoglobin subunit β, are constituents of the protein gel band SEB5, whereas the SEB6 fraction is composed of ITIH4 and the C3 component. Both SEB5 and SEB6, protein fractions were found to be up-regulated in the majority of BC cases. It is important to note that benign breast disease patients exhibited significant up-regulation of the SEB5 and down-regulation of the SEB6 fraction. X2 analysis indicated that the differential expression pattern of SEB5 and SEB6 protein fractions is also not helpful to discriminate BC patients from benign breast disease patients (Table 2). Up-regulation of SEB5 and SEB6 were more obvious in BC patients regardless of their chemotherapy status. Contrary to this, SEB5 and SEB6 fractions were observed to be down-regulated in large proportions of HCV positive BC patients. SEB5 protein fraction was predominantly up-regulated in different types of BC patients. In contrast, majority of the duct carcinoma patients exhibited down-regulation of ITIH4 and C3 component containing fraction (SEB6) while up-regulation was significant in all other cases (Table 3).
X2 analysis revealed that there was significant variation in the expression of SEB6 protein fraction among BC patients groups categorized according to the treatment type and status of HCV infection. However, differential expression of SEB5 was not associated with type of BC, status of chemotherapeutic treatment and HCV infection.
Down-regulation of TTR
SEB2 was found to contain transthyretin (TTR). This acute phase protein was found to be significantly down-regulated in the sera of BC patients (68.75%). Benign breast disease patients exhibited both up-(53.86%) and down-regulation (46.15%) with slightly higher frequency of up-regulation (Table 2). TTR was down-regulated in the majority of the BC patients, irrespective of whether the patients have been treated chemotherapeutically or not. Meanwhile, all studied HCV-positive BC patients had a lowered expression level of TTR as compared to the normal controls. X2 analysis indicated that TTR differential expression had significant variation among the BC patient groups, established on the basis of treatment type and HCV infection. However, no association was found between TTR differential expression and type of breast carcinoma (Table 3). Similarly, we were unable to discriminate BC patients from benign breast disease patients on the basis of TTR differential expression.
Expression of modified TTR
SEB3 also contained TTR with the same molecular weight and pI but with a different score of peptides match (Table 1). This could be the post-translationally modified proteolytically truncated, cysteinylated and/or glutathionylated form of TTR36 as it migrates as a separate band on SDS-PAGE. This modified form of TTR was observed in a small fraction of the total cases included in our study. It was not expressed in the HCV-positive BC patients and its expression was slightly higher in the case of infiltrating/invasive duct carcinoma as compared to the other types of breast carcinomas. X2 analysis indicated that expression of the modified TTR does not vary significantly (P > 0.05) among the BC patients and patients of benign breast diseases (Tables 2 and 3).
Expression of hemoglobin subunit β
Protein identified from gel band SEB4 is hemoglobin subunit β and it does not seem to be involved in pathobiology of BC as well as other breast complications. In the present study, only 1 non-chemotherapeutically treated BC patient and 2 patients suffering from benign breast diseases (ie, 1 mammary dysplasia and 1 fibroadenoma patient) showed elevated levels of this protein (Table 2). Meanwhile, majority of BC and benign breast disease patients were found to have no change in the expression level of this protein. Hence, the expression pattern of hemoglobin subunit β cannot be considered as the candidate biomarker of BC and benign breast diseases (Table 2). Hemoglobin subunit β was the part of SEB5 fraction whereas SEB4 is the band cut from the sample BC58 (lane 5, Fig. 2). It may represent a post-translationally modified form of hemoglobin subunit β that appears as a separate band in few samples. However, in most of the cases, hemoglobin subunit β was found to be part of SEB5 fraction.
Down-regulation of serum albumin and the complement C4-A containing fraction
Serum albumin and the complement C4-containing fraction (SEB7) was down-regulated in the majority of BC and benign breast disease patients (Table 2). Furthermore, detailed analysis revealed that all groups of BC patients, including treated, HCV infected and type of carcinoma-based groups, had down-regulation of the SEB7 fraction. X2 analysis showed that SEB7 differential expression data was not enough to discriminate BC patients from benign breast disease patients or to distinguish any of the BC patients group from others (Tables 2 and 3).
Altered expression of ApoA-1 and Ig κ chain c-region containing fraction
ApoA-1 and Ig κ chain c-region were identified in SEB8 fraction. Although a major proportion of breast cancer and benign breast disease patients exhibited up-regulation of this protein fraction, breast cancer patients cannot be discriminated from benign breast disease patients on the basis of the expression of these proteins (P = 0.099; Table 2). Expression pattern of this protein fraction seems to be influenced by the status of chemotherapeutic treatment and HCV infection (P = 0.032). Moreover, it is interesting to note that non-chemotherapeutically treated patients were found to have predominant up-regulation (65.63%) whereas a higher proportion of chemotherapeutically treated and HCV positive breast cancer patients showed down-regulation of this protein fraction (Table 3). Similarly, the apoA-1 and Ig κ chain c-region containing protein fraction was significantly up-regulated in infiltrating/invasive ductal carcinoma patients and patients whose carcinoma type was not identified, but predominantly down-regulated in in-situ duct carcinoma patients (Table 3). X2 analysis has revealed a 23.9% chance that the deviation from normal is due to chance only (P = 0.239). Hence, type of breast cancer did not contribute differentially towards the altered expression of this protein fraction (Table 3).
Effect of chemotherapy on the expression of various proteins
A small fraction of the BC patients (n = 8) investigated in the present study had undergone 2–6 cycles of chemotherapy. In these patients, all identified protein fractions were significantly down-regulated with the exception of SEB3 and SEB4. SEB3 (modified TTR) was expressed in a small fraction of chemotherapeutically treated patients whereas there was no change in the expression level of the SEB4 fraction as compared to the normal controls (Table 3).
