Abstract
Psoriasis is a common skin disorder of multifactorial origin. Genomewide scans for disease susceptibility have repeatedly demonstrated the existence of a major locus, PSORS1 (psoriasis susceptibility 1), contained within the major histocompatibility complex (MHC), on chromosome 6p21. Subsequent refinement studies have highlighted linkage disequilibrium (LD) with psoriasis, along a 150-kb segment that includes at least three candidate genes (encoding human leukocyte antigen–C [HLA-C], α-helix–coiled-coil–rod homologue, and corneodesmosin), each of which has been shown to harbor disease-associated alleles. However, the boundaries of the minimal PSORS1 region remain poorly defined. Moreover, interpretations of allelic association with psoriasis are compounded by limited insight of LD conservation within MHC class I interval. To address these issues, we have pursued a high-resolution genetic characterization of the PSORS1 locus. We resequenced genomic segments along a 220-kb region at chromosome 6p21 and identified a total of 119 high-frequency SNPs. Using 59 SNPs (18 coding and 41 noncoding SNPs) whose position was representative of the overall marker distribution, we genotyped a data set of 171 independently ascertained parent–affected offspring trios. Family-based association analysis of this cohort highlighted two SNPs (n.7 and n.9) respectively lying 7 and 4 kb proximal to HLA-C. These markers generated highly significant evidence of disease association (P<10-9), several orders of magnitude greater than the observed significance displayed by any other SNP that has previously been associated with disease susceptibility. This observation was replicated in a Gujarati Indian case/control data set. Haplotype-based analysis detected overtransmission of a cluster of chromosomes, which probably originated by ancestral mutation of a common disease-bearing haplotype. The only markers exclusive to the overtransmitted chromosomes are SNPs n.7 and n.9, which define a 10-kb PSORS1 core risk haplotype. These data demonstrate the power of SNP haplotype-based association analyses and provide high-resolution dissection of genetic variation across the PSORS1 interval, the major susceptibility locus for psoriasis.
Introduction
Psoriasis (MIM *177900) is a common, inflammatory skin disorder of unknown etiology (Camp 1998). The most prevalent form of the disease, chronic plaque psoriasis, is characterized by red scaly lesions that appear predominantly on extensor surfaces and are due to keratinocyte hyperproliferation, inflammatory-cell dermal infiltration, and new vessel formation (Barker 1991; Bos and De Rie 1999). Although rarely fatal, the disease is debilitating through its significant impact on quality of life (Nevitt and Hutchinson 1996). Approximately 3% of the population in developed countries have psoriasis, with health care costs exceeding $3 billion per year in the United States alone (Sander et al. 1993).
Although environmental agents, including infection and drug exposure, are risk factors in the development of psoriasis, twin and family studies point to a strong genetic component. The recurrence risk of psoriasis in first-degree relatives of affected subjects is ∼10 times greater than that seen in the general population (Lomholt 1963; Hellgren 1967). Associations with psoriasis—in particular, between psoriasis and the HLA-Cw6 antigen—have been reported in a wide range of ethnic groups, supporting the presence of a susceptibility gene or genes within the major histocompatibility complex (MHC) (Tiilikainen et al. 1980). More recently, linkage-based genomewide scans have identified chromosomal regions outside the MHC that harbor putative psoriasis-susceptibility loci (for review, see Elder et al. 2001; Capon et al. 2002). However, the majority of these studies generated highly significant evidence of linkage on chromosome 6p21 (PSORS1), supporting its role as the major susceptibility locus for psoriasis (Nair et al. 1997; Trembath et al. 1997; Lee et al. 2000; Veal et al. 2001). As an initial attempt to refine the PSORS1 genomic interval, microsatellite maps of varying density have been used as a framework for linkage disequilibrium (LD)–based fine mapping (Balendran et al. 1999; Oka et al. 1999; Nair et al. 2000). Taken together, these studies support a 150-kb minimal LD region, within the proximal segment of the MHC class I region, as the most likely location for PSORS1 (for review, see Capon et al. 2002). This interval contains five known genes, three predicted transcripts, and a number of ESTs (Oka et al. 1999). PSORS1 positional candidate genes have also been analyzed, and significant disease associations have been identified for the genes encoding human leukocyte antigen–C (HLA-C [MIM *142840]), octamer-binding transcription factor–3 (OTF3 [MIM *164177]), α-helix–coiled-coil–rod homologue (HCR [MIM *605310]), and corneodesmosin (CDSN [MIM *602593]) (Allen et al. 1999; Mallon et al. 1999; Tazi Ahnini et al. 1999; Asumalahti et al. 2000, 2002; Gonzalez et al. 2000). In particular, significant associations have been detected for nonconservative coding polymorphisms within HCR (e.g., HCR269; see Asumalahti et al. 2000) and CDSN (e.g., CDSN1243; see Allen et al. 1999). However, data interpretations are compounded by a lack of detail on the background pattern of LD both across the region and between the associated alleles (Jenisch et al. 1999; Chia et al. 2001; O’Brien et al. 2001). The stochastic nature of LD conservation is a major confounding factor in the design of disease-association studies, and the use of dense marker maps has been proposed as a possible means to improve resolution (Weiss and Terwilliger 2000; Cardon and Bell 2001).
SNPs—inherited, biallelic, 1-bp differences that are present in the human genome at a density of 1–10 per 1,000 nt—can significantly improve resolution in genetic studies (International SNP Map Working Group 2001). SNPs are more abundant throughout the genome and are less prone to mutation, as compared to STRs (i.e., microsatellites) (International SNP Map Working Group 2001).
In the present study, we have generated a high-density SNP map of a large genomic segment that includes the 150-kb minimal PSORS1 region but extends to 35 kb on either side of the interval boundaries. We have used the reference sequence developed by the MHC Sequencing Consortium (1999), and we have undertaken systematic SNP discovery by direct sequencing of eight affected individuals carrying PSORS1 risk haplotypes. By genotyping 59 SNPs, we have determined the allelic associations and fine structure of the haplotypes in the region. Family-based association studies and haplotype analysis highlight a 10-kb genomic segment, defined by two SNPs that lie 7 and 4 kb proximal to HLA-C, as contributing to psoriasis susceptibility at the PSORS1 locus.
Subjects and Methods
Subjects
A total of 171 previously described family-based trios of European origin (Asumalahti et al. 2002), each including an affected offspring and both parents, were ascertained, through information obtained from specialist dermatology clinics, at St John’s Institute of Dermatology, London, and from a network of collaborating sites throughout England and Scotland (Balendran et al. 1999). One hundred forty-nine parent-offspring units were derived from a family data set previously analyzed by Veal et al. (2001), and a further 22 trios, with and without a family history of psoriasis, were recruited for the purpose of this study. All probands had severe and extensive chronic plaque psoriasis, as assessed by two experienced dermatologists (J.N.W.N.B. and D.B.), using established clinical criteria (Camp 1998). A further cohort of case (n=77) and control (n=77) individuals of Gujarati Indian descent were ascertained as described elsewhere (Asumalahti et al. 2002). All subjects were genotyped for HLA-C. Analysis of LD conservation was determined for an unaffected control population of 90 unrelated U.K.-based subjects of European origin. For sequence analysis, we included eight psoriatic individuals with known microsatellite MHC haplotypes from a cohort described by Balendran et al. (1999). The ascertainment of subjects and the study were undertaken after receipt of approval from the Guy’s and St Thomas’ hospitals ethics committee of King's College, London.
SNP Identification
DNA from eight individuals who were known to be heterozygous for microsatellite-defined, high-risk PSORS1 haplotypes, was used to identify sequence variants in the 220-kb target region (Balendran et al. 1999). To recognize SNPs and thereby assess patterns of LD conservation, we also resequenced additional genomic fragments across a 700-kb area that was delimited by the tumor necrosis factor–β (TNFβ) and DDR genes.
