Abstract
Research towards biomarkers that predict patient outcome in colorectal cancer (CRC) is rapidly expanding. However, none of these biomarkers have been recommended by the American Association of Clinical Oncology or the European Group on Tumor Markers. Current staging criteria result in substantial under-and over-treatment of CRC patients. Evasion of apoptosis, a characteristic feature of tumorigenesis, is known to correlate with patient outcome. We reviewed the literature on immunohistochemistry-based studies between 1998 and 2011 describing biomarkers in this pathway in CRC and identified 26 markers. Most frequently described were p53, Bcl-2, survivin, and the Fas and TRAILR1 receptors and their ligands. None of the studies reviewed provided sufficient support for implementing a single marker into current clinical practice. This is likely due to the complex biology of this pathway. We suggest focusing on the combination of key markers within the apoptosis pathway that together represent an ‘apoptotic tumor profile’, which better reflects the status of this pathway in a tumor.
Keywords: biomarkers, colorectal cancer, apoptosis, immunohistochemistry, prognosis
Introduction
Colorectal cancer (CRC) is currently one of the major contributors to cancer-related deaths worldwide.1,2 The amount of data emerging from studies aimed at optimizing the diagnostic process and treatment of this disease is rapidly increasing. This makes the process of tumor development in CRC one of the most thoroughly studied and best characterized models of tumorigenesis. By emphasizing the need of early detection and development of new and improved treatment regimens, an increased understanding of the disease led to decreased mortality rates of nearly 5 percent over the last decade.3–10 However, CRC-related morbidity and mortality affects approximately 800,000 individuals each year worldwide.2 The survival of CRC patients largely depends on disease stage at the time of diagnosis and varies widely between stages. In clinical practice, however, treatment allocation and outcome prediction is still solely based on the International Union Against Cancer (UICC) Tissue Node Metastasis (TNM) classification.11 Addition of several pathology-based tumor characteristics is currently used to identify high-risk stage II patients that may benefit from adjuvant chemotherapy. These include perforation of the bowel wall at presentation, tumor invasion at the T4 level, venous tumor invasion, lymph node yield less than 10, and poor or no differentiation of the tumor cells.12 There is substantial evidence that even with the addition of these risk factors of poor outcome, TNM classification falls short in daily practice and may cause over-or, even worse, under-treatment of patients.11,13–18
In an attempt to improve treatment outcomes for CRC patients, both the American Society of Clinical Oncology’s Tumor Markers Expert Panel (ASCO TEMP-2006) and its European counterpart, The European Group on Tumor Markers (EGTM-2007), have reviewed the available literature to determine the clinical applicability of a number of widely studied biomarkers.19–21 Their conclusions were clear and consistent: despite the overwhelming amount of literature, no biomarkers have been recommended for clinical use. Therefore, to improve current staging criteria, new biomarkers must be identified and validated for clinical use. Pepe et al22 have developed a five-step program that can be used for the development of new biomarkers. The first step is biomarker discovery in a preclinical, exploratory setting. Subsequently, the clinical value of these biomarkers must be determined and verified in a large retrospective study. These results then need to be the validated and eventually confirmed by a prospective randomized controlled trial. It is not until these steps are completed successfully that biomarkers are ready for introduction into clinical practice. The first step, which involves identifying or discovering new biomarkers, can be accomplished by studying the process of tumorigenesis and its related pathways. Cancer cells harbor at least six features that distinguish them from normal cells, one of which is the characteristic ability to evade programmed cell death or apoptosis.23 In normal tissues, apoptosis plays a pivotal role in the maintenance of tissue homeostasis and the development of the immune system.24,25 Disturbance of this process in tumor cells results in the impaired removal of mutated cells and contributes to tumor progression. In addition, evasion of apoptosis enables malignant cells to escape from tumor immune surveillance and to acquire resistance to cancer therapy. In previous retrospective studies, the status of the apoptotic pathway in a tumor was shown to be of prognostic value in colorectal cancer patients.26–37 Therefore, we focused on this pathway in our search for new potential prognostic biomarkers in colorectal cancer. In this review, we provide an overview of studies designed to determine the prognostic value of biomarkers within the apoptotic pathway in colorectal cancer. Furthermore, we will discuss some of the difficulties and controversies that can arise when studying this tightly regulated and complex process. The goal is to identify key biomarkers in the apoptotic pathway that may be used clinically to determine cancer prognosis. We first discuss the route of apoptosis to identify key proteins in this process and then link this information to studies that examined the prognostic value of these proteins in colorectal cancer. Since immunohistochemistry (IHC) is still the most widely applied and available technique in pathology to determine the expression status of tumor-associated proteins and to study the clinical prognostic relevance of biomarkers, we limited our search to IHC studies.