Combined effect of HBV/HCV infection and BC
The current study included two lobular carcinoma patients and one spindle cell neoplasm patient. One patient was found to be HBV-positive and had also been treated chemotherapeutically. SEB1, 5 and 7 were up-regulated while SEB2 and 6 were down-regulated in the patient suffering from spindle cell neoplasm. Lobular carcinoma patients exhibited down-regulation of SEB6 and SEB7, while out of 2 lobular carcinoma patients, one exhibited up- and other down-regulation of SEB1, 2 and 5. In HBV-positive and chemotherapeutically-treated BC patient, SEB1, 2, 5 and 7 were down-regulated but SEB6 was up-regulated. Modified TTR (SEB4) expression was documented in one lobular carcinoma patient as well as in the patient suffering from spindle cell neoplasm. Modified TTR was not found in HBV-positive and chemotherapeutically-treated patients and there was no effect on the expression of hemoglobin subunit β (SEB4) in the groups with a low number (n = 1 or 2) of representative samples (Table 3).
While collecting samples we came across 7 HCV positive BC patients. The HCV-positive patients were separated from the main cohort of BC patients and analyzed separately. In HCV-positive BC patients all but SEB3 and SEB4 protein fractions were found to be significantly down-regulated. SEB3 was not expressed in any of the HCV-positive BC patients, while there was no change in the expression level of SEB4 as compared to the normal controls. There was only one HBV-positive BC patient. In this patient, all but SEB3 and SEB4 protein fractions were down-regulated. Modified TTR (SEB3) was not expressed and there was no change in the expression level of hemoglobin subunit β (SEB4) as compared to normal controls (Table 3).
Discussion
Our results have documented increased or decreased levels of various acute phase proteins including haptoglobin, SAA, ITIH4 and TTR. A variable expression pattern is also observed for some components of complement system including C3, C4 and C8. Differential expression of multiple proteins has been reported in the sera of colon cancer37 and metastatic oral cancer patients.38 In addition, a protein fraction containing apoA-1 and Ig kappa chain c-region is also pre-dominantly up-regulated in breast cancer and benign breast disease patients.
An acute phase reaction is the non-specific response to any infection, inflammation or trauma in the body mediated by inflammation linked cytokines, mainly IL6 in animals. Consequently, there is change in the concentration of a group of acute-phase proteins in the serum.39–41 The complement system, on the other hand, is an important part of humoral immune system and plays an important role in both innate and acquired immunogenicity. It consists of various proteins acting in different domains of an inter-linked network.42 The complement system takes care of the invading micro-organisms, transformed cells and molecular aggregates from tissues and biological fluids.43
Densities of the corresponding protein bands in normal controls determined by GelQuant.Net software were used as reference to compare the expression level of various proteins in breast cancer and benign breast disease patients. Normalization of protein expression data was particularly helpful for the samples like BC55 (Fig. 1), where it was difficult to justify whether the observed differential expression of proteins was the outcome of protein-loading errors or not. Further support was provided by serum albumin quantification data (not shown). To determine the significance level of variation among different groups of BC patients and benign breast disease patients, we performed a X2 analysis on the data. This analysis was helpful to evaluate the inherent relationship, if present, between altered expression level of any protein and type of BC or benign breast diseases. In short, the X2 test was performed to determine whether there is any contribution of either type of breast complication (ie, breast cancer/benign breast disease) or type of breast cancer (like infiltrating ductal carcinoma, in situ ductal carcinoma or lobular carcinoma). The P-value is the probability of deviation of the observed from expected due to chance alone or the contribution of some other factor.
Our results indicate that the BC patients are not distinguishable from benign breast disease patients (P = 0.670) on the basis of differential expression of haptoglobin and the C8-containing fraction (SEB1). Similarly, there is no relationship between type of BC and expression of the SEB1 fraction (P = 0.102). However, expression of this fraction (SEB1) varies significantly (P = 0.005) depending on the status of chemotherapeutic treatment and HCV/HBV infection in BC patients.
Our results correspond to earlier studies that have reported elevated serum levels of haptoglobin in various types of infections, inflammation and a wide range of carcinomas including BC,44 epithelial ovarian carcinoma,45 esophageal squamous cell carcinoma,46 urogenital tumor,47 leukemia,48 lung cancers and bladder cancers.49,50 It is involved in the formation of the membrane attack complex on bacterial cell membranes. Alpha and beta subunits are responsible for the complement mediated bacterial killing but the gamma subunit appears to bind with retinol.51
In our study, the TTR (fraction SEB2) was the only identified protein that was found to be down-regulated in the majority of BC and benign breast diseases. It is a 55-kDa homotetrameric protein which plays an important role in the transport of thyroid hormones in blood36,52 and metabolism of retinol.53 A scientific literature search has revealed that decreased levels of TTR have been reported in cases of severe liver diseases, malnutrition and acute inflammation54–56 as well as in ovarian cancer,57–59 lung cancer,60 cholangiocarcinoma,61 prostate cancer62 and advanced circular and endometrial carcinoma patients,58 but not in BC. Ours, therefore, is the first report to show that this protein is differentially expressed in breast diseases also. We also noticed a modified form of TTR (SEB3 fraction) being expressed in the sera of a few patients included in the study. In the published literature, TTR and its three post-translationally modified forms have been reported as serum markers in patients of mycosis fungoides, a type of cutaneous T-cell lymphoma.63 Post-translationally modified forms of TTR were also found in the serum of late-stage ovarian cancer, prostate, breast and colon cancer patients.36 We suggest that this protein may be a component of a cellular pathway that is common to the pathogenesis of different types of carcinoma.