PCR primers that amplify 2–2.5-kb fragments were designed on the basis of the consensus MHC sequence, which has been deposited in GenBank. PCRs were performed in 96-well microtiter plates (Abgene) on MJ Research DNA Engine Tetrads with a 50-μl reaction volume that contained 50 ng DNA, 45 mM Tris HCl (pH 8.8), 11 mM NH4SO4, 4.5 mM MgCl2, 6.7 mM β-mercaptoethanol, 4.4 μM EDTA (pH 8.0), 1 mM deoxyribonucleoside triphosphates, 113 μg/ml BSA, 10 pmol each primer, and 1 U Taq polymerase (Jeffreys et al. 1988). Amplified products were purified with the MultiScreen PCR system (Millipore), were sequenced using Big Dye Terminators (Applied Biosystems), and were electrophoresed through 5% LongRanger (FMC) polyacrylamide gels on an ABI 377 automated sequencer (Applied Biosystems). Sequence runs were aligned using ABI SeqEd 1.03, and SNPs were identified by visual inspection of chromatograms. Only SNPs presenting a minor-allele frequency >0.25 were genotyped. (For a detailed description of the entire SNP and primer resource, including position and nucleotide substitution, see tables A1–A7, in appendix AA1–A7, in appendix A, available online only.)
SNP Genotyping
Genomic DNA was extracted from whole blood as described elsewhere (Trembath et al. 1997). DNAs were plated out in family order in deep-well microtiter plates (Abgene), each of which included control samples from the eight individuals analyzed by resequencing. Plated DNA samples were amplified using sequence-analysis primers, and resultant products were blotted onto Hybond N (Amersham).
For each SNP, a pair of allele-specific oligonucleotides (ASOs) was synthesized. All probes were 18-mers, with the SNP located 8 nt from the 5′ end. ASOs were end-labeled to probe dot blots of the corresponding PCR products, as described by Jeffreys et al. (2000). In brief, 3 ng ASOs/ml were hybridized to blots at 53°C for 1 h, in a buffer that contained 3 M tetramethylammonium chloride (TMAC), 0.6% SDS, 10 mM sodium phosphate (pH 6.8), 1 mM EDTA, 0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% BSA, 4 μg yeast RNA/ml, and 10 μg single-stranded herring sperm DNA/ml. Filters were washed in 3 M TMAC, 0.6% SDS, 10 mM sodium phosphate (pH 6.8), and 1 mM EDTA at 56°C for 20 min; were rinsed in 3 × saline sodium citrate at room temperature; and were autoradiographed. Alleles were assigned by direct examination of autoradiographs compared for each allele, and Genetics Analysis Suite 2.0 (Alan Young, Oxford 1993–1995) was used to confirm Mendelian inheritance and to generate files for statistical analyses.
Statistical Analyses
Family-based association analysis was performed using TRANSMIT 2.5 (Clayton 1999). This package tests for association through the examination of transmission rates for markers and multilocus haplotypes, under “phase known” and “phase unknown” conditions. The tests are based on a score vector, which is averaged over all possible configurations of parental haplotypes and transmissions, consistent with the observed data (Clayton 1999). Excessive transmission of each haplotype is assessed using an asymptomatic χ2 test with 1 df. All markers were analyzed independently. To compensate for differences, reflecting allele frequency, in marker informativity, we also performed a three-locus haplotype analysis. The significance of this data set was assessed by simulation (with a program written, by C.D.V., in Visual Basic 6.0 [Microsoft]), generating 10,000 replicates of the data set with normal Mendelian inheritance from the parental genotypes. Transmission was assessed for each marker per replicate, and the number of times that particular χ2 values occurred was noted and compared to expected frequency.
Full-length haplotypes and frequencies were also computed using TRANSMIT. Because of computational limitations, overlapping segments of 10–15 SNPs were first determined. These were then pieced together, in succession, by the identification of unique SNPs in overlapping regions, until full-length haplotypes were generated. A subselection of markers that defined the haplotypes with a frequency >1% were then analyzed with PHASE 1.0 (Stephens et al. 2001), to identify and confirm haplotypes for each individual. This approach uses coalescent theory to predict haplotypes in population data.
Analysis of LD conservation among controls was performed using ad hoc software written, by Alec Jeffreys, in True Basic 4.1. The program estimates maximum-likelihood haplotype frequencies from the unphased diploid genotype, assuming that all markers are in Hardy-Weinberg equilibrium. On the basis of these haplotype frequencies, the level of LD between each pair of SNPs is assessed using the D′ measure of complete association (Jeffreys et al. 2001). LD conservation was also calculated in the untransmitted chromosomes, as found by PHASE and representing nondisease or control chromosomes, using D′ values calculated by a simple program written, by C.D.V., Visual Basic 6.0 (Microsoft).
Results
Resequencing and SNP Identification within the PSORS1 Interval
A high-density SNP map was generated by the resequencing of 83 PCR fragments (fig. 1) in each of eight affected individuals who carried a high-risk PSORS1 haplotype, as described by Balendran et al. (1999). Sequence analysis identified 119 SNPs, with a minor-allele frequency greater than the 25% arbitrary threshold value. Of these, a subset of 59 SNPs (fig. 1)—including all coding SNPs reported to show significant association with psoriasis (see the “Introduction” section)—yielded robust genotypes in >80% of the data set and provided representative distribution for all resequenced segments.
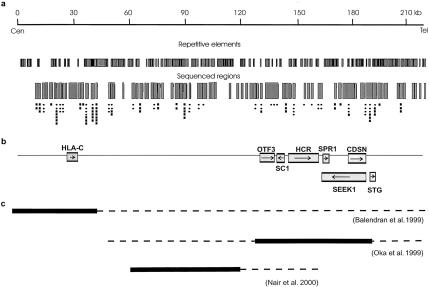
Figure 1.
The target region, spanning 220 kb in the MHC class I interval. a, Position of repetitive elements and sequenced areas, plotted against distance (upper line). The positions of SNPs identified by resequencing are represented by circles (for SNPs genotyped in the cohort with psoriasis) and squares (for nontyped SNPs). b, Positions of known genes. Arrows indicate the direction of transcription. c, Outcome of PSORS1 refinement studies. Black boxes indicate published minimal intervals. Dotted lines have been added to illustrate how much farther these regions may extend according to more-conservative interpretations of data.
LD-Conservation Patterns
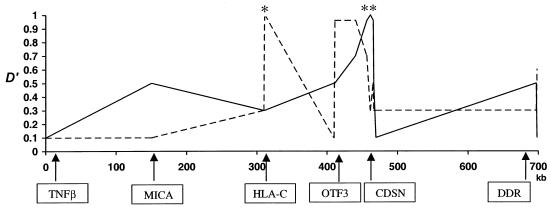
To reduce potential confounding effects from ascertainment bias, we investigated LD-conservation patterns in a sample (n=90) of healthy, unrelated individuals. Figure 2 shows the results from the analysis of diploid LD between 15 SNP loci. LD appears to be maintained between HLA-C and OTF3 (D′>0.8) but sharply decreases in the region between OTF3 and CDSN. We observed no evidence for LD between HLA-C and MICA (D′<0.2).
Figure 2.
Patterns of LD conservation, as measured by analysis of 15 SNPs spanning 700 kb, at the boundary between MHC class I and class III loci. D′ decay from HLA-C SNP 290 (dashed line) and from CDSN1243 (continuous line) is plotted against distance and location of analyzed genes. The asterisk (*) and double asterisk (**) symbols denote the positions of HLA-C SNP 290 and of CDSN1243, respectively.
This pattern of LD was also observed in the family cohort with psoriasis, by using the population of parental untransmitted chromosomes, with a D′ value of 0.8 between SNPs n.15 (6 kb distal from HLA-C) and n.43 (within HCR). D′ was 0.15 for LD between SNPs n.15 and n.58 (within CDSN).