Data Collection and Analysis
In order to review the literature on prognostic biomarkers related to the pathway of apoptosis and determined using IHC in CRC patients, we performed a search of the PubMed, Embase, and Web of Science databases. We used broad search terms, as recommended in the Stroup guidelines,38 to identify publications of interest published between January 1998 and June 2011. Key search terms included colorectal cancer, biomarker, apoptosis, prognosis, and immunohistochemistry. The following search strategy (simplified) shows how some of these terms were combined in our Web of Science Search; “TS = ((colorectal or colon or colonic or rectal or rectum) SAME (neoplasm or cancer or tumor or carcinoma)) AND TS = ((prognostic or tumor or cancer or neoplasm or biological or intracellular or signaling or intracellular signaling) SAME (marker or protein or peptide)) AND TS = ((prognosis or prognostic or morbidity or mortality or recurrence or relapse or (disease SAME progression))) AND TS = (immunohistochemistry or immunolabeling or immunocytohistochemistry). After amalgamating the results from the three medical databases and discarding the duplicates, this strategy yielded a total of 2923 unique citations. To extract papers for review, we screened the results for title and abstract. We used the following criteria to determine whether a study was considered eligible for the review:
The study contained data for a marker directly involved in the pathway of apoptosis;
The study was performed in primary tumors from CRC patients;
The study was performed using IHC;
The study contained an analysis of the relationship between expression of the marker and clinical outcome. We selected only studies that used logistic regression or survival curve-based statistical analysis methods to evaluate the impact of a marker;
A full publication in English with details of the method used was available.
Results
Overall, we were able to identify 26 potentially prognostic biomarkers that are directly involved in the apoptotic pathway, which will be discussed in detail below (Fig. 1). These markers were all studied using IHC in the 124 eligible publications that remained after applying our selection criteria from the total of 2923 publications. Expression patterns of these apoptotic (bio)markers were related to patient outcome using logistic regression or survival curve-based analysis methods. Most of the papers of the over 800 were excluded because they described the expression of markers related to the pathway of apoptosis in other types of cancers than colorectal cancer, despite the fact that our search terms included colorectal cancer as a major search term. Over 900 citations were excluded because they did not describe the marker in primary colorectal cancer lesions but rather in metastatic lesions. Table 1 provides an overview of our selection criteria and the corresponding number of citations that were excluded based on these criteria.
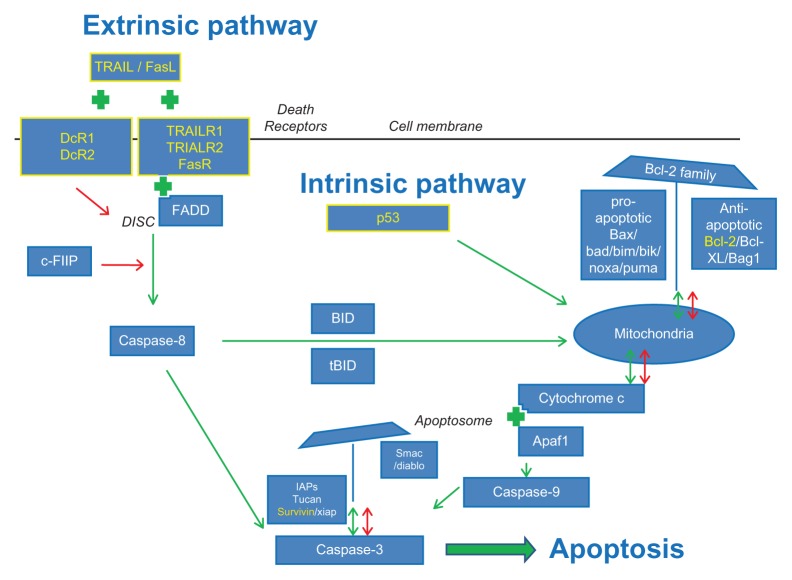
Figure 1.
The pathway of apoptosis. A simplified schematic view of the intrinsic and extrinsic pathway of apoptosis and their regulators as described in this review. A green arrow indicates a positive (stimulating or activating) effect of a regulator on a component in the pathway it points to. A green plus sign indicates that there is an interaction between two components. The combination of these two components exerts a stimulating effect on the progression of the apoptotic cascade. A red arrow indicates an inhibitory effect of a regulator on another member or on an activating step within the apoptotic pathway. The markers highlighted in yellow were found to have the most clinical prognostic significance in colorectal cancer patients based on the studies that resulted from our search of the literature. These markers, their functions, and whether they are of true prognostic value are discussed in detail in this review.
