Abstract
Genomewide association studies may offer the best promise for genetic mapping of complex traits. Such studies in outbred populations require very densely spaced single-nucleotide polymorphisms. In recently founded population isolates, however, extensive linkage disequilibrium (LD) may make these studies feasible with currently available sets of short tandem repeat markers, spaced at intervals as large as a few centimorgans. We report the results of a genomewide association study of severe bipolar disorder (BP-I), using patients from the isolated population of the central valley of Costa Rica. We observed LD with BP-I on several chromosomes; the most striking results were in proximal 8p, a region that has previously shown linkage to schizophrenia. This region could be important for severe psychiatric disorders, rather than for a specific phenotype.
Introduction
The elucidation of the genetic basis of common, genetically complex diseases remains extremely difficult. Standard linkage analysis has so far proved unsatisfactory for this task (Shork et al. 1998; Risch 2000). There is currently intense interest in searching for susceptibility genes for such disorders, using association analysis (Risch and Merikangas 1996), either through direct investigation of variants in candidate genes or through linkage disequilibrium (LD) mapping. LD mapping involves searching for association between a disease and alleles or haplotypes at mapped marker loci. This association reflects sharing of genome segments that surround a disease gene among affected individuals who are descended from a common ancestor. Such shared genome segments are, on average, shorter in older populations than in younger populations, reflecting the greater number of meioses separating affected individuals from their common ancestors. In older populations, including most outbred populations, LD will generally be detectable only over short distances (⩽100 kb), and therefore genomewide LD mapping studies may not be feasible before a very dense map of SNPs and improved methods for high-throughput SNP genotyping are available. In recently founded and genetically isolated populations, however, LD may be detectable over much longer genomic segments (i.e., spaced over intervals as large as a few centimorgans) than in older and heterogeneous populations (Mohlke et al. 2001; Service et al. 2001; Varilo et al. 2001). Over these distances, it is possible to conduct genomewide genotyping, using currently available sets of STR markers that have already been optimized for automated genotyping. This class of markers is much more informative for detecting LD than are SNPs. The higher mutation rate of STRs, compared with SNPs, potentially inhibits the detection of LD; however, this is a negligible factor over the relatively few meioses that have occurred since the founding of young isolates. For the reasons noted above, in recently founded population isolates, genomewide LD mapping of complex traits may be feasible with the use of STR markers spaced at intervals as large as a few centimorgans.
LD mapping in recently founded isolates may be particularly powerful if it is feasible to construct haplotypes among affected individuals. This approach has already been used in such populations to map loci for several Mendelian disorders (Houwen et al. 1994). For rare Mendelian disorders, the expectation is that all affected individuals from a population will share a common haplotype in the region of the disease locus. In contrast, we anticipate heterogeneity in disease etiology for a complex trait, even in a population isolate. Therefore our expectation is that not all affected individuals will share ancestry and a common haplotype in the vicinity of a trait locus; the goal of LD-based analysis is to identify haplotypes shared more frequently by these individuals than would be expected by chance.
Here, we report the results of a genomewide LD-mapping study of bipolar disorder (BP), a common syndrome that consists of episodes of mania and depression and is characterized by complex patterns of inheritance. Although several genomewide linkage studies of BP have been undertaken and have suggested several possible chromosomal localizations, there is still no unequivocal evidence for any single location for BP. In addition, these studies have used a wide range of definitions of the affected phenotype, and therefore their results are difficult to compare with one another (Prathikanti and McMahon 2001). We investigated a particularly severe and heritable form of BP (BP-I) and further limited our studies to probands with BP-I who had at least two psychiatric hospitalizations for this condition. We used a dense set of STR markers to genotype 109 probands with BP-I and their parents, a group drawn from a young population isolate, that of the central valley of Costa Rica (CVCR). The 2.6 million residents of the CVCR descend mainly from a small group of Spanish and Amerindian founders who lived in the 16th and 17th centuries; by the beginning of the 18th century, the CVCR had a single population that then grew rapidly, without subsequent immigration, for almost 200 years (Escamilla et al. 1996). Its population history is similar to those of other isolates that have been the focus of recent genetic-mapping efforts (Peltonen et al. 2000; Chapman and Thompson 2001; Shifman and Darvasi 2001).
The recent founding, isolation, and rapid expansion of the CVCR population are reflected in the extensive LD that has been observed in this population. A previous genomewide survey (Service et al. 2001) of background LD (BLD) (i.e., LD between markers independent of a shared phenotype) in the CVCR population showed that LD was significantly detectable in 310 of 1,012 adjacent marker pairs. These 310 marker pairs were an average of ∼3 cM (SD=1.7) apart. For several reasons, disease-related LD in this population is expected to display a more powerful signal than that of BLD. First, if markers are spaced at an average density of 3 cM (the average extent of BLD observed in the CVCR population), a putative disease locus should be no more than 1.5 cM distant from the nearest marker. Using the markers available for the genome screen described here, we were able to achieve an average density of 3.5 cM (i.e., a disease locus should still be an average of <2 cM from the nearest marker). In addition, although the samples of control chromosomes used in the investigation of BLD are essentially randomly chosen with respect to LD, a sample of affected individuals in a population isolate is, by definition, chosen to be enriched for LD in the region of disease-related variants. Finally, disease-related LD may be assessed via haplotype-based tests that are substantially more powerful than the two-point methods used to assess BLD. These factors suggested that the ∼1,000 STR markers that we used would be appropriate for an initial genomewide screen for LD for BP-I in the CVCR. The results of this screen are described here.