Association Studies
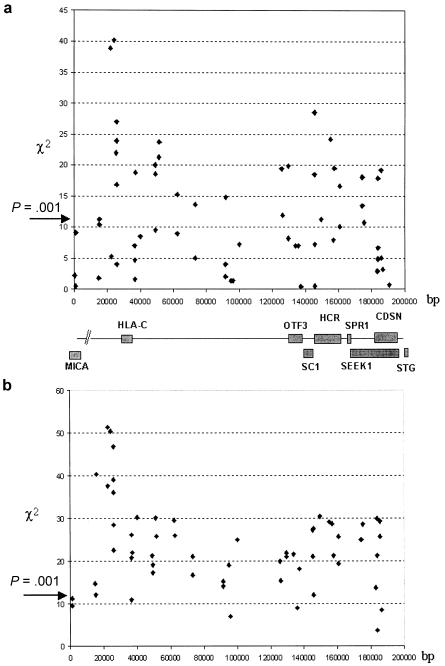
Figure 3a illustrates the results of single-marker transmission/disequilibrium testing for the 59 SNPs genotyped in the 171 parent–affected offspring trios. Significant association with psoriasis was observed throughout the resequenced regions, with a total of 26 SNPs generating χ2 values >10.8 (P<.001). Two SNPs—n.7 and n.9, respectively lying 7 and 4 kb proximal to HLA-C—yielded P values <10−9, exceeding by several orders of magnitude the significance displayed by any other marker in the data set. For SNPs distal to n.7 and n.9, >95% of loci demonstrated a significant decrease in χ2 values (P>10-6).
Figure 3.
Output of TRANSMIT family-based association analysis undertaken using all 171 parent–affected offspring trios. a, Results of single-marker analysis. χ2 values at each marker are plotted against distance and position of known genes. b, Results of three-marker analysis. χ2 values for consecutive haplotypes are plotted against distance and position of known genes.
Three-SNP haplotype analysis was also performed (fig. 3b). The pattern of association is maintained, with the haplotypes that contained SNPs n.7 and n.9 generating extreme P values (P=7.93×10-13). In simulations of 590,000 genotypes (10,000 replicates for each of the 59 SNP-based genotypes), no similar χ2 values were observed. To further characterize the pattern of haplotype association centromeric to SNPs n.7 and n.9, we genotyped three additional coding sequence variants that are found in MICA, the next non-HLA gene beyond HLA-C. This revealed a sharp decrease in χ2 values, indicating that the peak of association is neither increased nor maintained beyond SNPs n.7 and n.9. Assessment of a further cohort of case and control individuals of distinct ethnic origin also yielded evidence that SNPs n.7 and n.9 exhibit greater association than SNPs n.43 (HCR269) and n.58 (CDSN1243) (see table 1).
Table 1.
Analysis of Associated in SNPs in the Gujarati Case/Control Population
|
No. (Frequency) among |
|||||
| Case Individuals |
Control Individuals |
||||
| SNP | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Pa |
| n.7 | 94 (60%) | 62 (40%) | 92 (75%) | 30 (25%) | .01 |
| n.9 | 98 (60%) | 66 (40%) | 100 (75%) | 34 (25%) | .01 |
| HCR269 (n.43) | 110 (77%) | 32 (23%) | 132 (86%) | 22 (14%) | .09 |
| CDSN1243 (n.58) | 69 (48%) | 73 (52%) | 76 (44%) | 78 (56%) | .1 |
By χ2 test.
PSORS1 Haplotype Analysis
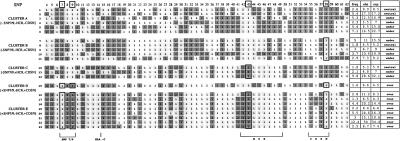
An extended haplotype analysis for the entire PSORS1 region is shown in figure 4. A total of 23 haplotypes, each with a frequency >1%, were observed. Nine haplotypes (haplotypes 15–23) were overtransmitted to affected offspring. All overtransmitted haplotypes exclusively included SNPs n.7 and n.9. Only clusters D and E were more frequent in the transmitted chromosomes—in the case of cluster E, representing a fourfold excess (45% in all transmitted chromosomes; 11% in all untransmitted chromosomes). We compared the presence of SNPs n.7 and n.9 with the previously genotyped presence of HLA-Cw6, in the affected offspring. Of the 120 subjects genotyped for each locus, 93 (77.5%) possessed each of the risk alleles, 6 (5%) carried only SNPs n.7 and n.9, and 1 (0.83%) carried HLA-Cw6 alone; 20 (16.7%) of the affected probands harbored other alleles at these loci.
Figure 4.
PSORS1 SNP haplotypes. Haplotypes are clustered on the basis of the presence of disease-associated alleles (indicated by a + or − sign before the locus name) at SNPs n.7 and n.9; at HCR269 (n.43) (Asumalahti et al. 2000), renamed as “HCR305” (Asumalahti et al. 2002); and at CDSN1243 (n.58) (Allen et al. 1999). The last four columns report haplotype frequencies (freq), observed (obs) versus expected (exp) transmissions, and haplotype status (neutral, overtransmitted [over], or undertransmitted [under]).
Discussion
We have developed an SNP-based map of the PSORS1 susceptibility locus, with the specific aim of increasing the resolution of mapping data for the major genetic discriminant for psoriasis. Interestingly, recent studies have demonstrated that most genomic regions can be decomposed into discrete blocks of low haplotype diversity that are separated by intervals corresponding to historical recombination breakpoints. Crossover events appear to be clustered between blocks, with little or no exchange within blocks (Daly et al. 2001; Jeffreys et al. 2001). Hence, reliable fine mapping of complex traits can be achieved by genotyping a limited number of SNPs within a critical interval, provided that such markers are representative of the region's composition of LD blocks (Daly et al. 2001; Rioux et al. 2001). Thus, in this study, we did not seek to identify or genotype all SNPs mapping to the target region; rather, we resequenced segments (total length 60 kb) of the PSORS1 interval, to generate an essential framework SNP map.
The most significant evidence for disease association was observed for SNPs n.7 and n.9, with probability values clearly exceeding any other markers studied, including coding SNPs that identified variants that have previously been implicated in psoriasis susceptibility (Allen et al. 1999; Asumalahti et al. 2000). By way of replication, these observations were supported in a case/control study of an Indian cohort with psoriasis. SNPs n.7 and n.9 lie in a noncoding region, respectively 7 and 4 kb centromeric to HLA-C. Although it is possible that these SNPs are themselves disease-causing variants, on the basis of the available data, we consider this to be unlikely. In a three-marker association analysis at these loci, we observed higher χ2 values, compared to those generated for a single-marker assessment. Such an incremental change in significance is likely to indicate that the strength of association between these SNPs and disease is influenced substantially by marker informativity. We might expect this to be the case for any SNP other than the disease-causing allele, whose informativity is, by definition, absolute (Martin et al. 2000). An alternative explanation would require that SNPs n.7 and n.9 each be independent susceptibility alleles, yet such a hypothesis would require both mutations to have occurred on the same haplotype background. Hence, we suggest that the disease-causing allele is in tight LD with SNPs n.7 and n.9 but that the resolution achieved by the present study should substantially facilitate the identification of these causal variants. The distribution of evidence for association at other loci across the interval is of some interest. In addition to SNPs n.7 and n.9, a further peak—extending from OTF3 to CDSN and being maximal (in single-marker analysis) for SNP n.43, located within HCR—is defined by three-marker analysis. This corresponds to the recently defined associated HCR haplotype HCR*WW (Asumalahti et al. 2002).
Since we genotyped only three PSORS1 SNPs proximal to SNP n.7, we sought to validate the centromeric boundary of the critical interval by genotyping three SNPs within MICA, the closest non-HLA gene. Haplotype analysis demonstrated that the three MICA SNPs are in LD with each other and freely associate with PSORS1 (data not shown). This is in agreement with the results generated by our survey of LD conservation in control individuals, showing D′ values <0.2 between MICA and HLA-C. Altogether, our data support the occurrence of recombination between these two genes. Moreover, our data define MICA as a conservative boundary for the critical region defined by the present study. This broader interval (MICA lies 150 kb distal to SNP n.6) contains only one additional functional gene—namely, the gene encoding the MHC class I antigen HLA-B. Associations between psoriasis and HLA-B have been repeatedly reported—in particular, HLA-B57 and HLA-B13 alleles have been implicated in disease susceptibility (Elder et al. 1994; Balendran et al. 1999). However, both HLA-B57 and HLA-B13 lie on ancestral haplotypes bearing HLA-Cw*0602 (Elder et al. 1994; Balendran et al. 1999), suggesting that HLA-B associations reflect LD with HLA-Cw*0602. Finally, the results of previous PSORS1 fine-mapping studies offered very little support for the inclusion of HLA-B within the critical region (Oka et al. 1999; Nair et al. 2000).