Table 1.
Selection of relevant studies on clinical prognosis of apoptosis-related markers.
| Exclusion criteria | Number of citations excluded |
|---|---|
| Exclusion criteria and the number of citations that were excluded based on these criteria (Total number of citations reviewed n = 2923) | |
| A. The study describes data on a marker not directly involved in apoptosis | 642 |
| B. The study was not performed in primary colorectal cancer patients | 985 |
| C. The study was not performed using immunohistochemistry | 707 |
| D. The study did not contain validated outcome results | 315 |
| E. An English full text version was not available | 150 |
| Total number of citations excluded based on these exclusion criteria | 2799 |
| Total number of citations included (2923–2799) | 124 |
Notes:Table 1 provides an overview of the exclusion criteria used to select the most relevant citations. The criteria were applied to the 2923 citations retrieved in our search of literature in three major online medical databases. The material and methods section provides further background on the postulation of these criteria and outline of our literature search. The number of citations excluded from further analyses based on each criterion is listed. Based on the criteria A, B, C, D, and E, 2799 citations were excluded from the selection. Therefore, 124 citations remained for in depth review of the prognostic value of the markers studied.
The general pathway of apoptosis is illustrated in Figure 1 and includes the markers discussed in this review. Although this figure represents a simplified version of the pathway, it shows that the process of apoptosis is highly regulated at multiple levels. Based on the stimulus presented, two pathways initiating the apoptotic process can be identified.39 The extrinsic pathway is triggered by external death signals that cause the formation of intracellular signaling complexes at the death receptors. This type of apoptosis is typically activated in immune responses.40 The second pathway, known as the intrinsic pathway, is activated by many different stimuli, including growth factor deprivation and DNA damage, caused by factors such as UV or gamma-irradiation or by chemotherapeutic agents. Exposure of cells to these stimuli initiates a set of intracellular death signals mediated by the p53 protein that activates the apoptotic process. Mitochondria play an important role in the intrinsic pathway with a major regulatory role for the Bcl-2 family members. Although already intimately connected via caspase-8 and Bid, both pathways converge at the level of the caspase cascade that eventually leads to the proteolytic activation of executioner members such as caspase-3.41 The function of caspase proteins is again highly regulated by a group of inhibitor of apoptosis proteins (IAPs).42 In general, executioner caspases will cleave several substrates, thus acting as a cellular disassembly machine. Cleavage of these substrates is eventually responsible for the morphological features that hallmark apoptotic cell death and include membrane blebbing, cell shrinkage, and chromatin condensation.26,41
To discuss the results of our literature review, markers are grouped and discussed based on their locations in the pathway as described in Figure 1, starting with the extrinsic pathway of apoptosis and ending with the IAPs. In addition, Table 2 provides a general overview of the number of studies identified that describe the prognostic value of a particular marker, grouped by their function and location in the pathway of apoptosis in the order at which they will be discussed in this review.
Table 2.
Overview of markers of the apoptosis pathway.