Subjects, Material, and Methods
Sample Collection
Individuals who experienced onset of BP-I at age ⩽50 years and had history of at least two psychiatric hospitalizations were recruited, independently from one another, from psychiatric hospitals and clinics (Escamilla et al. 1996). Details of recruitment and diagnostic procedures are described elsewhere (Escamilla et al. 1996). In brief, all subjects were interviewed by a bilingual psychiatrist in Costa Rica, using the Diagnostic Interview for Genetic Studies (Nurnberger et al. 1994). The interview results and abstracts of hospital records were reviewed by two independent psychiatrists at the University of California, San Francisco, to arrive at a consensus best-estimate diagnosis. Fifty-eight probands were women, and 51 were men. Both parents of 48 probands were available for genotyping, and the remaining 61 probands had one parent available, making a total of 266 genotyped individuals (109 probands and 157 parents). Eighty-one probands had all eight great-grandparents from the CVCR; 11 and 17 had seven and six great-grandparents from the CVCR, respectively. The majority of subjects in the current sample were not available in earlier LD analyses of chromosome 18 markers in this population (Escamilla et al. 1999, 2001). The first LD study on chromosome 18 (Escamilla et al. 1999) used a sample of 113 people that included 48 affected individuals. Of those 113, 56 are genotyped in the current sample, including 26 affected individuals. In the second LD analysis of chromosome 18, work was divided into two phases (Escamilla et al. 2001). The phase I portion genotyped 162 people (69 affected individuals), and the phase II portion (a follow-up of regions identified in phase I) genotyped 566 individuals (227 affected individuals). In the present study, 103/162 people from the phase I portion are genotoyped (41/69 affected individuals). Everyone genotyped in the present study was also genotyped in the phase II portion of the LD analysis of selected regions on chromosome 18 (Escamilla et al. 2001). Informed consent was obtained from all subjects in the study.
Genotyping
The 1,186 fluorescently labeled microsatellite markers were mainly from the Généthon collection (Dib et al. 1996), and a single genetic map provided the location of all the markers in relation to one other (Broman et al. 1998). The genotype data from 150 markers were discarded because of such problems as PCR failure and difficulty in scoring or because of large-scale Mendelian errors (null alleles). Of the remaining 1,036 markers, the vast majority (n=807) were selected from the ABI Prism Linkage Mapping Set HD-5 (Applied Biosystems). The order and sex-averaged distance of the markers were based on the Marshfield map (Broman et al. 1998). Sixteen intervals had a size of 7–10 cM; no intervals were >10 cM. PCR was performed using standard conditions in PE 9700 PCR machines. PCR products were detected using an ABI 377 sequencer and were analyzed by use of GENESCAN and GENOTYPER software. All genotypes were independently double scored. Data were checked for Mendelian inheritance by use of the UNKNOWN program. For all markers, at least 100 probands of the total 109 had acceptable genotypes (average 108 probands). The average observed heterozygosity of the markers was 77%.
Each marker was tested for Hardy-Weinberg equilibrium (HWE) by comparing the observed homozygosity in the parental chromosomes with the values expected on the basis of the allele frequencies that were calculated from our sample.
Population Homogeneity Assessment
The method of Pritchard et al. (2000) uses multilocus genotype data from unlinked markers to infer population structure. We applied this method to the genotypes of probands at 120 marker loci separated from one another by at least 20 cM. In the application of this Markov chain Monte Carlo method, we used 10,000 replications, for the burn-in period of the chain, and 100,000 replications, for parameter estimation. The number of populations present in the sample (K) is unknown, and we ran the analysis at K=1, K=2, and K=3. For each value of K, multiple chains were run and compared, to assess chain convergence and consistency of estimates. From these results, the best estimate of K was found by calculating the posterior probabilities, as described by Pritchard et al. (2000)
Statistical Analysis of LD
LD was evaluated between markers and disease by two statistical tests, a modified version (Escamilla et al. 1999) of the two-point method of Terwilliger (1995) (LD-T) and a haplotype-based method, ancestral haplotype reconstruction (AHR) (Service et al. 1999). We used both tests for the following reason: AHR is apparently more powerful than LD-T (Service et al. 1999); however, LD-T, as a two-point test, is not sensitive to BLD. The LD-T evaluates the likelihood that a particular allele at a single locus is overrepresented on disease chromosomes (transmitted), compared with nondisease chromosomes (nontransmitted). This overrepresentation is quantified by a single parameter, λ. No correction for multiple alleles is necessary with the LD-T, because the likelihoods for individual alleles are weighted by allele frequency and are combined into a single likelihood-ratio test. The AHR method compares the observed distribution of haplotypes in affected individuals with the distribution expected among individuals who bear a disease mutation inherited from a common ancestor (Service et al. 1999). Nontransmitted chromosomes of parents of probands were used as controls. The probability model for AHR assumes a multinomial distribution of the observed counts of haplotypes. The expected haplotype probabilities are calculated by assuming a given founder haplotype, the position of the disease locus (x), the proportion of chromosomes in the sample of affected individuals that is likely to have descended from this founder haplotype (α), the separation time (in generations) from the common founding haplotype (g), and the marker-allele frequencies. The likelihood of this putative founder chromosome giving rise to the observed sample of disease haplotypes can be easily calculated under the assumptions of the multinomial distribution and the independence of chromosomes. These calculations are repeated for each of the putative founder chromosome types, weighted by the probability of observing that haplotype in the population, and are summed to create an overall likelihood for the chromosomal segment. When this approach is used, no correction for multiple haplotypes at the same markers is needed. The likelihood is maximized over x, g, and α and is compared with the null likelihood. Under the null hypothesis, marker-allele and haplotype frequencies (assuming a one-step Markov process) are estimated, and α is set to zero. We calculated the ratio of the log likelihoods (LR) under the alternative hypothesis and under the null hypothesis. Under the null hypothesis, −2*LR has half its weight concentrated on zero and the other half on a distribution that can be approximated by max (X1,X2), where X1 and X2 are independent χ2 variables with 1 df (see Service et al. 1999). AHR was used in overlapping haplotype windows, consisting of three markers each, on all autosomes; the model for recombination probabilities used in haplotype-based tests that assume descent from a common ancestor cannot be applied straightforwardly to markers on sex chromosomes. The likelihood was evaluated at five steps (estimates of x) between each marker, at 15 estimates of g (ranging from 10 to 1,000), and at 50 estimates of α (ranging from 0.02 to 1.0). AHR was modified from the form presented by Service et al. (1999) to allow for LD between markers under the null hypothesis (McPeek and Strahs 1999). The currently observed LD is used as an estimate throughout the population history.