Analysis of extended haplotypes (including all 59 SNPs genotyped in the cohort that we studied) demonstrated that the associated alleles at SNPs n.7 and n.9 are unique to the overtransmitted chromosomes and define a 10-kb genomic segment (delimited by SNPs n.6 and n.10) as a core risk haplotype. Database analysis of this restricted region identified a number of repeat motifs, together with a pseudogene (not expressed) for the ubiqitin-specific protease–8 gene. All other associated alleles, including those within HCR and CDSN, are found to be present on clusters (A–C) that do not exhibit any evidence of overtransmission. Previous studies have suggested that disease associations seen with these genes, both located in the telomeric end of the MHC class I region, are due to LD with HLA-Cw6 (Jenisch et al. 1999; Chia et al. 2001; O’Brien et al. 2001). To minimize ascertainment bias, we calculated pairwise LD between SNPs, by studying a control population and using the untransmitted chromosomes from parents within the trio cohort. D′ scores of 0.80–1.0, indicating strong LD, were observed for a block from HLA-C to HCR; however, these values markedly decreased between this block and CDSN (D′=0.15 between SNPs n.15 and n.58). These data indicate that recombination has occurred between the regions in the past and, as such, argue against the hypothesis that association between CDSN and psoriasis is due to LD with HLA-C. Haplotype analysis indicates that the overtransmitted haplotype in cluster D was probably generated by recombination between HLA-C and HCR. However, notably, the clustered haplotype retains the associated SNP, n.58 (CDSN1243). Finally, 5% of affected individuals that harbor SNPs n.7 and n.9 do not possess HLA-Cw6, yet these chromosomes include associated alleles that are within CDSN. We must assume that this risk chromosome has been derived after a further recombination event between HCR and CDSN.
Cumulatively, these observations provide genetic evidence that a region in close proximity to HLA-C is the PSORS1 susceptibility locus. Indeed, HLA-Cw*0602 has long been recognized as the marker that is most significantly associated with psoriasis and as the marker that confers the highest disease risk (Mallon et al. 1999; Asumalahti et al. 2000). Case/control studies have reported significant associations, with the Ala-73 and Asp-9 variant, that define an antigen-binding pocket unique to HLA-Cw6/Cw7 molecules (Roitberg-Tambur et al. 1994; Asahina et al. 1996), with HLA-Cw*0602 proving a more discriminatory indicator of disease susceptibility (Mallon et al. 1997). Data from the present study have identified haplotypes spanning HLA-C on at least one undertransmitted haplotype (namely, haplotype 3), leading us to consider that variants within the coding sequence of the gene encoding HLA-C are unlikely to be causal in the conferral of disease susceptibility. Rather, the disease allele may lie within a regulatory region, potentially influencing expression of HLA-C or other genes in this interval. It is plausible that structural differences in regulatory elements of genes within the MHC class I region may have locus- and tissue-specific effects on expression levels and on their resultant differential binding affinities for transcription factors. To date, the expression of HLA-C in normal lesional and nonlesional skin has been poorly characterized.
Altered activity of HLA-C represents a plausible biological pathway for psoriasis susceptibility, since it contributes to the process of self-recognition by the immune system (Cerundolo and Braud 1996). Interestingly, it has been suggested that psoriasis may be triggered by direct activation of CD8 and/or natural killer (NK) T cells, both of which bear receptors for MHC class I molecules (Bos and De Rie 1999). In particular, it has been proposed that activating receptors may trigger NK cells’ cytotoxicity and contribute to the damage of epidermal basal membrane. Conversely, inhibitory receptors, some of which are specific to HLA-C, would prevent the lysis of cells bearing self–MHC class I molecules (Nickoloff et al. 1999). Clearly, detailed functional analysis (including expression of HLA-C and response to key peptides) appear justified in light of high-resolution genetic studies that have been achieved through a combination of resequencing and SNP-based genotyping.
Acknowledgments
We wish to thank all the families with psoriasis, whose participation made this project possible. We would also like to thank Prof. Sir Alec Jeffreys for access to software for the analysis of diploid LD. This research was supported by Wellcome Trust grant 056713/Z/99/Z and a Medical Research Council U.K. Cooperative Group grant. F.C. is the recipient of a Wellcome Trust Travelling Research Fellowship.