| Marker | Function in the pathway | Number of studies | References |
|---|---|---|---|
| Overview of the markers reviewed, the total number of citations including the references that described the prognostic relevance of these markers | |||
| Intrinsic pathway | |||
| p53 | Triggers the intrinsic pathway | 31 | 31,71,73–76,76,90,113,113–136* |
| Bcl-2 | Anti-apoptotic Bcl-2 family member | 38 | 29,31,35,73,83–90,94,113,114, 116,118,123,125,128,129,132, 134,137–151 |
| Bcl-XL | Anti-apoptotic Bcl-2 family member | 1 | 90 |
| Bag1 | Enhancing anti-apoptotic function of Bcl-2 | 2 | 90,151 |
| Apaf-1 | Formation of the apoptosome | 8 | 90,113,118,121,152–155 |
| Bax | Pro-apoptotic Bcl-2 family member | 8 | 90,123,123,146,147,156–159 |
| Bad | Pro-apoptotic Bcl-2 family member | 1 | 160 |
| Bid | Pro-apoptotic Bcl-2 family member | 2 | 90,160 |
| Bim | Pro-apoptotic Bcl-2 family member | 1 | 161 |
| Noxa | Pro-apoptotic Bcl-2 family member | 1 | 161 |
| Puma | Pro-apoptotic Bcl-2 family member | 1 | 161 |
| Caspase-8 | Initiator caspase of instrinsic pathway | 2 | 60,155 |
| Extrinsic pathway | |||
| FasR | Death receptor | 2 | 56,57 |
| TrailR1 | Death receptor | 2 | 57,59 |
| TrailR2 | Death receptor | 2 | 57,59 |
| DcR1 | Decoy death receptor | 1 | 57 |
| DcR2 | Decoy death receptor | 1 | 57 |
| FasL | Death receptor ligand | 1 | 56 |
| TRAIL | Death receptor ligand | 3 | 57,59,60 |
| c-Flip | Inhibitor of extrinsic apoptosis induction | 2 | 56,60 |
| Caspase-9 | Initiator caspase of the extrinsic pathway | 1 | 155 |
| Cascade regulator | |||
| IAP | Inhibitor of the caspase cascade | 3 | 90,162,163 |
| Survivin | Inhibitor of the caspase cascade | 8 | 35,90,94–97,99,108 |
| Tucan | Inhibitor of the caspase cascade | 1 | 90 |
| XIAP | Inhibitor of the caspase cascade | 2 | 90,164 |
| Smac/Diablo | Inhibitor of the IAPs, pro-apoptotic | 3 | 90,163,165 |
Notes: Table 2. Listed are biomarkers related to the pathway of apoptosis that emerged from the review of the literature. The biomarkers were studied by immunohistochemical analyses and their expression was related to clinical outcome in colorectal cancer patients. For each marker, the number of publications in which these markers were studied and their primary function within the pathway of apoptosis are listed.
The systematic review of Munro et al, in which the available literature on the prognostic value of p53 expression until 2005 was reviewed extensively, was used as a starting point for our search in of the IHC literature on p53 expression. We therefore only included studies published since this review.
The death receptor family and the extrinsic pathway of apoptosis
The most studied signaling pathway in apoptosis is the extrinsic pathway, activation of which is primarily facilitated by the death receptors (DRs). Based on our search, we were able to identify 8 individual biomarkers in this part of the apoptotic pathway for which the prognostic relevance was studied in CRC patients. These markers included Fas receptor (FasR), TRIAL1, TRAILR2, TRAILR3, TRAILR4, FasL, TRAIL, and c-Flip. (Table 2). A unique feature of the extrinsic pathway is that DRs can induce apoptosis independently of the p53 tumor suppressor gene. DRs are members of the tumor necrosis factor receptor superfamily, of which eight family members have been characterized.43,44 The most common receptors are FasR (CD95, DR2) with its ligand FasL and TRAILR1 and -R2 (DR4 and DR5) with their ligand tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL). All DRs contain cysteine-rich extracellular domains that allow them to recognize their ligands with great specificity. They also harbor intracellular sub-domains better known as the death domains (DDs).45 DDs allow them to interact with adapter molecules such as FADD (Fas-associated death domain).46 Signal transduction within the extrinsic pathway starts with the binding of the ligands to the DRs, followed by the formation of multi-protein signaling complexes called death inducing signaling complexes (DISCS) at the intracellular domains of the DRs.47,48 The DISC complex auto-activates pro-caspase 8 through an interaction with a FADD protein. Activated caspase-8 will eventually activate the effector caspase-3 by proteolytic cleavage.48–51 Downregulation of any of the DRs or downstream apoptotic proteins can cause severe limitations in the induction of apoptosis through the extrinsic pathway. There are two other mechanisms involved in the regulation of the extrinsic signaling pathway. First, TRAIL can also bind to two decoy receptors (DcR) in addition to the DRs, including DcR1 and DcR2 (also known as TRAILR3 and TRAILR4, respectively). However, neither decoy receptor can transduce an apoptosis-stimulating signal upon TRAIL binding. The sensitivity of a cell to TRAIL-mediated apoptosis may, therefore, be a function of the ratio of DcR to DR. If there is significant upregulation of the DcRs or downregulation of the DRs, TRAIL will bind to the DcRs instead of the DRs, and the apoptotic signaling is interrupted.52 Second, cellular Flice-like inhibitory protein (c-FLIP) is, similarly to FADD, a DD-containing protein and