These LD analyses were used to identify promising regions for follow-up investigations, rather than to produce definitive localization, and we therefore applied a low significance threshold. Previous simulations (Service et al. 1999) had shown AHR to have a false-positive rate that was in accordance with the rate predicted by a χ2 distribution; however, those simulations were performed under conditions of no BLD between markers in control chromosomes. In the application of AHR to our data, we were concerned that BLD could inflate the false-positive rate of this test, and we therefore applied a higher threshold for AHR (P⩽.01) than for LD-T (P⩽.05). The P values presented here are not corrected for multiple tests. We view these results as a guide for further investigation, rather than statistically significant associations.
By definition, regions with a high likelihood-ratio statistic have a higher likelihood under the model of descent from a common disease-bearing ancestor than under the null hypothesis of no disease locus present. Although the observed data fit the alternative hypothesis better than the null hypothesis, it is not necessarily true that the data have a good fit to the alternative hypothesis. We formulated a goodness-of-fit test by calculating the expected number of haplotypes of each type at the maximum-likelihood estimates of the parameters for both case and control chromosomes. For the goodness-of-fit test, we collapsed the haplotypes into six categories and compared observed counts with expected counts in each. The categories were formed by considering the possible similarity of a three-marker haplotype to the most likely founding haplotype: exactly like the founding haplotype, different from the founding haplotype at flanking markers (four possibilities), and different from the founding haplotype at each marker.
Results
Independent Samples from the Population Isolate
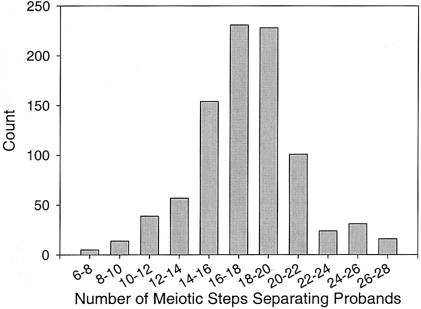
From available genealogical information, we calculated the relatedness among the probands with BP-I. With 109 probands, there are a total of 5,886 possible pairwise relationships ([109×108]÷2) to consider. We had genealogical information to identify 900 of these 5,886 pairwise connections. These known pairwise connections were an average of 16 meiotic steps apart (see fig. 1). There were 47 probands for whom we have not identified relationships to other probands in the study. For the majority (35) of these 47 probands, we had no genealogical information further than the great-grandparental generation; therefore, the apparent lack of relatedness to other probands is probably a result of incomplete information. In fact, for 20 probands, we do not have genealogical information on 1 or 2 of their great-grandparents. The incomplete information connecting probands precludes the use of linkage tests in such a population sample, because it is not possible simply to use this pedigree structure in a standard linkage program.
Figure 1.
Distribution of the number of meiotic steps connecting the 109 probands, calculated on the basis of available genealogical information.
Statistical Analysis of LD
We genotyped the 109 probands with BP-I, each of whom had at least one available parent, for 1,036 STR markers spaced at an average distance from each other of 3.5 cM. Analysis of these genotype data, by use of the method of Pritchard et al. (2000), showed no evidence for cryptic population substructure in this sample. HWE was violated (with a significance level <.01), for a total of 20 markers (data not shown). Reexamination revealed only one marker that could be explained by the presence of a potential null allele. This marker, D18S54, was subsequently discarded from any further analysis. For the other markers, HWE violation could not be explained other than by chance, and, after Bonferroni correction for multiple tests, only one marker (D6S434) was significant at the .05 level.
The LD-T analysis showed 21 markers with evidence of LD at a significance level of ⩽.05 (table 1), including three markers (D2S303, D9S1847, and D19S904) showing P<.01. The markers displaying association were widely dispersed throughout the genome, except two regions of ∼5 cM, each of which contained two markers showing LD to BP-I (on chromosomes 2 and 8p).
Table 1.