Appendix A: Supplementary Data
Table A1.
Reference Sequence Scheme, Illustrating How to Trace the Position of the PSORS1 SNPs and Primers Described in the Following Tables
Table A2.
PCR Primers Used for SNP Identification[Note]
| Primer | Primer Sequence | Target Region |
| P1F1 | 5′-TCCCCAATGCTCCTTATCAC-3′ | |
| P1R1 | 5′-ATCCTGCCTTTTCACCTCTC-3′ | 9521–11856 |
| P1F2 | 5′-TTACAGACATGAGCCACTGC-3′ | |
| P1R2 | 5′-AAAAGGGGGAAGAGAAGGAG-3′ | 11123–13644 |
| P1F3 | 5′-TCCAAACCATTCTCACGACC-3′ | |
| P1R3 | 5′-TACCATCCTGTTTCCACTCC-3′ | 13374–15689 |
| P1F4 | 5′-CTTAGTTCTCACCTCTGCCC-3′ | |
| P1R4 | 5′-ATGACATTCCTCTCTCCACC-3′ | 15332–17929 |
| P1F5 | 5′-CCATTCCTCCTTTAGCAACC-3′ | |
| P1R5 | 5′-CCGCATGACACAAGTTTACC-3′ | 17568–19958 |
| P2F1 | 5′-ACAAAACACAGAACCCAACC-3′ | |
| P2R1 | 5′-TACCATCTCCCACCAGTCAG-3′ | 19823–22392 |
| P2F2 | 5′-GCTTCTTGCCATCCTCATCC-3′ | |
| P2R2 | 5′-CTCACCCGTCACAAAAAGTC-3′ | 21135–23664 |
| P2F3 | 5′-AAAAAAAAGGGGGGGGGGAG-3′ | |
| P2R3 | 5′-GCGGGGATTTTGGCCTAAAC-3′ | 27735–30423 |
| P2F4 | 5′-GAGGAAATGGAGGGGAAGAC-3′ | |
| P2R4 | 5′-GAATCAGGGAAGGGAGAGAG-3′ | 30949–33423 |
| P2F5 | 5′-CTCACCCACCCCATAATTCC-3′ | |
| P2R5 | 5′-TTCTCCAGCCCAGTCCTTTC-3′ | 32365–34730 |
| P2F6 | 5′-AGTGAGTAGAAGGAAGGGTG-3′ | |
| P2R6 | 5′-AGGGAATGGGTGGAAAGATG-3′ | 34545–37168 |
| P2F7 | 5′-ACATCTTTCCACCCATTCCC-3′ | |
| P2R7 | 5′-ATTCCCCTCTATTCCCTGCC-3′ | 37148–39683 |
| P3F1 | 5′-CAGGGAATAGAGGGGAATGG-3′ | |
| P3R1 | 5′-CAGATGGGGTTTTGGTGTAG-3′ | 39665–42177 |
| P3F2 | 5′-CCCTCTGAGACGAAACTTCC-3′ | |
| P3R2 | 5′-GTGTTTGCTCTTGCTTCTCC-3′ | 41947–44296 |
| P3F3 | 5′-CATCCTGATACCAAAGCCTG-3′ | |
| P3R3 | 5′-CTCCTAATGCTATCCCTCCC-3′ | 44847–47533 |
| P3F4 | 5′-GGCAGGAGAATGGCATGAAC-3′ | |
| P3R4 | 5′-ATTCCCCACATCACAGGCAC-3′ | 47161–49592 |
| P3F5 | 5′-GGAGAGAGTAAAGCCCACAG-3′ | |
| P3R5 | 5′-GAGCCAAAGCAGGAATGAAC-3′ | 48951–51606 |
| P3F6 | 5′-GTTGCTTTATTGCGTCACTG-3′ | |
| P3R6 | 5′-ACACATAATACCCACCACCC-3′ | 53927–56492 |
| P4F1 | 5′-AGAGTTTTGCCTGACTCCAC-3′ | |
| P4R1 | 5′-TTTCCTCCTTCCTTCCTTCC-3′ | 61221–63861 |
| P4F2 | 5′-ACAAGGAAGGAAGGAAGGAG-3′ | |
| P4R2 | 5′-TACAAGGATGAGGGAATGGC-3′ | 63838–66234 |
| P4F3 | 5′-AGCTTGTCTAAGTCCTGCTC-3′ | |
| P4R3 | 5′-GTAATCCCTGTTTCTCCCCC-3′ | 65408–67720 |
| P4F4 | 5′-TTACGAGACAGCCCATACAC-3′ | |
| P4R4 | 5′-ATTCTCTTGCCTCAGCATCC-3′ | 67717–70148 |
| P4F5 | 5′-TAATCCTGTTCACGCCTTTC-3′ | |
| P4R5 | 5′-TTCATTTCCCTCACTCCCTC-3′ | 68899–71532 |
| P4F6 | 5′-GGCAATGATTACGGAACCAC-3′ | |
| P4R6 | 5′-TTGGAGGGACAAACAGGGAG-3′ | 71480–74003 |
| P4F7 | 5′-AGGACAAGTGAACCAGCCAG-3′ | |
| P4R7 | 5′-TGAAAGGAGGCAGAGGGTAG-3′ | 72877–75464 |
| P4F8 | 5′-CACCTCAATCCACAATCCAC-3′ | |
| P4R8 | 5′-TTTCCCACTTACAGCCAGAC-3′ | 75045–77468 |
| P4F9 | 5′-TTGGCAACACCCTCACAGAC-3′ | |
| P4R9 | 5′-AGAACGCAATCTCGGCTCAC-3′ | 76968–79659 |
| P5F1 | 5′-GATCTCAGCTCACCATCAAC-3′ | |
| P5R1 | 5′-AAAAGAAGGGAGGGAGGAAG-3′ | 79963–82569 |
| P5F2 | 5′-TAAACCCATGAGCAGACACC-3′ | |
| P5R2 | 5′-CAAAACCCACCAAAACCAAG-3′ | 82296–84992 |
| P5F3 | 5′-GCCTTATTTTACTCAGCCCC-3′ | |
| P5R3 | 5′-TGATCCAGCAATCCTACCAC-3′ | 85121–87497 |
| P5F4 | 5′-ACCTTCATACTCCTTCCAGC-3′ | |
| P5R4 | 5′-ACAAATCTCTCCCTTTCCCC-3′ | 87529–90138 |
| P5F5 | 5′-AGAAAACAGCCATTGCTTCC-3′ | |
| P5R5 | 5′-TCCTATTGCCCCCCTAAAAC-3′ | 89704–92197 |
| P5F6 | 5′-ACCTGTAGTCCCAGCTACTC-3′ | |
| P5R6 | 5′-TAACACCTCTTCCCACCCAC-3′ | 93907–96508 |
| P5F7 | 5′-GAAGCAGCAAAAGTCCCAAG-3′ | |
| P5R7 | 5′-CAGTGAGCCAATATCGCACC-3′ | 95963–98440 |
| P5F8 | 5′-GGCAGGAAGGAAGGTTAGAG-3′ | |
| P5R8 | 5′-CGTGAGGATATGGCAAGAAG-3′ | 97454–99955 |
| P6F1 | 5′-CCCAACCTCTCAACCTCTTC-3′ | |
| P6R1 | 5′-TCTCTACTCACCTGTCTCCG-3′ | 100022–102520 |
| P6F2 | 5′-ATGAAAATACCCACCACCCC-3′ | |
| P6R2 | 5′-TCACTGCAACCTCAAACTCC-3′ | 100758–103367 |
| P6F3 | 5′-AGAGACTCCATCTCACACAC-3′ | |
| P6R3 | 5′-GACTACCTTCCACATCAGCC-3′ | 103025–105387 |
| P6F4 | 5′-GTGCTAAGTCCCTCATTGCC-3′ | |
| P6R4 | 5′-TGCTCACTGTAACCTCCACC-3′ | 104681–107084 |
| P6F5 | 5′-AATCACTGCCCTAAAGCAAC-3′ | |
| P6R5 | 5′-GATTACAGATGCCCACCACC-3′ | 106539–108899 |
| P6F6 | 5′-CAACACAGCAAGACTCCATC-3′ | |
| P6R6 | 5′-ATTTTCCTCTCTCAGCCTCC-3′ | 108202–110795 |
| P6F7 | 5′-GTGTCTCACGCCTGTAATCC-3′ | |
| P6R7 | 5′-TCCCCATCATTCTTTCAGCC-3′ | 110619–112985 |
| P6F8 | 5′-GCCAGATCACACCATTGCAC-3′ | |
| P6R8 | 5′-AAACACCACGCTCACACCAC-3′ | 111818–114503 |
| P6F9 | 5′-TCCCCACAAAACCATTGAAG-3′ | |
| P6R9 | 5′-TAAGTCCAGTCCGAGACAAG-3′ | 117007–119418 |
| P7F1 | 5′-TCACTTCATGCTATTGTGCC-3′ | |
| P7R1 | 5′-TGTTTCCTTCCCTTCTCTCC-3′ | 121898–124511 |
| P7F2 | 5′-GGACTCCCTTCTTAGCACAC-3′ | |
| P7R2 | 5′-CCCCAATTTCCCCACTCAAC-3′ | 124419–126981 |
| P7F3 | 5′-TCTCATTGAACACCACTCCC-3′ | |
| P7R3 | 5′-AATCCCCTCACACAGAATCC-3′ | 125680–128161 |
| P7F4 | 5′-TCAGATTTCGCCTTCTCGCC-3′ | |
| P7R4 | 5′-TCTCACCCTTTTTCTCCCCC-3′ | 129683–132349 |
| OTF3F1 | 5′-CTCCATAATCTCCTTCCCTCC-3′ | |
| OTF3R1 | 5′-CCCATCCCTACCTCAGTAAC-3′ | OTF3 |
| P7F5 | 5′-GGGGAGAAAAAGGGTGAGAG-3′ | |