can competitively bind to FADD in the DISC formation process instead of the DD domain of the DRs (Fig. 1). This protein, particularly in the c-FLIPL isoform, shows strong structural similarities to pro-caspase-8, and may be a potentially strong inhibitor of the extrinsic apoptotic pathway. There are two other features that are unique to the extrinsic pathway, but contribute considerably to the complexity of its regulation. The first feature is an indirect link with the intrinsic pathway, which can be activated through the formation of tBid, a truncated form of the BID protein. In a subset of cells known as type II cells, DISC formation occurs less frequently, resulting in lower caspase-8 activation and subsequently truncation of the Bid protein into tBid.53 tBid induces oligomerization of Bax or Bad, upon which the mitochondria release cytochrome c; this eventually induces apoptosis further down the intrinsic pathway. Because of the mitochondrial involvement apoptosis regulation in Type II cells is subject to regulation by the Bcl-2 family proteins. This regulation, which will be discussed in detail in this review, provides the cell with an apoptosis-evading mechanism such as downregulation of DR expression that may occur in a cell during tumorigenesis.54 A second feature unique to the extrinsic pathway is explained by the so-called ‘Fas-counterattack hypothesis’.55 In normal tissue homeostasis, the Fas/FasL-induced extrinsic pathway of apoptosis plays a major role in immune surveillance. Activated T lymphocytes express FasL and upon recognition of a tumor cell as a target (via MHC-presented peptides on the tumor cell surface), a tumor cell expressing the FasR may be eliminated by induction of apoptosis. However, it is known that tumor cells can also express FasL and thus are able to counterattack cells from the immune system.55 By downregulation of FasR expression as well as by upregulation of FasL expression, tumor cells can escape immune surveillance.
In summary, the key players of the extrinsic pathway are the DRs, specifically TRAILR1, R2, and FasR and their ligands. Tumor cells attempt to disrupt signaling through these DRs to overcome apoptosis, which has been widely studied in many types of cancers. Our search identified 12 studies in which one or more of these DRs and their ligands were studied. In 5 of the 12 studies, one of the DR pathway-related markers (FasR, FasL, TRAILR1, and TRAIL) was found to be of significant prognostic value (Table 3).56–60 Hypothetically, based on the biology of the process of tumorigenesis, downregulation of DR expression or upregulation of expression of their ligands indicate a more aggressive tumor type, and hence worse clinical outcome parameters. Interestingly, most studies reported that upregulation of the expression of Fas and TRAIL was significantly related to worse outcome parameters. The expression of FasL and FasR was studied by both Korkolopoulou et al56 and Strater et al.58 In a smaller study by Korkolopoulou et al56 involving 90 patients, normal cells did not express FasL, but tumor cells showed significant upregulation, which was related to a significantly lower overall survival (OS). Tumor cells also showed expression of the Fas receptor with a mainly cytoplasmatic and granular staining pattern. According to the authors, this indicates that although the Fas receptor was present, it had no true functional properties. Therefore, according to the authors, the seemingly contradictory result of a worse outcome despite upregulation of DR expression could be explained by the Fas-counterattack hypothesis. In the second study by Strater et al,58 overexpression of the Fas receptor correlated with a significantly better disease-free survival (DFS). Unfortunately, this study did not describe the exact location of FasR expression in the cell. Therefore, it is difficult to determine whether their results confirm Korkolopoulou results or actually oppose them. We were therefore not able to determine whether the Fas-counterattack hypothesis has true clinical value in CRC.
Table 3.
Extrinsic pathway of apoptosis.
| Marker | Reference | Population size | Tumor type | Disease stage | Expression | Outcome parameter | Hazard ratio | P-value |
|---|---|---|---|---|---|---|---|---|
| Overview of the literature on statistically significant, prognostic markers within the extrinsic part of the apoptotic pathway | ||||||||
| FasR | Strater58 | 128 | Colon | II–III | Up | DFS | 0.4 | 0.034 |
| FasL | Korkolopoulou56 | 90 | Colon | I–IV | Up | OS | 3.491 | 0.005 |
| TRAILR1 | Van Geelen59 | 376 | Colorectal | III | Up | REC | 2.19 | 0.03 |
| Strater57 | 129 | Colon | II–III | Up | OS | 2.22 | 0.04 | |
| DFS | 2.59 | 0.003 | ||||||
| TRAIL | McLornan60 | 253 | Colorectal | II–III | Up | OS | 1.210 | 0.026 |
Notes:Table 4. This table provides the references of the studies that describe the prognostic value of FasR, FasL, TRAILR1, or TRAIL, identified in this review. For each marker the important study characteristics are listed.
Abbreviations: DFS, disease-free survival; OS, overall survival; REC, recurrence; Up, upregulation of the expression of the marker.