Markers for Which the LD-T P Value Was ⩽.05
| Marker | Distancefrom pter | λ | P | AssociatedAllele |
| D1S227 | 238.5 | .250 | .037 | 2 |
| D2S2268 | 1.9 | .450 | .039 | 4 |
| D2S2150 | 40.5 | .303 | .035 | 1 |
| D2S303 | 88.2 | .433 | .009 | 1 |
| D2S286 | 94.0 | .355 | .040 | 1 |
| D4S413 | 158.0 | .183 | .044 | 5 |
| D5S2049 | 160.9 | .427 | .039 | 2 |
| D8S1825 | 15.4 | .469 | .022 | 3 |
| D8S520 | 20.6 | .252 | .049 | 1 |
| D9S1847 | 144.7 | .352 | .002 | 7 |
| D10S602 | 4.3 | .325 | .027 | 4 |
| D10S548 | 45.7 | .399 | .015 | 1 |
| D10S208 | 60.6 | .355 | .015 | 6 |
| D10S1773 | 134.2 | .322 | .050 | 3 |
| D11S1320 | 141.9 | .336 | .021 | 4 |
| D12S352 | .0 | .376 | .018 | 5 |
| D15S642 | 122.2 | .339 | .028 | 5 |
| D17S787 | 75.0 | .199 | .046 | 6 |
| D17S1862 | 97.6 | .221 | .044 | 4 |
| D19S1150 | 39.0 | .295 | .020 | 1 |
| D19S904 | 78.1 | .282 | .008 | 4 |
The AHR analyses showed 14 regions with evidence (P⩽.01) for an overrepresented three-marker haplotype (henceforth termed a “shared segment”) on the chromosomes transmitted to affected individuals (table 2). Three of these regions are within 5 cM of markers showing LD to BP-I by use of the LD-T (D8S503, D8S520, and D17S788). In 4 of the 14 regions, consecutive shared segments overlapped with each other: on 2p, we observed an overlap of five such shared segments; on 2q and 8p, we observed an overlap of four segments; and, on 17p, we observed overlap of three segments. We manually examined haplotypes in these four regions. On 8p, the overrepresentation of extended haplotypes among the chromosomes transmitted to patients, compared with nontransmitted chromosomes, is particularly apparent, notably in the segment between D8S503 and D8S520. In this segment, >39% of transmitted chromosomes have either the 3-1 or 3-5 haplotype, compared with only 19% among nontransmitted chromosomes (table 3). Compared with what is observed on 8p, the overrepresentation on chromosomes of particular haplotypes among affected individuals, compared with nontransmitted chromosomes, is not as visually obvious in the other three regions with evidence of overlapping haplotype. Haplotype data from chromosome 17p are presented as examples (table 4).
Table 2.
Regions for Which the AHR Result Was Significant at a Level ⩽.01
| ClosestMarker | Length of Three-Marker Haplotype(cM) | Estimate of Distancefrom pterof Disease Locus | Associated Three-Marker Haplotype | αa | gb | Pc |
| D1S2841 | 7.2 | 107.4 | 5 2 6 | .08 | 27 | .000987 |
| D2S2241 | 4.4 | 157.5 | 9 5 4 | .10 | 29 | .002732 |
| D2S156 | 4.9 | 164.5 | 2 4 1 | .12 | 31 | <.000001 |
| D2S369 | 4.3 | 202.9 | 5 2 7 | .06 | 17 | .001217 |
| D2S325 | 1.6 | 205.0 | 5 1 3 | .08 | 17 | .000390 |
| D4S395 | 4.9 | 94.5 | 4 6 3 | .08 | 10 | .006305 |
| D6S1575 | 2.6 | 61.6 | 6 2 2 | .08 | 14 | .003544 |
| D8S503 | 5.2 | 17.1 | 3 3 1 | .16 | 10 | .000057 |
| D8S520 | 5.8 | 20.6 | 1 10 3 | .10 | 21 | .000894 |
| D8S1778 | 8.7 | 111.1 | 4 4 4 | .06 | 13 | .005137 |
| D9S257 | 5.6 | 91.9 | 2 6 4 | .04 | 10 | .000325 |
| D17S1529 | 6.0 | 3.9 | 3 8 2 | .06 | 10 | .000377 |
| D17S1828 | 4.1 | 10.0 | 2 5 8 | .06 | 40 | .005015 |
| D17S788 | 6.8 | 73.6 | 1 5 6 | .12 | 10 | .001897 |
Maximum likelihood estimate of the proportion of disease chromosomes inherited from a common ancestor.
Maximum likelihood estimate of the number of generations since that common ancestor.
Based on χ2 approximation.
Table 3.
Distribution of Two-Marker Haplotypes for D8S503-D8S520 Chromosomes Transmitted and Nontransmitted from Parents to Probands
| Haplotypea | Transmitted(%) | Nontransmitted(%) |
| 1-1 | 3.43 | 4.72 |
| 2-5 | 4.57 | 5.51 |
| 3-1 | 22.86 | 12.60 |
| 3-2 | 2.86 | 3.15 |
| 3-4 | 4.00 | 5.51 |
| 3-5 | 16.57 | 7.87 |
| 3-6 | 4.00 | 4.72 |
| 5-4 | .57 | 6.30 |
| 5-5 | 4.00 | 6.30 |
| 6-2 | 4.00 | 5.51 |
| 6-4 | 1.71 | 4.72 |
| 6-5 | 4.57 | 4.72 |
| 6-6 | 1.14 | 7.09 |
| Other (n = 29) | 25.71 | 21.26 |
Haplotype counts of at least 5 for either transmitted or nontransmitted categories are reported here; counts <5 are grouped in the category “other.”