Earlier studies have reported α and β subunits of hemoglobin as potential serum biomarkers for the diagnosis and prognosis of ovarian cancer.64 In our study, hemoglobin subunit β (SEB4) was identified to be up-regulated in only 3 of the total 61 patients (ie, BC and benign breast disease patients).
We have observed up-regulation of the SAA protein-containing fraction (SEB5) in majority of the BC and benign breast disease patients. SAA protein is an immunomodulator and is involved in various physiological and pathological processes including inflammation, arthereosclerosis and thrombosis.65 Its levels significantly increase in response to tissue damage and inflammation. Elevated levels of this protein have also been reported in rheumatoid arthritis, primary biliary cirrhosis, chronic active hepatitis and various carcinoma including chronic lymphocytic leukemia, Hodgkin’s disease, myelomatosis, lung, bladder, colorectal, and gastric cancers as well as BC.66,67 Higher levels of albumin are also found in benign lesions of breast while low albumin contents in the breast tumor cytosol are linked to a higher tendency of axillary node involvement in BC.68 Human serum albumin and hemoglobin α and β subunit levels are known to be elevated in ovarian carcinoma and are reported to be potential serum markers for the diagnosis and prognosis of ovarian cancer.64,69
In the current study we have noticed the complement C3, ITIH4 protein fraction to be significantly up-regulated in a major fraction of BC patients while down-regulation was predominant in benign breast diseases. Alternatively, serum albumin and the complement C4-A protein fraction was down-regulated in a majority of the BC and benign breast diseases patients. ITIH4 is a 35 KDa glycoprotein and is the fifth member inter-alpha-trypsin inhibitor (ITI). It belongs to the super family of Kunitz type protease inhibitors,57 is mainly synthesized by liver, and is secreted into blood where it is cleaved by plasma kallikrein into smaller fragments.70 Its suggested roles include the stabilization of extracellular matrices71 and inhibition of apoptosis.72 It is well documented that ITIH4 serum levels are elevated in various types of carcinoma including BC, epithelial ovarian carcinoma and germ cell ovarian carcinoma. A cleavage fragment of ITIH4 is up-regulated in early stages of ovarian cancer.57 Conversely, ITIH4 levels are not raised in patients of nasopharyngeal carcinoma and osteosarcoma.73 Our results are in accordance with the earlier studies.
Significantly higher levels of complements (CH50, C3, C4 and C1q) in subjects with cancer (n = 200) versus healthy subjects (n = 90) have been reported by some workers and suggested to be related to the stage of the carcinoma. This is because nearly normal complement levels are reached at remission stage.74 Similar profiles are seen in the total hemolytic complement (CH50) and its fractions C3 and C4 in localized juvenile periodontitis75 and in patients with carcinoma of the oral cavity, uterine cervix, and breast.76 Lymphogenous leukemia patients had also shown wide variations in serum complement levels. Recently, it has been shown that C3 and C4 are up-regulated in response to epirubicin and docetaxel chemotherapy.77 In our study, we have observed altered expression levels of various complement system components (ie, C3, C4-A and C8). Only 8 of the breast cancer patients included in our study have undergone chemotherapy. A large-scale retrospective study with appropriate follow-up of patients is required to document effect of chemotherapy on expression of various proteins.
ApoA-1 is encoded by the APOA1 gene and is the major component of high-density lipoprotein (HDL) in plasma. ApoA-1 binds with the ATP-binding cassette transporter A1 (ABCA1) to mediate the efflux of cholesterol and proinflammatory phospholipids from tissues to liver for excretion.78–80 ApoA-1 has been shown to possess anti-inflammatory and anti-oxidant properties.81 It is known that lipid transport, inflammation and oxidative stress are the key factors associated with the pathogenesis of cancer.82–84 Down-regulated expression of ApoA-1 has been observed in ovarian cancer.58,85 Moreover, it was found that human apoA1, when overexpressed in transgenic mice tumor growth, was inhibited whereas survival rate was enhanced in a mouse model of ovarian cancer.86 It has been suggested that the apoA-1 protein mediates anti-tumor effects by binding and removing lysophosphatidic acid (LPA), a bioactive lysophospholipid.86,87 Lysophospholipids are known to activate the process of proliferation in cancer cells in general and have also been implicated in pathogenesis of breast cancer.87,88 It has been reported that low density lipopolysaccharides (LDL) increase in women suffering from breast cancer and fibrocystic disease, and the ratio of apoA-1 to apoB has been found as the best predictor of cancer recurrence.89
Similarly, immunoglobulins are key components of the immune system and are composed of two heavy and two light chains. There are two subtypes (ie, kappa ‘κ’ and lambda ‘λ’) of light chains. Understanding immunological defense mechanism gives a better account of breast cancer progression.90,91 Ig κ chain has been used for monitoring prognosis and response to therapy in breast cancer and other cancers.92,93 Higher expression of Ig κ chain has proven to be an indicator of better metastasis free survival of breast cancer patients.93 Further an increased expression of κ chain c region mRNA has been documented in human cervical epithelial cells dysplasia and carcinoma.94
In the present study we have observed both up- and down-regulation of apoA1 and Ig κ chain c region containing protein fraction although majority of breast cancer and benign breast disease patients were found to have up-regulated expression. We were unable to differentiate between breast cancer and benign breast disease patients on the basis of the expression of this protein fraction. We have found significant up-regulation of this protein fraction in patients suffering from patients suffering from unidentified type of carcinoma, infiltrating/invasive ductal carcinoma and non-chemotherapeutically treated patients whereas majority of the chemotherapeutically treated, HCV positive and in-situ duct carcinoma patients exhibited down-regulation of this protein fraction. At present we are unable to suggest about the role/mechanism of apolipoprotein in the pathogenesis of breast cancer. However, results are encouraging and demand a detailed follow-up study to address correlation of down- or up-regulated expression of apoA-1 with breast cancer and benign breast diseases.