With respect to DR4 and its ligand TRAIL, we could identify three studies reporting the prognostic value of these biomarkers in CRC.57,59,60 In all studies, upregulation of expression of DR4 or its ligand were related to worse outcome parameters, such as higher levels of recurrence and shorter OS. This apparent contradiction with expectations based on biology of tumorigenesis can, according to Van Geelen et al, be explained by the fact that DR4 is also known to have effects on cell proliferation through the activation of nuclear factor kappa B (NF-κB), as described in a number of studies.60–63
In conclusion, we were able to identify five studies reporting FasL, FasR, TrailR1, or TRAIL 2 as significant prognostic markers in colorectal cancer patients. Conclusions varied, which may be due to differences in patient selection and/or study methods. However, more importantly, their conclusions were in contradiction with what is expected based on the biology of the apoptosis pathway. This can be explained by the fact that the functionality of this extrinsic part of the apoptotic pathway in the included studies was only investigated based on changes in protein expression patterns of the tumor cells. The involvement of the immune system was not considered in most of these studies. The process of tumorigenesis attracts many cells that are part of the immune system into the tumor microenvironment. The presence of these cells such as activated CD8+ T cells or Foxp3+ regulatory T cells has been shown to be of prognostic relevance in CRC.64,65 Moreover, activated T cells produce CD95 ligand and can thereby trigger apoptosis in target cells such as tumor cells.66 As described above, some tumors cells may be able to counteract this mechanism and remove attacking antitumor T cells by increasing their own CD95 L expression. However, this counterattack theory has not yet been conclusively demonstrated in vivo. Therefore, until additional preclinical, exploratory research has been performed to clarify how the pathway of apoptosis and the immune system interact, none of the related markers appear suitable for clinical prognostic application.
p53 Tumor suppressor gene and the intrinsic pathway of apoptosis
The p53 tumor suppressor gene, likely the most well-known protein within the intrinsic pathways of apoptosis, encodes for a transcription factor that regulates the expression of genes involved in the pathway of apoptosis, as well as angiogenesis, cell cycle progression, and genomic maintenance.67,68 Within the intrinsic pathway, it exerts its function at the beginning of the intrinsic apoptotic pathway. p53 causes cell cycle arrest at the G1 phase in response to DNA damage; in case the DNA damage turns out to be irreparable, the p53 protein will activate the appropriate cellular signaling cascades to execute apoptosis. In 50% of human colorectal cancers, p53 is absent or mutated, which has major implications for the execution of apoptosis in colorectal cancer.69 Mutations in p53 can be determined using IHC since mutated proteins accumulate in the nucleus due to their increased half-life.70 Different mutations have varying effects and can implicate either loss or gain of function of the p53 protein. Two research groups carried out major systematic reviews on the relationship between p53 abnormalities and outcome in colorectal cancer patients.71,72 Munro et al71 reviewed a total of 168 IHC-based studies as well as mutation-based studies. Russo et al72 pooled data from studies analyzing p53 DNA mutations only. Together, these studies reported on p53 expression and mutations in relation to survival in 18,766 patients. Their key finding was that abnormal expression of p53, detected using IHC, was related to an increased risk of death. They concluded that mutations in exon 5 were associated with an adverse outcome, predominantly in proximal, right-sided tumors. Both studies suggested an impact of mutated p53 on clinical outcome, though this relationship was only modest despite the overwhelming amount of data analyzed. The results of these studies were taken into account in the review of prognostic biomarkers in CRC by the ASCO Clinical Oncology’s Tumor Marker Expert Panel in 2006. The ASCO panel’s recommendation was that with current methods of detection, using either mutation analysis or IHC, p53 status was a poor guide for predicting prognosis in colorectal cancer patients.19 Since the published literature on IHC-based p53 studies until 2005 has already been thoroughly reviewed by Munro et al, we limited our search to reports that have been published since. We identified an additional 30 studies reporting on the expression of p53 determined using IHC that were published after Munro’s review in 2005. Four of these 30 papers confirmed the results of Munro et al and claimed p53 to be a significant, prognostic marker in colorectal cancer.73–76 Table 3 provides an overview of their study characteristics. From this table, it can be concluded that although p53 has been widely studied, the studies reporting on statistically significant prognostic relevance show differences in population size, tumor type selection, and disease stage selection. When we examined these publications in more detail, we also noticed that they varied in their IHC methods. Previously, a comparative study by Baas et al77 demonstrated the monoclonal antibody DO7 to be superior over 5 other antibodies for detecting p53 gene protein in archival tissue of colorectal carcinomas, suggesting that this antibody should be used as a gold standard for IHC of p53. This particular antibody was used in 3 out of the 4 studies listed.73,75,76 Furthermore, the sample sizes of all studies