| P7R5 | 5′-CAGGTGGTGGTGTGAAAAGG-3′ | 132331–134922 |
| P7F6 | 5′-ATCCTGCCTTTTCACACCAC-3′ | |
| P7R6 | 5′-AAATACCCCCCTTCCCTTCC-3′ | 134897–137515 |
| P7F7 | 5′-ATTTCCATCTCTCCCCTTCC-3′ | |
| P7R7 | 5′-GCAGCAGAATCACTTGAACC-3′ | SC1 exons 2–3 |
| SC1F1 | 5′-CTTTCTGATGATATCCCAGG -3′ | |
| SC1R1 | 5′-GTCGGCTTAACCAATTTTGC-3′ | SC1 exon 1 |
| P8F1 | 5′-GACCTAAACCCCTACTTCCC-3′ | |
| P8R1 | 5′-CCATCAGAATGAACCCACAC-3′ | 141457–143929 |
| P8F2 | 5′-TAAAAAGGGACGGGAGTAGG-3′ | |
| P8R2 | 5′-AGGGAGGAATGGGCATAGAG-3′ | 142428–145073 |
| P8F3 | 5′-GCTTACGACCTCTCTATGCC-3′ | |
| P8R3 | 5′-GACTTCACACGCCTCTTCAC-3′ | 145043–147362 |
| PG8EX1-2F | 5′-TTGTCTCTGCCTGGGAAACT-3′ | |
| PG8EX1-2R | 5′-TCATGGAGGGTCTTTTCTGC-3′ | HCR exons 1–2 |
| PG8EX2F | 5′-CAGAGGGGTCCTCTTTTCCT-3′ | |
| PG8EX2R | 5′-GAAGCTACTGCCCAGCTCTC-3′ | HCR exon 2 |
| PG8EX3-6F | 5′-GAGCTCAGGAGGTAGGCAGA-3′ | |
| PG8EX3-6R | 5′-GGGAATACCGGGAGAAAAGA-3′ | HCR exons 3–6 |
| PG8EX7-8F | 5′-CCCTGCATGGCATTCTTAC-3′ | |
| PG8EX7-8R | 5′-TAAATCTGCCCAGCCCTGTA-3′ | HCR exons 7 and 8 |
| PG8EX9-13F | 5′-AGGCATGGAGGATCAGTGAC-3′ | |
| PG8EX9-13R | 5′-TGAACACACTTTGAGGGGAAA-3′ | HCR exons 9–13 |
| PG8EX14F | 5′-TCTGGCCTGACTCACACC-3′ | |
| PG8EX14R | 5′-CCCTCTCTAGCTCCTGCAA-3′ | HCR exon 14 |
| PG8EX14-16F | 5′-AGGAACTGGAGGATGGTTCA-3′ | |
| PG8EX14-16R | 5′-TTCCCAAACATTTCCAAAGC-3′ | HCR exons 14–16 |
| P8F4 | 5′-TTTTCTCCCACTCCTTCTCC-3′ | |
| P8R4 | 5′-TGCCTGTAATCCAGCTACTC-3′ | 150070–152458 |
| P8F5 | 5′-TGTCTCTCCTACCAACTTCC-3′ | |
| P8R5 | 5′-TTTTCCTCATCCTCTCCACC-3′ | 152739–155170 |
| P8F6 | 5′-TACTCCCTGTCCCCACTTTC-3′ | |
| P8R6 | 5′-TCCCTCACCATCCTCTTTCC-3′ | 154737–157124 |
| MHC4F | 5′-GAACAGATGAGATCACGCC-3′ | |
| EMHC4R | 5′-GTGTTAGTTAAAGCAGC-3′ | SEEK1 exons 4–6, SPR |
| P9F1 | 5′-TCAAGCCATTCTCCTGCCTC-3′ | |
| P9R1 | 5′-ACCCCTCTTCATCCCTAACC-3′ | 163222–165594 |
| P9F2 | 5′-TTTCTCTCCCACATCCTCAC-3′ | |
| P9R2 | 5′-CATTCTCCTACCTCAGCCTC-3′ | 167242–169777 |
| P9F3 | 5′-AGGTCGGGGGAAAATTAGGG-3′ | |
| P9R3 | 5′-AAAGGAAAGGTGGGGATGGG-3′ | SEEK1 exon 3 |
| P9F4 | 5′-TGCCCACACAATCAACATTC-3′ | |
| P9R4 | 5′-ACCTTTCCTTCCTTCTTCCC-3′ | 171305–173809 |
| P9F5 | 5′-AGGGAAGAAGGAAGGAAAGG-3′ | |
| P9R5 | 5′-TGATGAGAGAAGCACAGAGC-3′ | SEEK1 exon 2 |
| P9F6 | 5′-AAATACACCTGAAGCACCAC-3′ | |
| P9R6 | 5′-CCCAAACCTCATCTCCTCAC-3′ | 176307–178627 |
| SEX1F | 5′-CCCACAGTGACTCCTGC-3′ | |
| SEX1R | 5′-AGATTCCAGAGCCCCTG-3′ | CDSN exon 1 |
| SEX2P1 | 5′-GTGAGGGAGGAAGCCAAG-3′ | |
| SEX2P2 | 5′-GGTTTAGTATTCCGCGGTAAG-3′ | CDSN exon 2 |
| SEX2P3 | 5′-CAGCTTTCAGTTCAGCAGC-3′ | |
| SEX2P4 | 5′-AAGGGACCCCTGGAGAG-3′ | CDSN exon 2 |
| SEX2P5 | 5′-GGGTAAAATCTATCCTGTGGG-3′ | |
| SEX2P6 | 5′-TTTCCAGCACTGCTGGAG-3′ | CDSN exon 2 |
| SEX2P7 | 5′-GCATGTCTGTCTCCTCCT-3′ | |
| SEX2P8 | 5′-TGGACCATTTCAACACAGT-3′ | CDSN exon 2 |
| P10F1 | 5′-CCCCACCCAAACTCCTTTTC-3′ | |
| P10R1 | 5′-TCCTGCCTTTACCCCTTTCC-3′ | SEEK1 exon 1, STG exon 1 |
| P10F2 | 5′-TAGGCATAGGGGAAGCAAAG-3′ | |
| P10R2 | 5′-AGATGATGAGAGAGGGGGAG-3′ | STG exon 2 |
| P10F3 | 5′-AACACCTCAACCTCCTCACC-3′ | |
| P10R3 | 5′-CAGTATGCCAGCAGTAGTCC-3′ | 190191–192610 |
| P10F4 | 5′-CAACAACAACAACAACACCC-3′ | |
| P10R4 | 5′-CTGTTCTCGAACTCCTGACC-3′ | 192483–194874 |
| P10F5 | 5′-CTTCCTCTGTCCCTCCTTTC-3′ | |
| P10R5 | 5′-CTCTTTACCCGTTGCTCGTC-3′ | 193933–196557 |
| P11F1 | 5′-CTGGGCGACAAGAGAGAAAC-3′ | |
| P11R1 | 5′-TTCCTACCACACACACCCAC-3′ | 200935–203479 |
| P11F2 | 5′-TCCTCCCCCCATCTTCTTTC-3′ | |
| P11R2 | 5′-ATTCTCGTGCCTCAGCTTTC-3′ | 203792–206355 |
| P11F3 | 5′-GCACGAGAATCACTTGAACC-3′ | |
| P11R3 | 5′-AAACATCTCAACCACACCAC-3′ | 206346–208803 |
| P11F4 | 5′-TCCCTGGATTAAGCCAGACC-3′ | |
| P11R4 | 5′-ACACACACACACACACACAC-3′ | 208639–211053 |
| P11F5 | 5′-CAGTGGCACAATCACAGCTC-3′ | |
| P11R5 | 5′-CATCAACCCCCCTCCTTTTC-3′ | 210465–213063 |
| P11F6 | 5′-AACTCCCCATTCTCCTTTCC-3′ | |
| P11R6 | 5′-GTGACTCACGCCTGTAATCC-3′ | 211539–214142 |
| P11F7 | 5′-GTTCAAGTGATTCTCCAGCC-3′ | |
| P11R7 | 5′-CCCTCCTCAATTCCATCTCC-3′ | 213946–216361 |
Note.— Primers in italic were also used for SNP identification in control individuals.
Table A3.
PCR Primers Used for SNP Identification in Controls Only
| Primer | Primer Sequence | Target Region |
| TNFAF | 5′-GTCTCGGTTTCTTCTCCATC-3′ | |
| TNFAR | 5′-TGGGTAGGAGAATGTCCAGG-3′ | TNFα promoter |
| TNFBF | 5′-GAGGCATAGGAGTGGGCTCC-3′ | |
| TNFBR | 5′-GCTGCTGCTGGTTCTGCTGC-3′ | TNFβ exons 2–3 |
| MICBF | 5′-GAGGAATAGGGTCAGGGAGG-3′ | |
| MICBR | 5′-TGGACCCTCTGCCGCTGATG-3′ | MICB exons 3–5 |
| MICAF | 5′-GGAAGGTTGGGACAGCAGAC-3′ | |
| MICAR | 5′-TCTTGGAGGTGAGGCAACTC-3′ | MICA exons 2–3 |
| HLACF1 | 5′-TTCTAAAGTCCCCAGTCACC-3′ | |
| HLACR1 | 5′-GCGGGGATTTTGGCCTAAAC-3′ | HLA-C exons 1–2 |
| GTFF | 5′-TCTGCCCTCAATTACCTCTC-3′ | |
| GTFR | 5′-GTTTCACCATGTTGGCCAGG-3′ | GTF2H4 exons 1–2 |
| DDRF | 5′-CCCTATTGTGTGCTCTGATG-3′ | |
| DDRR | 5′-CATTCCTGGAGAACAAGGAG-3′ | DDR1 exon 3 |
Table A4.