On the basis of the observations discussed above we conclude that TTR, and proteins present in SEB5, SEB6 and SEB7 fractions, may be the part of a common pathway related to the pathogenesis of breast diseases including cancer and benign breast diseases. These include mammary dysplasia, chronic mastitis, fibroadenoma, and non-specific inflammation. Further, as it is known through earlier studies that benign breast diseases increase the risk of developing BC,95 it is possible that variations in the expression level of studied proteins are responsible for oncogenic transformation and changes at the molecular level that are otherwise not detectable by histopathological study.
The present study has indicated multiple proteins as the potential BC biomarkers. As other protein fractions did not contain a single protein except TTR and modified TTR, we recommend TTR down-regulation as a potential candidate biomarker of BC. Similarly, the presence or the absence of modified TTR may be related to type, stage or grade of BC and medication.
Conclusions
A correlation of TTR has been found with the pathogenesis of breast carcinoma. About 68.75% of the analyzed data have shown down-regulation of TTR. In addition, a modified form of TTR is also present in 27.87% of the analyzed BC patients. We suggest that quantification-based assays may be more helpful and conclusive for proper validation of TTR as a biomarker for BC and pre-cancerous BC complications. Further studies are to be conducted on the role of transthyretin in BC. Predominant up-regulation of SEB5 and SEB6 protein bands have revealed the possible role of SAA, albumin, C3 and ITIH4 in BC. Likewise, down-regulation of C4-A and the serum albumin-containing fraction might play an important role during the course of BC. TTR levels are also regulated by nutritional status, inflammation and presence or the absence of different hepatic diseases.96 A detailed study is required in this regard with careful selection of controls to validate the specificity of TTR. There is a need to explore the hierarchy of events and identify the major proteins involved in due course. Once the very first molecule responsible for triggering the pathway is identified, this will be helpful for the diagnosis of BC and related complications like mastitis and dysplasia at the earliest possible stage. The information obtained can be further employed to design better therapeutic strategies for the treatment of the disease. Our results suggest that the use of multiple biomarkers including all or few of the acute phase proteins (ie, albumin, haptoglobin, ITIH4, SAA, TTR) and complement system components (C3, C4-A, and C8 gamma chain) may be a better option for diagnosis and follow up of a disease. We have not quantified TTR using specific ELISA. Present results are based on densitometric analysis. TTR quantification data will be helpful to perform a logistic regression analysis and determine the prediction score of TTR for predicting an individual’s risk for breast cancer.
The major aim of our study was not to explore the combined effect of HBV/HCV infection and breast cancer. We collected samples from breast cancer patients and screened for HBV/HCV/HIV infection. Few breast cancer and benign breast disease patients were found to be HBV/HCV positive. Following the exclusion criteria of study we excluded HBV/HCV positive patients from the main cohort of the patients. But it was interesting to notice the combined effect of HBV/HCV infection and breast disease. At present, interpretation results may seem to be exaggerated due to the low number of samples, but this pilot study can serve as a starting point for a future study designed to address the issue.
Our results have predicted the involvement of multiple proteins including immunoglobulins (Ig κ chain c region), complement system components (C3, C4-A, C8 γ chain), acute phase proteins (haptoglobin, TTR, albumin, ITIH4, SAA) and an amyloidosis-associated protein (apoA-1) in the pathogenesis of BC and benign breast diseases. These proteins are known key components associated with chronic inflammation. On the basis of available clinical and experimental data, it has been suggested that chronic inflammation significantly contributes to cancer development. Inflammation is known as the 7th hallmark of cancer.97 Persistent humoral immune response greatly enhances the recruitment and activation of innate immune cells in neoplastic microenvironments. Activated immune cells in turn regulate various cancer-associated pathways including tissue remodelling, pro-angiogenic and pro-survival pathways. Moreover, examination of individuals with chronic inflammatory disorders has shown that suppressed cellular immunity, increased humoral immunity and humoral immunity associated cytokines together contribute towards the suppression of anti-tumor immune responses. As a result of this suppression, the process of angiogenesis is accelerated, increasing the risk of cancer development in inflamed tissues.97,98 On the basis of de novo organspecific cancer development studies conducted in transgenic mouse models it has been proposed that tumor promotion and progression might be facilitated through immunoglobulins and immune complexes mediated inflammation. All of this evidence supports the hypothesis that increased local humoral and innate immune system activation along with the suppressed cellular immunity and failed cytotoxic T cell anti-tumor immunity alter risk of cancer development. These components can be powerful targets for anti-cancer immunotherapeutic drugs.91,99,100 Results of our study support the role of inflammation mediators in the pathogenesis of breast cancer.
Supplementary Data
Acknowledgements
The authors would like to acknowledge cooperation of the patients and the healthy volunteers who donated their blood samples for the study.
Footnotes
Author Contributions
The study was conceived and planned by FHN. SE did all the bench work in Pakistan and wrote the first draft. MA was involved in densitometric and statistical analysis of the data. LC-MS/MS protein identification was performed by ARA and MO in Germany. GA, GAM and A-u-R were part of the team involved in clinical diagnosis and collection of samples. The manuscript was mainly prepared by FHN, SE and MA. All authors reviewed and approved the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Funding
Partial financial support for this study was provided to SE through a scholarship by the Higher Education Commission (HEC), Pakistan.