rather small at an average of 103 patients, and a closer look at these populations showed that most also seems highly selected on clinical parameters. For example, Jurach et al73 only included stage II and III rectal cancer patients and the small cohort of Torsello et al74 consisted only of patients under age 40. The only relatively large study by Lim et al75 that analyzed the results of 231 stage I, II, and III CRC patients, showed a correlation between upregulation of p53 expression and poor OS. This correlation was more pronounced in their stage III patient selection, but disappeared when only the adjuvant-treated patients were analyzed. Their results appeared to be confirmed by the study of Noske et al.76 However, in this study, the prognostic value of p53 was only present in a multivariate analysis when expression was analyzed in combination with p21 expression, a major downstream cell cycle inhibitor. The expression of p53 alone in univariate analysis was only borderline-significant at a P-value of 0.045 in this cohort of 116 stage II/III patients. As a single marker, p53 expression showed no independent statistical significance with respect to the prediction of outcome. Hence, neither of the four studies, although they claimed p53 to be an independent prognostic predictor of outcome in CRC, were able to add more significance to the conclusions drawn by Munro et al.71 Therefore, their results are likely not sufficiently significant to alter the recommendations of the ASCO of 2006 with respect to the applicability of p53 as a prognostic biomarker in colorectal cancer.19
Bcl-2 family members and the intrinsic apoptotic pathway
Downstream of p53, the mitochondria play a major role in the initiation and execution of the intrinsic pathway of apoptosis. The B-cell CLL/lymphoma 2 (Bcl-2) family members are mainly responsible for regulating the intrinsic pathway and can be categorized into two groups. The first group consists of antiapoptotic proteins that are structural and functional homologs of Bcl-2. The most important members of this group are Bcl-2 itself and its splice variant Bcl-2 XL.78,79 They are mainly bound to the mitochondrial outer membrane (MOM) by their transmembrane (TM) domain, where they stabilize the MOM to prevent cytochrome c release into the cytosol of the cell under normal homeostatic circumstances.80 Therefore, they can be considered anti-apoptotic proteins.81
The second group of Bcl-2 family members has proapoptotic capacities. These members include Bcl-2 associated X protein (BAX) and proteins such as Bad, Bid, Bim, Bik, Noxa, and Puma, which are, based on their structures, also known as BH3-only proteins.78 These proteins are typically bound to the cytoskeleton or cytosol, but upon stimulation they interact with and inhibit their anti-apoptotic counterparts such as Bcl-2.78 The relative ratio or balance between the expression of both groups of Bcl-2 family members will determine whether stimulation of the intrinsic pathway of apoptosis results in apoptosis as is graphically pointed out in Figure 1. If the pro-apoptotic factors predominate, cytochrome c will be released into the cytosol where it binds to the apoptosis-activating factor 1 (Apaf-1) to form an apoptosome. More downstream in the apoptotic pathway, this apoptosome will form a complex together with an initiator caspase, caspase-9. This caspase will subsequently activate the executioner caspases, caspase-3 and -7.78,82 Deregulation of apoptosis during tumor development can be caused by a disturbance in the homeostatic balance of the Bcl-2 family members.
Our search resulted in 55 studies describing the prognostic relevance of markers related to the Bcl-2 protein family. In most of these studies, 38 in total, the expression of Bcl-2 was examined using IHC. In only 9 out of these 38 studies, a statistically demonstrated prognostic relevance of this marker could be established (Table 5).73,83–90 The 9 studies generally used the same methods to determine Bcl-2 expression and all but one were performed on whole paraffin-embedded tissue sections. Furthermore, they were very consistent in their conclusions: in all studies, upregulation of Bcl-2 was related with better survival, as shown for either disease-free, overall, or recurrence-free survival. This is contradictory to is assumed given the anti-apoptotic function of Bcl-2: upregulated Bcl-2 expression should be more likely to be a marker of a more aggressive tumor phenotype. The most plausible explanation for this paradoxical finding is the fact that Bcl-2 not only has an anti-apoptotic function, it can also exert a distinct negative influence on cell cycle progression, which can eventually slow down tumor growth. This may explain the survival benefit for several patients with upregulated Bcl-2 expression.83–85,89 Whether the anti-cell cycle progression or the anti-apoptotic role of Bcl-2 predominates during tumorigenesis may depend on disease stage. In early carcinogenesis, the anti-apoptotic function of Bcl-2 plays a large role, causing genetic alterations to accumulate. In later stages, Bcl-2 functions more as a cell cycle progression inhibitor, lowering the rate of tumor proliferation. This hypothesis is supported by the inverse correlation between Bcl-2 and the percentage of cells in S-phase found by Buglioni et al.83 The studies describing the prognostic relevance of Bcl-2 would therefore be more informative if disease stage and the expression of other family members were taken into account. This would provide us additional insight in the biological function and effects on the apoptotic pathway of Bcl-2, which will tremendously improve the interpretation of the results.