Table 4.
Distribution of Two-Marker Haplotypes for D17S1529-D17S831 Chromosomes Transmitted and Nontransmitted from Parents to Probands
| Haplotypea | Transmitted(%) | Nontransmitted(%) |
| 2-1 | 2.84 | .00 |
| 2-3 | 5.11 | 3.15 |
| 2-4 | 3.41 | 3.15 |
| 2-5 | 4.55 | 6.30 |
| 2-6 | 4.55 | 5.51 |
| 3-3 | 3.41 | 1.57 |
| 3-5 | 4.55 | 2.36 |
| 5-3 | 11.93 | 6.30 |
| 5-6 | 2.84 | 4.72 |
| 6-3 | 3.98 | 4.72 |
| 6-4 | 2.84 | 2.36 |
| 6-5 | 2.84 | 3.94 |
| 6-6 | 5.11 | 3.15 |
| 7-3 | 2.84 | .00 |
| 8-2 | 5.68 | 2.36 |
| Other (n = 48) | 33.52 | 50.39 |
Haplotype counts of at least 5 for either transmitted or nontransmitted categories are reported here; counts <5 are grouped in the category “other.”
The LD evidence in the AHR test derives from reconstruction of a single most likely ancestral haplotype with consequent estimation of (1) the proportion of chromosomes descended from this ancestral haplotype among the affected individuals (parameter a) and (2) the number of generations that have elapsed since this haplotype occurred in a common ancestor of the affected individuals (parameter g). For each region highlighted by the AHR analysis, we determined whether the model of descent from a single ancestral haplotype fits the observed distribution of haplotypes in the chromosomes from affected individuals. For 7 of the 14 regions (D1S2841, D4S395, D6S1575, D8S520, D9S257, D17S1828, and D17S788), we observed reasonable goodness of fit in the chromosomes transmitted to affected individuals (P>.10). For the regions that had poor goodness of fit, the observed LD is not fully explained by the model of a single ancestral susceptibility haplotype, and the parameter estimates may be inaccurate. The results in these regions, however, are still consistent with identical-by-descent sharing of haplotypes among the affected individuals because of a shared susceptibility gene in these regions.
We also evaluated goodness of fit of the nontransmitted chromosomes to the distribution hypothesized by AHR. This distribution is formed under the assumption that LD is present between adjacent markers (modeled as a one-step Markov process) but that linkage equilibrium is present between nonadjacent markers; this simplifying assumption may not hold for all genome regions. Of the 14 regions shown in table 2, 5 (D4S395, D8S520, D9S257, D17S1828, and D17S788) showed reasonable goodness of fit to this model (P>.10). The lack of fit for other regions is probably a result of LD between nonadjacent markers.
Discussion
The results described here demonstrate the application of genomewide LD analysis for the mapping of susceptibility loci for complex traits. We confirmed our initial hypothesis that independently identified affected individuals in a recently founded genetic isolate would share common ancestry. Genealogical information, although incomplete, illustrates that the CVCR is a young and inbred population. Indeed, such close relationship between independently ascertained probands is consistent with the absence of demonstrable population substructure in this sample and with the extensive genomewide LD observed in this population (Service et al. 2001), and it confirms that the sample is suitable for genomewide LD mapping.
Our data indicate that, in a few genome regions, the probands display haplotype sharing that is greater than would be expected by chance. Because these haplotypes are as long as several centimorgans, they are detectable with the use of currently available sets of mapped STR markers that have already been optimized for automated genotyping. We anticipate that the type of analysis that we report here could be used for initial mapping of loci for a wide range of complex traits, in any of several populations with demographic histories similar to that of the CVCR (e.g., in subisolates in Finland in which LD has been observed for distances of several centimorgans [Varilo et al. 2001]).
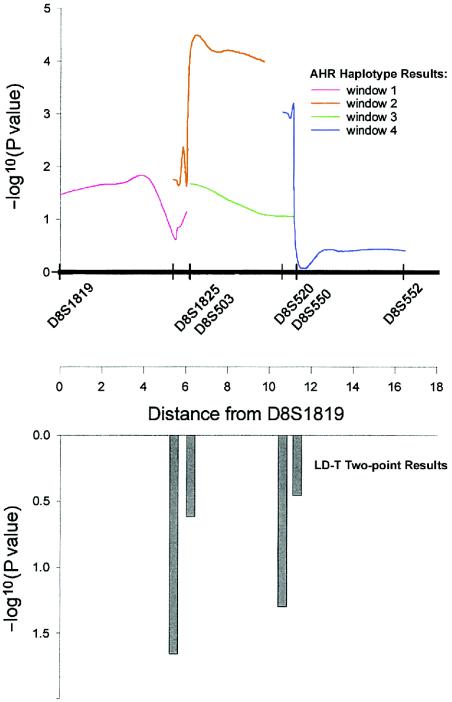
The genome screen for BP-I of this CVCR sample has identified several candidate regions for further genetic investigation, with the strongest evidence being for a 22-cM region on chromosome 8p. All of the overlapping AHR analyses of this region provided evidence of association with the disease (fig. 2), and four of the six markers in this segment showed evidence of association when analyzed by use of LD-T (fig. 2B, two markers with P<.05). In addition, the two LD tests (LD-T and AHR) identified the same alleles as being overrepresented among the patients. The AHR analysis suggests that the most likely localization of a BP-I gene is between D8S503 and D8S520. Direct observation of haplotypes supports this conclusion, because the haplotypes that were most common among the affected individuals are observed more than twice as frequently in that group as in the control individuals. It has been suggested that the length of a region showing evidence of association should be considered, as well as the magnitude of any single peak within the region, with the idea that broader peaks are more likely to contain a true locus (Terwilliger et al. 1997). The mapping evidence over contiguous markers that cover several centimorgans makes this region particularly attractive for follow-up. The segment in which we identified LD for BP-I overlaps with those in which possible schizophrenia loci have been identified through several independent linkage studies (Blouin et al. 1998; Kaufmann et al. 1998; Brzustowicz et al. 1999; Kendler et al. 2000; Gurling et al. 2001). These earlier studies consisted mainly of analyses of affected sib pairs or multiple small pedigrees, and, consequently, the candidate region suggested by these studies is wide (covering as much as ∼50 cM); however, the BP-I candidate segment suggested by our LD analyses is close to the center of this region.