References
- 1.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80(6):827–41. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6(6):229–39. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Cancer Society. Breast Cancer Facts and Figures 2011–2012. Atlanta, Georgia: American Cancer Society, Inc; 2011. [Google Scholar]
- 4.Gilani GM, Kamal S, Akhter AS. A differential study of breast cancer patients in Punjab, Pakistan. J Pak Med Assoc. 2003;53(10):478–81. [PubMed] [Google Scholar]
- 5.Bhurgri Y, Pervez S, Kayani N, et al. Cancer profile of Larkana, Pakistan (2000–2002) Asian Pac J Cancer Prev. 2006;7(4):518–21. [PubMed] [Google Scholar]
- 6.Sohail S, Alam SN. Breast cancer in pakistan—awareness and early detection. J Coll Physicians Surg Pak. 2007;17(12):711–2. [PubMed] [Google Scholar]
- 7.Junaidi I. Breast cancer claims 40,000 lives a year in Pakistan. Dawn. Jan 10, 2012. Available at: http://dawn.com/2012/2001/2010/breast-cancer-claims-400000-lives-a-year-in-pakistan-400002/
- 8.Saleem S. Death from breast cancer more likely for Pakistani patients: US expert. The Express Tribune. 2011 Jan 22; Available at: ribune.com.pk/story/107232/death-from-breast-cancer-more-likely-for-pakistani-patients-us-expert/ [Google Scholar]
- 9.Junaid SK. 90,000 breast cancer cases annually in Pakistan. Daily Times. 2007. Available at: http://www.dailytimes.com.pk/default.asp?page=2007%2005C2010%2005C2028%2005Cstory_2028-2010-2007_pg2007_2043.
- 10.Mathelin C, Tomasetto C, Cromer A, Rio MC. Proteomics and breast cancer. Gynecol Obstet Fertil. 2006;34(12):1161–9. doi: 10.1016/j.gyobfe.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 12.Houssami N, Irwig L, Simpson JM, McKessar M, Blome S, Noakes J. The influence of knowledge of mammography findings on the accuracy of breast ultrasound in symptomatic women. Breast J. 2005;11(3):167–72. doi: 10.1111/j.1075-122X.2005.21643.x. [DOI] [PubMed] [Google Scholar]
- 13.Shin BK, Wang H, Hanash S. Proteomics approaches to uncover the repertoire of circulating biomarkers for breast cancer. J Mammary Gland Biol Neoplasia. 2002;7(4):407–13. doi: 10.1023/a:1024038132381. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48(8):1296–304. [PubMed] [Google Scholar]
- 15.Ross JS, Linette GP, Stec J, et al. Breast cancer biomarkers and molecular medicine. Expert Rev Mol Diagn. 2003;3(5):573–85. doi: 10.1586/14737159.3.5.573. [DOI] [PubMed] [Google Scholar]
- 16.Adourian A, Jennings E, Balasubramanian R, et al. Correlation network analysis for data integration and biomarker selection. Mol Biosyst. 2008;4(3):249–59. doi: 10.1039/b708489g. [DOI] [PubMed] [Google Scholar]
- 17.Magni F, Van Der Burgt YE, Chinello C, et al. Biomarkers discovery by peptide and protein profiling in biological fluids based on functionalized magnetic beads purification and mass spectrometry. Blood Transfus. 2010;8( Suppl 3):s92–7. doi: 10.2450/2010.015S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misek DE, Kim EH. Protein biomarkers for the early detection of breast cancer. Int J Proteomics. 2011;2011:343582. doi: 10.1155/2011/343582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pegram MD, Konecny G, Slamon DJ. The molecular and cellular biology of HER2/neu gene amplification/overexpression and the clinical development of herceptin (trastuzumab) therapy for breast cancer. Cancer Treat Res. 2000;103:57–75. doi: 10.1007/978-1-4757-3147-7_4. [DOI] [PubMed] [Google Scholar]
- 20.Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. J Clin Oncol. 2005;23(28):7212–20. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 21.Hopp TA, Weiss HL, Parra IS, Cui Y, Osborne CK, Fuqua SA. Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res. 2004;10(22):7490–9. doi: 10.1158/1078-0432.CCR-04-1114. [DOI] [PubMed] [Google Scholar]
- 22.Skliris GP, Munot K, Bell SM, et al. Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyl transferase inhibitors in a cell line model. J Pathol. 2003;201(2):213–20. doi: 10.1002/path.1436. [DOI] [PubMed] [Google Scholar]
- 23.Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. 2010;17(4):R245–62. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 24.Koff A, Giordano A, Desai D, et al. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257(5077):1689–94. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 25.Buckley MF, Sweeney KJ, Hamilton JA, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8(8):2127–33. [PubMed] [Google Scholar]
- 26.Anderson NG, Anderson NL. Twenty years of two-dimensional electrophoresis: past, present and future. Electrophoresis. 1996;17(3):443–53. doi: 10.1002/elps.1150170303. [DOI] [PubMed] [Google Scholar]
- 27.Cho WC. Research progress in SELDI-TOF MS and its clinical applications. Sheng Wu Gong Cheng Xue Bao. 2006;22(6):871–6. doi: 10.1016/S1872-2075(06)60061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colantonio DA, Chan DW. The clinical application of proteomics. Clinica chimica acta. 2005;357(2):151–8. doi: 10.1016/j.cccn.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 29.Fung ET, Gavin E. Analysis of mass spectrometry profiles of the serum proteome. Clin Chem. 2005;51(7):1309. doi: 10.1373/clinchem.2005.050799. author reply 1309. [DOI] [PubMed] [Google Scholar]