SNPs Genotyped in Trios[Note]
| SNP Position | Nucleotide Change | |
| SNP1 | nt 6988 in X92841 (MICA) | G/T |
| SNP2 | nt 7055 in X92841 (MICA) | A/G |
| SNP3 | nt 7607 in X92841 (MICA) | A/G |
| SNP4 | 14871 | A/C |
| SNP5 | 15370 | C/T |
| SNP6 | 15488 | A/G |
| SNP7 | 22222 | A/G |
| SNP8 | 22531 | C/T |
| SNP9 | 24118 (USP8 pseudogene) | C/T |
| SNP10 | 25663 (USP8 pseudogene) | A/T |
| SNP11 | 25747 (USP8 pseudogene) | A/G |
| SNP12 | 25783 (USP8 pseudogene) | A/G |
| SNP13 | 25812 (USP8 pseudogene) | C/T |
| SNP14 | 25875 (USP8 pseudogene) | C/G |
| SNP15 | 36669 | C/G |
| SNP16 | 36755 | C/T |
| SNP17 | 36844 | A/G |
| SNP18 | 36989 | A/G |
| SNP19 | 40057 | A/T |
| SNP20 | 49153 | A/G |
| SNP21 | 49285 | A/C |
| SNP22 | 49330 | A/T |
| SNP23 | 51271 | A/G |
| SNP24 | 51510 | C/T |
| SNP25 | 62363 | A/G |
| SNP26 | 62500 | A/C |
| SNP27 | 73366 | A/C |
| SNP28 | 73400 | C/T |
| SNP29 | 91729 | A/G |
| SNP30 | 91818 | A/G |
| SNP31 | 91838 | A/G |
| SNP32 | 95000 | C/T |
| SNP33 | 96000 | A/G |
| SNP34 | 100245 | C/T |
| SNP35 | 125818 | G/T |
| SNP36 | 126540 | A/G |
| SNP37 | 129684 (OTF3) | A/G |
| SNP38 | 129955 (OTF3) | C/T |
| SNP39 | 134127 (OTF3) | G/T |
| SNP40 | 135940 (OTF3) | C/T |
| SNP41 | 137260 (SC1) | A/G |
| SNP42 | 145593 (HCR) | C/T |
| SNP43 | 145611 (HCR) | C/T |
| SNP44 | 145763 (HCR) | C/T |
| SNP45 | 145778 (HCR) | C/G |
| SNP46 | 149523 (HCR) | C/G |
| SNP47 | 155337 (HCR) | G/T |
| SNP48 | 157000 (HCR) | A/G |
| SNP49 | 157683 (HCR) | G/T |
| SNP50 | 160749 | A/G |
| SNP51 | 160808 | A/G |
| SNP52 | 174310 | C/T |
| SNP53 | 174499 (SEEK1) | C/T |
| SNP54 | 175545 | C/T |
| SNP55 | 183299 (CDSN) | C/T |
| SNP56 | 183895 (CDSN) | A/G |
| SNP57 | 183916 (CDSN) | G/T |
| SNP58 | 183923 (CDSN) | C/T |
| SNP59 | 185679 | A/G |
| SNP60 | 185798 | C/T |
| SNP61 | 186692 | A/G |
| SNP62 | 190455 | A/G |
Note.— SNPs in italic were also genotyped in control individuals.
Table A5.
SNPs Genotyped in Controls Only
| SNP Position | Nucleotide Change | |
| SNP I | nt 5617 in Z15026 (TNFα) | C/T |
| SNP II | nt 7934 in Z15026 (TNFβ) | G/T |
| SNP III | nt 8034 in U65416 (MICB) | A/G |
| SNP IV | nt 71855 in AP000508 (HLA-C) | A/G |
| SNP V | nt 71943 in AP000508 (HLA-C) | A/G |
| SNP VI | 126093 | G/T |
| SNP VII | 171542 | C/T |
| SNP VIII | 175962 | C/T |
| SNP IX | 176224 | C/T |
| SNP X | 185779 | A/C |
| SNP XI | 187630 | A/G |
| SNP XII | nt 15117 in AC004211 (GTF2H4) | C/T |
| SNP XIII | nt 15403 in AC004211 (GTF2H4) | A/G |
| SNP XIV | nt 1896029 in NT_001520 (DDR1) | C/T |
| SNP XV | nt 1896258 in NT_001520 (DDR1) | C/T |
| SNP XVI | nt 1896315 in NT_001520 (DDR1) | A/G |
Table A6.
Additional SNPs Identified in Patients
| SNP Position | Nucleotide Change | |
| SNP63 | 9585 | C/G |
| SNP64 | 9852 | G/T |
| SNP65 | 10541 | C/G |
| SNP66 | 11758 | A/G |
| SNP67 | 11761 | A/G |
| SNP68 | 17888 | A/G |
| SNP69 | 22238 | A/G |
| SNP70 | 22333 | A/G |
| SNP71 | 22382 | C/T |
| SNP72 | 22449 | A/T |
| SNP73 | 22483 | C/T |
| SNP74 | 33003 | A/G |
| SNP75 | 33353 | G/T |
| SNP76 | 34592 | A/C |
| SNP77 | 37001 | C/T |
| SNP78 | 37095 | C/T |
| SNP79 | 37212 | C/T |
| SNP80 | 39386 | G/T |
| SNP81 | 39640 | A/G |
| SNP82 | 40057 | A/T |
| SNP83 | 42009 | C/T |
| SNP84 | 42010 | C/T |
| SNP85 | 44911 | C/T |
| SNP86 | 45033 | C/T |
| SNP87 | 45037 | C/T |
| SNP88 | 45051 | C/T |
| SNP89 | 45060 | G/T |
| SNP90 | 45106 | A/G |
| SNP91 | 45113 | A/G |
| SNP92 | 51359 | G/T |
| SNP93 | 51389 | C/T |
| SNP94 | 65572 | A/T |
| SNP95 | 65638 | A/G |
| SNP96 | 65657 | A/C |
| SNP97 | 75098 | C/T |
| SNP98 | 75355 | C/T |
| SNP99 | 84116 | A/G |
| SNP100 | 84557 | C/T |
| SNP101 | 89782 | A/G |
| SNP102 | 89828 | A/G |
| SNP103 | 89848 | A/C |
| SNP104 | 89863 | A/C |
| SNP105 | 89977 | C/G |
| SNP106 | 132496 | C/T |
| SNP107 | 141824 | G/T |
| SNP108 | 142507 | A/C |
| SNP109 | 142549 | C/G |
| SNP110 | 169204 | A/C |
| SNP111 | 169237 | C/T |
| SNP112 | 171542 | C/T |
| SNP113 | 174730 | A/G |
| SNP114 | 185679 | A/G |
| SNP115 | 186637 | C/T |
| SNP116 | 186896 | A/G |
| SNP117 | 187120 | A/G |
| SNP118 | 187630 | A/G |
| SNP119 | 187669 | C/T |
| SNP120 | 192527 | A/C |
| SNP121 | 203832 | C/T |
| SNP122 | 203987 | A/T |
| SNP123 | 203996 | G/T |
Table A7.