Figure 2.
Estimates of LD on chromosome 8p. A, Haplotype results of AHR tests. The −log10 (P value) from four different tests, each using three markers, are plotted against genetic distance from marker D8S1819. Estimates of α were between 0.06 and 0.16, and estimates of g were between 10 and 20. B, Two-point results from the LD-T tests. The –log10 (P value) is plotted from tests for which the estimate of λ was greater than the null hypothesis of zero. (Both D8S1819 and D8S552 had λ estimated to be zero.) Estimates of λ were 0.25–0.46
Other regions highlighted by the LD analysis show less consistent evidence for BP-I localization than does 8p, but they still warrant follow-up investigations. Marker D2S156 showed the strongest genomewide statistical evidence of LD of any marker, but the segment surrounding this locus also displays substantial LD on nontransmitted chromosomes (i.e., BLD), and there is no striking visual evidence of overrepresentation of particular haplotypes on disease chromosomes (data not shown). Similarly, on chromosome 17p, evidence for association of BP-I extended over a 10-cM region, encompassing five markers, but visual inspection does not point toward overrepresentation of particular haplotypes in transmitted chromosomes, compared with nontransmitted chromosomes (table 4). No markers in 17p, however, are as closely spaced as markers on 8p, and therefore we would not expect to detect as much haplotype conservation. It is precisely in these situations that a method like AHR may be able to detect LD that is not readily identified by visual inspection.
As with most common disorders—even those in an isolate—it is likely that several different susceptibility alleles determine the genetic risk of BP-I at the population level and that these alleles vary in their contribution to the total risk. The parameter α provides a rough assessment of the relative contributions that various genome segments make to risk, through estimation of the proportion of disease chromosomes that share a common ancestor. The demographic history of the study population can dramatically affect such estimates. This history explicitly connects two parameters estimated in the reconstruction of ancestral haplotypes in a sample of patients: the proportion of individuals who share an ancestral chromosome (α), and the number of generations separating the patients from the introduction of that chromosome into the population (g). A similar diversity of haplotypes, at the level of resolution of the genome screen, could be observed in the sample under widely varying values of α and g. If the ancestor was far in the past (large g), many different haplotypes would be expected, which would be consistent with a larger estimate for α. Alternatively, the ancestor could have been recent, in which case one would expect more haplotype conservation. To match the observed haplotype diversity, then, α must be lower. The estimates of α and g are not independent, and higher estimates of g may be associated with higher α. In fact, very similar expected distributions of haplotypes can be generated with a high g and high α and with a low g and low α.
In our view, the results of genomewide screening for LD for complex traits are best used to guide follow-up studies of the highlighted chromosomal regions, and they do not necessarily indicate statistically significant associations. Several statistical issues remain unresolved in the determination of appropriate significance levels for such genomewide studies (Kruglyak 1997). It is not clear what adjustment should be used to correct for the multiple tests that are performed in genomewide LD mapping. Resampling methods are not feasible because of the very extensive computing time that they would require, and a simple correction, such as the Bonferroni procedure, is far too conservative when tests are not independent. As our group and others have demonstrated, recently founded isolates, such as the population of the CVCR, display LD between a high proportion of markers that are separated by several centimorgans from each other. Clearly, in these populations, the LD tests used to map the disease phenotype are far from independent. The level of dependence between such tests in a genomewide screen depends on the degree of relatedness of the population studied, and it is important to develop correction procedures that will take into account the expected level of dependence in a population. Furthermore, expectations for genomewide significance in traditional mapping studies for simple Mendelian traits were based on the expectation of finding a single major locus responsible for the trait under study. A more stringent control for false positives could be applied in such a situation. In the case of a complex trait, however, one expects that there will be multiple loci affecting the trait, and a very strict control of the number of false positives, such as the Bonferroni method, will result in a considerable loss of power to identify secondary signals. Current research in multiple-comparison procedures is focusing on corrections that will handle dependent tests and multiple true signals. For example, a promising measure of multiple-hypothesis–testing error called the “false discovery rate” (Benjamini and Hochberg 1995) is currently being investigated by members of our group for use in LD mapping studies (C. Sabatti, S.K.S, and N.B.F., unpublished data).