- 30.Molloy MP, Witzmann FA. Proteomics: technologies and applications. Brief Funct Genomic Proteomics. 2002;1(1):23–39. doi: 10.1093/bfgp/1.1.23. [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Zafar S, von Ahsen N, Oellerich M, et al. Proteomics approach to identify the interacting partners of cellular prion protein and characterization of Rab7a interaction in neuronal cells. J Proteome Res. 2011;10(7):3123–35. doi: 10.1021/pr2001989. [DOI] [PubMed] [Google Scholar]
- 34.Damodaran S, Wood TD, Nagarajan P, Rabin RA. Evaluating peptide mass fingerprinting-based protein identification. Genomics Proteomics Bioinformatics. 2007;5(3–4):152–7. doi: 10.1016/S1672-0229(08)60002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asif AR, Armstrong VW, Voland A, Wieland E, Oellerich M, Shipkova M. Proteins identified as targets of the acyl glucuronide metabolite of mycophenolic acid in kidney tissue from mycophenolate mofetil treated rats. Biochimie. 2007;89(3):393–402. doi: 10.1016/j.biochi.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Fung ET, Yip TT, Lomas L, et al. Classification of cancer types by measuring variants of host response proteins using SELDI serum assays. Int J Cancer. 2005;115(5):783–9. doi: 10.1002/ijc.20928. [DOI] [PubMed] [Google Scholar]
- 37.Kemik O, Sumer A, Kemik AS, et al. The relationship among acute-phase response proteins, cytokines and hormones in cachectic patients with colon cancer. World J Surg Oncol. 2010;8:85. doi: 10.1186/1477-7819-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Jiang J, Arellano M, et al. Quantification of Serum Proteins of Metastatic Oral Cancer Patients Using LC-MS/MS and iTRAQ Labeling. Open Proteomics J. 2008;1:72–8. doi: 10.2174/1875039700801010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyoumi S, Tamion F, Petit J, et al. Induction and modulation of acute-phase response by protein malnutrition in rats: comparative effect of systemic and localized inflammation on interleukin-6 and acute-phase protein synthesis. The J Nutr. 1998;128(2):166–74. doi: 10.1093/jn/128.2.166. [DOI] [PubMed] [Google Scholar]
- 40.Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochemical J. 1990;265(3):621–36. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tilg H, Dinarello CA, Mier JW. IL-6 and APPs: anti-inflammatory and immunosuppressive mediators. Immunol Today. 1997;18(9):428–32. doi: 10.1016/s0167-5699(97)01103-1. [DOI] [PubMed] [Google Scholar]
- 42.Botto M, Kirschfink M, Macor P, Pickering MC, Wurzner R, Tedesco F. Complement in human diseases: Lessons from complement deficiencies. Mol Immunol. 2009;46(14):2774–83. doi: 10.1016/j.molimm.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 43.Oksjoki R, Kovanen PT, Meri S, Pentikainen MO. Function and regulation of the complement system in cardiovascular diseases. Front Biosci. 2007;12:4696–708. doi: 10.2741/2419. [DOI] [PubMed] [Google Scholar]
- 44.Awadallah SM, Atoum MF. Haptoglobin polymorphism in breast cancer patients form Jordan. Clin Chim Acta. 2004;341(1–2):17–21. doi: 10.1016/j.cccn.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, Annamalai L, Guo C, et al. Circulating haptoglobin is an independent prognostic factor in the sera of patients with epithelial ovarian cancer. Neoplasia. 2007;9(1):1–7. doi: 10.1593/neo.06619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An JY, Fan ZM, Zhuang ZH, et al. Proteomic analysis of blood level of proteins before and after operation in patients with esophageal squamous cell carcinoma at high-incidence area in Henan Province. World J Gastroenterol. 2004;10(22):3365–8. doi: 10.3748/wjg.v10.i22.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunzendorfer U, Jung K, Ohlenschlager G. Transferrin, C3 complement, haptoglobin, plasminogen and alpha 2-microglobulin in patients with urogenital tumors. Eur Urol. 1980;6(4):232–6. doi: 10.1159/000473339. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell RJ, Carzino R, Janardhana V. Associations between the two serum proteins haptoglobin and transferrin and leukaemia. Hum Hered. 1988;38(3):144–50. doi: 10.1159/000153775. [DOI] [PubMed] [Google Scholar]
- 49.Beckman G, Eklund A, Frohlander N, Stjernberg N. Haptoglobin groups and lung cancer. Hum Hered. 1986;36(4):258–60. doi: 10.1159/000153638. [DOI] [PubMed] [Google Scholar]
- 50.Benkmann HG, Hanssen HP, Ovenbeck R, Goedde HW. Distribution of alpha-1-antitrypsin and haptoglobin phenotypes in bladder cancer patients. Hum Hered. 1987;37(5):290–3. doi: 10.1159/000153720. [DOI] [PubMed] [Google Scholar]
- 51.Parker CL, Sodetz JM. Role of the human C8 subunits in complement-mediated bacterial killing: evidence that C8 gamma is not essential. Molecular Immunol. 2002;39(7–8):453–8. doi: 10.1016/s0161-5890(02)00121-9. [DOI] [PubMed] [Google Scholar]
- 52.Power DM, Elias NP, Richardson SJ, Mendes J, Soares CM, Santos CR. Evolution of the thyroid hormone-binding protein, transthyretin. Gen Comp Endocrinol. 2000;119(3):241–55. doi: 10.1006/gcen.2000.7520. [DOI] [PubMed] [Google Scholar]
- 53.Wei S, Episkopou V, Piantedosi R, et al. Studies on the metabolism of retinol and retinol-binding protein in transthyretin-deficient mice produced by homologous recombination. J Biol Chem. 1995;270(2):866–70. doi: 10.1074/jbc.270.2.866. [DOI] [PubMed] [Google Scholar]