Additional SNPs Identified in Controls
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the MHC sequence [accession numbers AC004204, AC006048, AC004185, AC006047, AC004195, and AC006163])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for psoriasis [MIM *177900], HLA-C [MIM *142840], OTF3 [MIM *164177], HCR [MIM *605310], and CDSN [MIM *602593])
References
- Allen MH, Veal C, Faassen A, Powis SH, Vaughan RW, Trembath RC, Barker JNWN (1999) A non-HLA gene within the MHC in psoriasis. Lancet 353:1589–1590 [DOI] [PubMed] [Google Scholar]
- Asahina A, Kuwata S, Tokunaga K, Juji T, Nakagawa H (1996) Study of aspartate at residue 9 of HLA-C molecules in Japanese patients with psoriasis vulgaris. J Dermatol Sci 13:125–133 [DOI] [PubMed] [Google Scholar]
- Asumalahti K, Latinen T, Itkonen-Vatjus R Lokki ML, Suomela S, Snellman E, Saarialho-Kere U, Kere J (2000) A candidate gene for psoriasis near HLA-C, HCR (Pg8), is highly polymorphic with a disease-associated susceptibility allele. Hum Mol Genet 9:1533–1542 [DOI] [PubMed] [Google Scholar]
- Asumalahti K, Veal C, Latinen T, Suomela S, Allen M, Outi E, Moser M, et al (2002) Coding haplotype analysis supports HCR as the putative susceptibility gene at the MHC PSORS1 locus. Hum Mol Genet 11:589–597 [DOI] [PubMed] [Google Scholar]
- Balendran N, Clough RL, Arguello JR, Barber R, Veal C, Jones AB, Rosbotham JL, Little AM, Madrigal A, Barker JNWN, Powis SH, Tremabth RC (1999) Characterization of the major susceptibility region for psoriasis at chromosome 6p21.3. J Invest Dermatol 113:322–328 [DOI] [PubMed] [Google Scholar]
- Barker JN (1991) The pathophysiology of psoriasis. Lancet 338:227–230 [DOI] [PubMed] [Google Scholar]
- Bos J, De Rie MA (1999) The pathogenesis of psoriasis: immunological facts and speculations. Immunol Today 20:40–46 [DOI] [PubMed] [Google Scholar]
- Camp RDR (1998) Psoriasis. In: Champion RH, Burton JL, Burns DA, Breathnach SM (eds) Textbook of dermatology. Blackwell Science, Oxford, pp 1589–1649 [Google Scholar]
- Capon F, Munro M, Barker J, Trembath R (2002) Searching for the MHC psoriasis susceptibility gene. J Invest Dermatol 118:745–751 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI (2001) Association study designs for complex traits. Nat Rev Genet 2:91–99 [DOI] [PubMed] [Google Scholar]
- Cerundolo V, Braud V (1996) Cell biology of MHC class I molecules. In: Browning M, McMichael A (eds) HLA and MHC: genes, molecules and function. Bios Scientific Publishers, Oxford, pp 193–223 [Google Scholar]
- Chia NV, Stuart P, Nair RP Henseler T, Jenisch S, Lim HW, Christophers E, Voorhees JJ, Elder JT (2001) Variations in HCR (Pg8) gene are unlikely to be causal for familial psoriasis. J Invest Dermatol 116:823–824 [DOI] [PubMed] [Google Scholar]
- Clayton D (1999) A generalization for the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet 65:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Elder JT, Henseler T, Christophers E, Voorhees JJ, Nair RP (1994) Of genes and antigens: the inheritance of psoriasis. J Invest Dermatol 103 Suppl 5:150S–153S [DOI] [PubMed] [Google Scholar]
- Elder JT, Nair RP, Henseler T, Jenisch S, Stuart P, Chia N, Christophers E, Voorhees JJ (2001) The genetics of psoriasis 2001: the odyssey continues. Arch Dermatol 137:1447–1453 [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Martinez Borra J, Del Río JS, Santos-Juanes J, Lopez-Vazquez A, Blanco-Gelaz M, López-Larrea C (2000) The OTF3 gene polymorphism confers susceptibility to psoriasis independent of the association of HLA-Cw*0602. J Invest Dermatol 115:824–828 [DOI] [PubMed] [Google Scholar]
- Hellgren I (1967) Psoriasis: the prevalence in sex, age, and occupational groups in total population in Sweden: morphology, inheritance and association with other skin and rheumatic diseases. Almqvist and Wiksell, Stockholm [Google Scholar]
- International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Ritchie A, Neumann R (2000) High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet 9:725–733 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Neumann R, Keyte J (1988) Amplification of human minisatellites by the polymerase chain reaction: towards DNA fingerprinting of single cells. Nucleic Acids Res 16:10953–10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenisch S, Koch S, Henseler T, Nair RP, Elder JT, Watts CE, Westphal E, Voorhees JJ, Christophers E, Kronke M (1999) Corneodesmosin gene polymorphism demonstrates strong linkage disequilibrium with HLA and association with psoriasis vulgaris. Tissue Antigens 54:439–449 [DOI] [PubMed] [Google Scholar]
- Lee YA, Ruschendorf F, Windemuth C, Schmitt-Egenolf M, Stadelmann A, Nurnberg G, Stander M, Wienker TF, Reis A, Traupe H (2000) Genomewide scan in German families reveals evidence for a novel psoriasis-susceptibility locus on chromosome 19p13. Am J Hum Genet 67:1020–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomholt G (1963) Psoriasis: prevalence, spontaneous course and genetics. GEC Gad, Copenhagen [Google Scholar]
- Mallon E, Bunce M, Wojnarowska F, Welsh K (1997) HLA-Cw*0602 is a susceptibility factor in type I psoriasis, and evidence Ala-73 is increased in male type I psoriatics. J Invest Dermatol 109:183–186 [DOI] [PubMed] [Google Scholar]
- Mallon E, Newson R, Bunker CB (1999) HLA-Cw6 and the genetic predisposition to psoriasis: a meta-analysis of published serologic studies. J Invest Dermatol 113:693–695 [DOI] [PubMed] [Google Scholar]
- Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ, Riley J, Finch KL, Stevens JF, Livak KJ, Slotterbeck BD, Slifer SH, Warren LL, Conneally PM, Schmechel DE, Purvis I, Pericak-Vance MA, Roses AD, Vance JM (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet 67:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, Elder JT (1997) Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6:1349–1356 [DOI] [PubMed] [Google Scholar]
- Nair RP, Stuart P, Heneseler T, Jenisch S, Chia NVC, Westphal E, Schork NJ, Kim J, Lim HW, Christophers E, Voorhees JJ, Elder JT (2000) Localization of psoriasis-susceptibility locus PSORS1 to a 60-kb interval telomeric to HLA-C. Am J Hum Genet 66:1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt GJ, Hutchinson PE (1996) Psoriasis in the community: prevalence, severity and patients' beliefs and attitudes towards the disease. Br J Dermatol 135:533–537 [PubMed] [Google Scholar]
- Nickoloff BJ, Wrone-Smith T, Bonish B, Porcelli SA (1999) Response of murine and normal human skin to injection of allogeneic blood-derived psoriatic immunocytes: detection of T cells expressing receptors typically present on natural killer cells, including CD94, CD158, and CD161. Arch Dermatol 135:546–552 [DOI] [PubMed] [Google Scholar]
- O’Brien KP, Holm SJ, Nilsson S, Carlén L, Rosenmüller T, Enerbäck C, Inerot A, Ståhle-Bäckdahl M (2001) The HCR gene on 6p21 is unlikely to be a psoriasis susceptibility gene. J Invest Dermatol 116:750–754 [DOI] [PubMed] [Google Scholar]
- Oka A, Tamiya G, Tomizawa M, Ota M, Katsuyama Y, Makino S, Shiina T, Yoshitome M, Lizuka M, Sasao Y, Iwashita K, Kawakubo Y, Sigai J, Ozawa A, Ohkido M, Kimura M, Bahram S, Inoko H (1999) Association analysis using refined microsatellite markers localizes a susceptibility locus for psoriasis vulgaris within a 111 kb segment telomeric to the HLA-C gene. Hum Mol Genet 8:2165–2170 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Roitberg-Tambur A, Friedmann A, Tzfoni EE, Battat S, Ben Hammo R, Safirman C, Tokunaga K, Asahina A, Brautbar C (1994) Do specific pockets of HLA-C molecules predispose Jewish patients to psoriasis vulgaris? J Am Acad Dermatol 31:964–968 [DOI] [PubMed] [Google Scholar]
- Sander HM, Morris LF, Phillips CM, Harrison PE, Menter A (1993) The annual cost of psoriasis. J Am Acad Dermatol 28:422–425 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi Ahnini R, Camp NJ, Cork NJ, Mee JB, Keohane SG, Duff GW, di Giovine FS (1999) Novel genetic association between the corneodesmosin (MHC S) gene and susceptibility to psoriasis. Hum Mol Genet 8:1135–1140 [DOI] [PubMed] [Google Scholar]
- Tiilikainen A, Lassus A, Karvonen J, Vartiainen P, Julin M (1980) Psoriasis and HLA-Cw6. Br J Dermatol 102:179–184 [DOI] [PubMed] [Google Scholar]
- Trembath RC, Clough RL, Rosbotham JL, Jones AB, Camp RDR, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop GM, Barker JNWN (1997) Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome-wide search in psoriasis. Hum Mol Genet 6:813–820 [DOI] [PubMed] [Google Scholar]
- Veal CD, Clough RL, Barber RC, Mason S, Tillman D, Ferry B, Jones AB, Ameen M, Balendran N, Powis SH, Burden AD, Barker JNWN, Trembath RC (2001) Identification of a novel psoriasis susceptibility locus at 1p and evidence of epistasis between PSORS1 and candidate loci. J Med Genet 38:7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs? Nat Genet 26:151–157 [DOI] [PubMed] [Google Scholar]