In our effort to further assess the observed significance levels, we applied a goodness-of-fit test to the results. Although this is a commonplace procedure in statistical analysis in other disciplines, such goodness-of-fit tests are rarely if ever applied in statistical genetics. These tests provide an additional guide for interpretation of mapping studies for complex traits. Using the results of these tests, the reader may judge not only the level of significance of the mapping findings but also whether the data are in reasonable agreement with the assumptions of the method of analysis; in the present study, these assumptions were rather restrictive (i.e., that disease chromosomes descend from a single ancestral haplotype).
An additional factor that complicates the interpretation of LD mapping results, is that the tests used to analyze LD usually treat affected individuals as independent from one another, assuming equidistant connections between them. As shown in figure 1, the subjects in the present study are not equidistantly related. The decision to treat individuals as independent serves to artificially decrease the variance of the parameter estimates, leading to an overstatement of the level of significance of statistical tests. Other researchers have attempted to address this issue by using a conditional coalescent approach (McPeek and Strahs 1999) in LD mapping; however, this correction applied uniformly to genomewide results does not take into account the variability across the genome in the degree of genetic similarity between related subjects.
LD mapping of disease genes may be influenced by the LD that exists throughout a population (i.e., by BLD). Although BLD varies between genome regions and between populations, we have shown that BLD is extensive in the nontransmitted chromosomes of this sample and that it extends, in many instances, across several centimorgans (Service et al. 1999). We expect BLD to be similarly extensive in other recently founded isolated populations. Although we have reformulated the AHR statistic to incorporate BLD between adjacent markers in control chromosomes, more extensive BLD, involving more than two loci, may still influence our results. The data from chromosome 2 are an example of the strong BLD that can exist on nontransmitted chromosomes in our sample. It is unclear whether the signal we are detecting around D2S156 is an artifact of BLD. Alternatively, this signal may reflect BLD on top of a disease-related association deriving from a haplotype overrepresented on chromosomes in affected individuals.
The chromosomal regions in which we observed LD for BP-I do not overlap with the regions suggested by pedigree-based linkage studies of BP (Prathikanti and McMahon 2001), including a study of our group that was conducted in extended Costa Rican pedigrees (McInnes et al. 1996). There are several possible explanations for this observation. First, the present study is not directly comparable to previous mapping studies, in that we employed a particularly narrow definition of the BP phenotype, and all of the probands were severely affected. Such a stringent definition of the affected phenotype is usually not feasible when patients are sampled from pedigrees but is possible when the probands are sampled from clinic populations. Second, none of the localizations in the present or previous studies is sufficiently unequivocal to rule out the possibility of false-positive results. Third, even true susceptibility loci may be specific to a particular study sample, especially if the sample is drawn from a population isolate. Finally, the expectations for pedigree- and population-based samples are different: in pedigrees, one may observe susceptibility alleles that are highly penetrant but are infrequent in the population; but, in population samples, one expects lower-penetrance alleles that are relatively prevalent. Alzheimer disease (AD) exemplifies these expectations; rare high-penetrance alleles have been observed in a small number of extended pedigrees, and the ApoE4 risk allele does not segregate in such pedigrees, although it is highly prevalent among patients with AD in many different populations (Tanzi and Bertram 2001).
Our results may also have been influenced by such factors as substructure, admixture, or nonrandom mating that could have affected the pedigree and evolutionary history of this patient sample. Although these possibilities are not suggested by the extensive genealogical information that we have obtained or by the empirical assessment of population homogeneity in our sample, their presence could explain the variability between results of the present study and those of previous association analyses of BP-I that we have conducted in the CVCR. Although the sample used in the present genome-screening study overlaps with the samples used in our previous studies, for the present study we used strict criteria for CVCR ancestry and included only the probands for whom at least one parent was available for genotyping (see “Subjects, Material, and Methods” section). In our previous studies, the majority of subjects did not fit these criteria. In addition, the markers used in the present study were chosen from a single genetic map and with the goal of providing equivalent coverage of all chromosomes, whereas the markers used in the previous studies, which were limited to chromosome 18, were chosen from several genetic maps (Escamilla et al. 1999, 2001). The differences in markers and samples could have been responsible for the fact that previous LD analyses of the CVCR population samples (probands with BP-I and available relatives) identified associations on chromosome 18 (Escamilla et al. 1999, 2001) that were not observed in the analysis of the present data set.
Clearly, genomewide LD mapping is in its early stages, and general application for complex traits will likely require both larger samples and denser marker maps. The LD genome screen presented here has highlighted multiple regions that have overrepresented haplotypes or alleles in 10%–40% of patients with BP-I. Each of these regions could harbor a BP-I–susceptibility gene, but no genome segment is associated with disease in the majority of patients. Prioritizing these regions for future work will require not only consideration of the LD evidence from LD-T and AHR but also evaluation of BLD in nontransmitted chromosomes and visual inspection of transmitted and nontransmitted haplotypes. Regions selected for further analyses will be saturated with additional markers in this patient sample and will also be investigated in independently collected samples of patients with BP-I from the CVCR and from other populations. The results reported here should encourage implementation of population-based mapping for other complex traits in population isolates.
Acknowledgments
We thank Marjan Ophoff, for compiling the information in figure 1; Joe DeYoung, for technical assistance; and Chiara Sabatti, for helpful comments on the manuscript. R.A.O. was partly supported by the TALENT stipend of the Netherlands Organization for Scientific Research. This work was supported by grants from Millennium Pharmaceuticals, Inc., and the U.S. National Institutes of Health.