- 54.Imanishi T. Clinical and experimental studies on the profiles of serum proteins in acute hepatic injury. Gastroenterol Jpn. 1981;16(5):493–505. doi: 10.1007/BF02774521. [DOI] [PubMed] [Google Scholar]
- 55.Marten NW, Sladek FM, Straus DS. Effect of dietary protein restriction on liver transcription factors. Biochemical J. 1996;317(Pt 2):361–70. doi: 10.1042/bj3170361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O. Reference distributions for the negative acute-phase proteins, albumin, transferrin, and transthyretin: a comparison of a large cohort to the world’s literature. J Clin Lab Anal. 1999;13(6):280–6. doi: 10.1002/(SICI)1098-2825(1999)13:6<280::AID-JCLA5>3.0.CO;2-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z, Bast RC, Jr, Yu Y, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64(16):5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 58.Kozak KR, Su F, Whitelegge JP, Faull K, Reddy S, Farias-Eisner R. Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics. 2005;5(17):4589–96. doi: 10.1002/pmic.200500093. [DOI] [PubMed] [Google Scholar]
- 59.Schweigert FJ, Sehouli J. Transthyretin, a biomarker for nutritional status and ovarian cancer. Cancer research. 2005;65(3):1114. author reply 1114. [PubMed] [Google Scholar]
- 60.Liu L, Liu J, Dai S, et al. Reduced transthyretin expression in sera of lung cancer. Cancer science. 2007;98(10):1617–24. doi: 10.1111/j.1349-7006.2007.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu L, Wang J, Liu B, et al. Serum levels of variants of transthyretin down-regulation in cholangiocarcinoma. J Cell Biochem. 2008;104(3):745–55. doi: 10.1002/jcb.21661. [DOI] [PubMed] [Google Scholar]
- 62.Wang D, Liang H, Mao X, Liu W, Li M, Qiu S. Changes of transthyretin and clusterin after androgen ablation therapy and correlation with prostate cancer malignancy. Transl Oncol. 2012;5(2):124–32. doi: 10.1593/tlo.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Escher N, Kaatz M, Melle C, et al. Posttranslational modifications of transthyretin are serum markers in patients with mycosis fungoides. Neoplasia. 2007;9(3):254–9. doi: 10.1593/neo.06805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woong-Shick A, Sung-Pil P, Su-Mi B, et al. Identification of hemoglobin-alpha and -beta subunits as potential serum biomarkers for the diagnosis and prognosis of ovarian cancer. Cancer Sci. 2005;96(3):197–201. doi: 10.1111/j.1349-7006.2005.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cunnane G, Grehan S, Geoghegan S, et al. Serum amyloid A in the assessment of early inflammatory arthritis. J Rheumatol. 2000;27(1):58–63. [PubMed] [Google Scholar]
- 66.Raynes JG, Cooper EH. Comparison of serum amyloid A protein and C-reactive protein concentrations in cancer and non-malignant disease. J Clin Pathol. 1983;36(7):798–803. doi: 10.1136/jcp.36.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moshkovskii SA. Why do cancer cells produce serum amyloid a acute-phase protein? Biochemistry (Mosc) 2012;77(4):339–41. doi: 10.1134/S0006297912040037. [DOI] [PubMed] [Google Scholar]
- 68.Soreide JA, Lea OA, Kvinnsland S. Cytosol albumin content in operable breast cancer. Correlations to steroid hormone receptors, other prognostic factors and prognosis. Acta Oncol. 1991;30(7):797–802. doi: 10.3109/02841869109091823. [DOI] [PubMed] [Google Scholar]
- 69.Abdullah-Soheimi SS, Lim BK, Hashim OH, Shuib AS. Patients with ovarian carcinoma excrete different altered levels of urine CD59, kininogen-1 and fragments of inter-alpha-trypsin inhibitor heavy chain H4 and albumin. Proteome Sci. 2010;8:58. doi: 10.1186/1477-5956-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saguchi K, Tobe T, Hashimoto K, et al. Cloning and characterization of cDNA for inter-alpha-trypsin inhibitor family heavy chain-related protein (IHRP), a novel human plasma glycoprotein. J Biochem. 1995;117(1):14–8. doi: 10.1093/oxfordjournals.jbchem.a124701. [DOI] [PubMed] [Google Scholar]
- 71.Huang L, Yoneda M, Kimata K. A serum-derived hyaluronan-associated protein (SHAP) is the heavy chain of the inter alpha-trypsin inhibitor. J Biol Chem. 1993;268(35):26725–30. [PubMed] [Google Scholar]
- 72.Stahler F, Roemer K. Mutant p53 can provoke apoptosis in p53-deficient Hep3B cells with delayed kinetics relative to wild-type p53. Oncogene. 1998;17(26):3507–12. doi: 10.1038/sj.onc.1202245. [DOI] [PubMed] [Google Scholar]
- 73.Mohamed E, Abdul-Rahman PS, Doustjalali SR, et al. Lectin-based electrophoretic analysis of the expression of the 35 kDa inter-alpha-trypsin inhibitor heavy chain H4 fragment in sera of patients with five different malignancies. Electrophoresis. 2008;29(12):2645–50. doi: 10.1002/elps.200700828. [DOI] [PubMed] [Google Scholar]
- 74.Verhaegen H, De Cock W, De Cree J, Verbruggen F. Increase of serum complement levels in cancer patients with progressing tumors. Cancer. 1976;38(4):1608–13. doi: 10.1002/1097-0142(197610)38:4<1608::aid-cncr2820380427>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 75.Anil S, Beena VT, Remani P, Mysore J, Vijayakumar T. Total hemolytic complement (CH50) and its fractions C3 and C4 in the sera of patients with localized juvenile periodontitis. Ann Dent. 1993;52(1):18–20. [PubMed] [Google Scholar]
- 76.Vijayakumar T, Ankathil R, Remani P, Beevi VM, Vijayan KK, Panicker CK. Total hemolytic complement (CH50) and its fractions (C3 and C4) in the sera of patients with carcinoma of the oral cavity, uterine cervix, and breast. J Clin Immunol. 1987;7(4):300–3. doi: 10.1007/BF00915551. [DOI] [PubMed] [Google Scholar]
- 77.Garantziotis S. Modulation of plasma complement by the initial dose of epirubicin/docetaxel therapy in breast cancer and its predictive value. Br J Cancer. 2011;104(3):542. doi: 10.1038/sj.bjc.6606068. author reply 543–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sethi AA, Amar M, Shamburek RD, Remaley AT. Apolipoprotein AI mimetic peptides: possible new agents for the treatment of atherosclerosis. Curr Opin Investig Drugs. 2007;8(3):201–12. [PubMed] [Google Scholar]
- 79.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7(5):365–75. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 80.Florentin M, Liberopoulos EN, Wierzbicki AS, Mikhailidis DP. Multiple actions of high-density lipoprotein. Curr Opin Cardiol. 2008;23(4):370–8. doi: 10.1097/HCO.0b013e3283043806. [DOI] [PubMed] [Google Scholar]
- 81.Tardif JC, Heinonen T, Noble S. High-density lipoprotein/apolipoprotein A-I infusion therapy. Current Atheroscler Rep. 2009;11(1):58–63. doi: 10.1007/s11883-009-0009-7. [DOI] [PubMed] [Google Scholar]
- 82.Hamed EA, Zakhary MM, Maximous DW. Apoptosis, angiogenesis, inflammation, and oxidative stress: basic interactions in patients with early and metastatic breast cancer. J Cancer Res Clin Oncol. 2012;138(6):999–1009. doi: 10.1007/s00432-012-1176-4. [DOI] [PubMed] [Google Scholar]
- 83.Cai F, Dupertuis YM, Pichard C. Role of polyunsaturated fatty acids and lipid peroxidation on colorectal cancer risk and treatments. Curr Opin Clin Nutr Metab Care. 2012;15(2):99–106. doi: 10.1097/MCO.0b013e32834feab4. [DOI] [PubMed] [Google Scholar]
- 84.Panis C, Victorino VJ, Herrera AC, et al. Differential oxidative status and immune characterization of the early and advanced stages of human breast cancer. Breast Cancer Res Treat. 2012;133(3):881–8. doi: 10.1007/s10549-011-1851-1. [DOI] [PubMed] [Google Scholar]
- 85.Su F, Lang J, Kumar A, et al. Validation of candidate serum ovarian cancer biomarkers for early detection. Biomarker insights. 2007;2:369–75. [PMC free article] [PubMed] [Google Scholar]
- 86.Su F, Kozak KR, Imaizumi S, et al. Apolipoprotein A-I (apoA-I) and apoA-I mimetic peptides inhibit tumor development in a mouse model of ovarian cancer. Proc Natl Acad Sci U S A. 2010;107(46):19997–20002. doi: 10.1073/pnas.1009010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu Y, Fang XJ, Casey G, Mills GB. Lysophospholipids activate ovarian and breast cancer cells. Biochem J. 1995;309(Pt 3):933–40. doi: 10.1042/bj3090933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kitayama J, Shida D, Sako A, et al. Over-expression of lysophosphatidic acid receptor-2 in human invasive ductal carcinoma. Breast Cancer Res. 2004;6(6):R640–6. doi: 10.1186/bcr935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lane DM, Boatman KK, McConathy WJ. Serum lipids and apolipoproteins in women with breast masses. Breast Cancer Res Treat. 1995;34(2):161–9. doi: 10.1007/BF00665788. [DOI] [PubMed] [Google Scholar]
- 90.Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 91.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19(2):209–16. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin k C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18(9):2695–703. doi: 10.1158/1078-0432.CCR-11-2210. [DOI] [PubMed] [Google Scholar]
- 93.Whiteside TL, Ferrone S. For breast cancer prognosis, immunoglobulin kappa chain surfaces to the top. Clin Cancer Res. 2012;18(9):2417–9. doi: 10.1158/1078-0432.CCR-12-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li M, Feng DY, Ren W, et al. Expression of immunoglobulin kappa light chain constant region in abnormal human cervical epithelial cells. Int J Biochem Biol. 2004;36(11):2250–7. doi: 10.1016/j.biocel.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 95.Hartmann LC, Sellers TA, Frost MH, et al. Benign breast disease and the risk of breast cancer. N Engl J Med. 2005;353(3):229–37. doi: 10.1056/NEJMoa044383. [DOI] [PubMed] [Google Scholar]
- 96.Lasztity N, Biro L, Nemeth E, Pap A, Antal M. Protein status in pancreatitis—transthyretin is a sensitive biomarker of malnutrition in acute and chronic pancreatitis. Clin Chem Lab Med. 2002;40(12):1320–4. doi: 10.1515/CCLM.2002.227. [DOI] [PubMed] [Google Scholar]
- 97.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22(2):231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 98.Bui JD, Schreiber RD. Cancer immunosurveillance, immunoediting and inflammation: independent or interdependent processes? Curr Opin Immunol. 2007;19(2):203–8. doi: 10.1016/j.coi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 99.Wang HY, Wang RF. Regulatory T cells and cancer. Curr Opin Immunol. 2007;19(2):217–23. doi: 10.1016/j.coi.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 100.Mitchell DA, Fecci PE, Sampson JH. Immunotherapy of malignant brain tumors. Immunol Rev. 2008;222:70–100. doi: 10.1111/j.1600-065X.2008.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.