References
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300 [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Honer WG, Chow EW, Little D, Hogan J, Hodgkinson K, Bassett AS (1999) Linkage of familial schizophrenia to chromosome 13q32. Am J Hum Genet 65:1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NH, Thompson EA (2001) Linkage disequilibrium mapping: the role of population history, size, and structure. Adv Genet 42:413–447 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Escamilla MA, McInnes LA, Service SK, Spesny M, Reus VI, Molina J, Gallegos A, Fournier E, Batki S, Neylan T, Matthews C, Vinogradov S, Roche E, Tyler DJ, Shimayoshi N, Mendez R, Ramirez R, Ramirez M, Araya C, Araya X, Leon PE, Sandkuijl LA, Freimer NB (2001) Genome screening for linkage disequilibrium in a Costa Rican sample of patients with bipolar I disorder: a follow-up study on chromosome 18. Am J Med Genet 105:207–213 [DOI] [PubMed] [Google Scholar]
- Escamilla MA, McInnes LA, Spesny M, Reus VI, Service SK, Shimayoshi N, Tyler DJ, Silva S, Molina J, Gallegos A, Meza L, Cruz ML, Batki S, Vinogradov S, Neylan T, Nguyen JB, Fournier E, Araya C, Barondes SH, Leon P, Sandkuijl LA, Freimer NB (1999) Assessing the feasibility of linkage disequilibrium methods for mapping complex traits: an initial screen for bipolar disorder loci on chromosome 18. Am J Hum Genet 64:1670–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla MA, Spesny M, Reus VI, Gallegos A, Meza L, Molina J, Sandkuijl LA, Fournier E, Leon PE, Smith LB, Freimer NB (1996) Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet 67:244–253 [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwen RH, Baharloo S, Blankenship K, Raeymaekers P, Juyn J, Sandkuijl LA, Freimer NB (1994) Genome screening by searching for shared segments: mapping a gene for benign recurrent intrahepatic cholestasis. Nat Genet 8:380–386 [DOI] [PubMed] [Google Scholar]
- Kaufmann CA, Suarez B, Malaspina D, Pepple J, Svrakic D, Markel PD, Meyer J, Zambuto CT, Schmitt K, Matise TC, Harkavy Friedman JM, Hampe C, Lee H, Shore D, Wynne D, Faraone SV, Tsuang MT, Cloninger CR (1998) NIMH Genetics Initiative Millenium Schizophrenia Consortium: linkage analysis of African-American pedigrees. Am J Med Genet 81:282–289 [PubMed] [Google Scholar]
- Kendler KS, Myers JM, O'Neill FA, Martin R, Murphy B, MacLean CJ, Walsh D, Straub RE (2000) Clinical features of schizophrenia and linkage to chromosomes 5q, 6p, 8p, and 10p in the Irish Study of High-Density Schizophrenia Families. Am J Psychiatry 157:402–408 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1997) What is significant in whole-genome linkage disequilibrium studies? Am J Hum Genet 61:810–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, Rojas E, Spesny M, Baharloo S, Blankenship K, Peterson A, Tyler D, Shimayoshi N, Tobey C, Batki S, Vinogradov S, Meza L, Gallegos A, Fournier E, Smith LB, Barondes SH, Sandkuijl LA, Freimer NB (1996) A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci USA 93:13060–13065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Strahs A (1999) Assessment of linkage disequilibrium by the decay of haplotype sharing, with application to fine-scale genetic mapping. Am J Hum Genet 65:858–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlke KL, Lange EM, Valle TT, Ghosh S, Magnuson VL, Silander K, Watanabe RM, Chines PS, Bergman RN, Tuomilehto J, Collins FS, Boehnke M (2001) Linkage disequilibrium between microsatellite markers extends beyond 1 cM on chromosome 20 in Finns. Genome Res 11:1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T (1994) Diagnostic interview for genetic studies: rationale, unique features, and training, NIMH Genetics Initiative. Arch Gen Psychiatry 51:849–859 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Prathikanti S, McMahon FJ (2001) Genome scans for susceptibility genes in bipolar affective disorder. Ann Med 33:257–262 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephen M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Schork NJ, Cardon LR, Xu X (1998) The future of genetic epidemiology. Trends Genet 14:226–272 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Service SK, Ophoff RA, Freimer NB (2001) The genome-wide distribution of background linkage disequilibrium in a population isolate. Hum Mol Genet 10:545–551 [DOI] [PubMed] [Google Scholar]
- Service SK, Temple Lang DW, Freimer NB, Sandkuijl LA (1999) Linkage-disequilibrium mapping of disease genes by reconstruction of ancestral haplotypes in founder populations. Am J Hum Genet 64:1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman S, Darvasi A (2001) The value of isolated populations. Nat Genet 28:309–310 [DOI] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L (2001) New frontiers in Alzheimer's disease genetics. Neuron 32:181–184 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Shannon WD, Lathrop GM, Nolan JP, Goldin LR, Chase GA, Weeks DE (1997) True and false positive peaks in genomewide scans: applications of length-biased sampling to linkage mapping. Am J Hum Genet 61:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varilo T, Paunio T, Parker A, Meyer J, Terwilliger JD, Peltonen L (2001) Significant linkage disequilibrium (LD) cover wide intervals in alleles of the late settlement subpopulations of Finland. Am J Hum Genet 69:197 [Google Scholar]