Table 5.
Bcl-2 as a clinical prognostic marker.
| Reference | Population size | Tumor type | Disease stage | Expression | Outcome parameter | Hazard ratio | P-value |
|---|---|---|---|---|---|---|---|
| Overview of the studies reporting on Bcl-2 as significant, prognostic marker within the pathway of apoptosis | |||||||
| Buglioni83 | 171 | Colorectal | I–IV | Down | DFS | 5.61 | 0.0009 |
| OS | 5.21 | 0.0063 | |||||
| Schwander84 | 160 | Colorectal | I–III | Up | REC | – | 0.0242 |
| Chatla85 | 158 | Colorectal | II–III | Down | OS | – | 0.0012 |
| Sinicrope89 | 63 | Colorectal | I–II | Up | RFS | 0.23 | 0.04 |
| 154 | Colon | II | Up | OS | 0.17 | 0.03 | |
| RFS | 0.45 | 0.04 | |||||
| Krajewska90 | 106 | Colorectal | II | Up | OS | 0.25 | 0.0009 |
| Leahy86 | 102 | Colorectal | I–III | Up | OS | 0.5 | 0.005 |
| Ilyas87 | 66 | Colorectal | II | Up | REC | 0.77 | 0.02 |
| Torsello73 | 58 | Colorectal (<40 years age) | IV | Down | OS | 3.02 | 0.015 |
| Elkablawy88 | 52 | Colorectal | I–IV | Up | OS | – | 0.016 |
Notes:Table 5. A list of 9 studies, identified by this review, describing a statistically significant, prognostic effect of Bcl-2 in colorectal cancer patients. For each maker, the important study characteristics are listed.
Abbreviations: DFS, disease-free survival; OS, overall survival; REC, recurrence; RFS, recurrence-free survival; Up, upregulation of marker expression; Down, downregulation of marker expression.
Inhibitors of apoptosis family proteins and the execution of apoptosis
The actual apoptotic cell death machinery, responsible for the execution of apoptosis and resulting in the morphological features characteristic of apoptosis, consists of a very complex cascade of interacting proteins. The key components include the caspase proteins, as described above. At many levels, regulation takes place to ensure appropriate functioning of the caspase machinery. Key regulators of the caspase cascade are the inhibitors of apoptosis proteins (IAPs) that exert their function through binding of activated caspases. Thus far, 8 IAPs have been identified in mammals, the most well-known being livin, X-linked inhibitor of apoptosis (XIAP), and survivin.26,91,92 All IAP family proteins have one or several specific Baculoviral IAP repeats (BIRs). They require at least one BIR to exert their anti-apoptotic function. The function of the IAPs is also strictly regulated by their own set of inhibitors such as Smac/Diablo and Omi/HtrA2.26 Under normal circumstances, when apoptotic stimuli are present, cells release Smac/DIABLO from their mitochondria into the cytosol, where the complex exerts its pro-apoptotic effect by interacting with the IAPs in order to release bound caspases into the cytosol.93 The most frequently studied IAP in our search results was survivin, likely because the role of survivin in apoptosis has been the subject of controversy over the last few years. Of the 8 studies describing the prognostic effects of survivin, 4 demonstrated a statistically prognostic effect of survivin (Table 6).35,90,94,99 Initially, it was thought that survivin and the other IAPs selectively bind active caspase-3/-7, and -9, promoting their degradation and thereby inhibiting apoptosis.100 Survivin, however, lacks the structural motif to bind to caspases and likely only inhibits activated caspase-9 with the help of XIAP.101–103 In contrast to other IAPs, survivin is undetectable in normal adult tissues, but abundantly expressed in transformed cell types and a variety of human cancers, such as cancers originating in the colon, stomach, pancreas, lung, prostate, and breast.104 Although all four studies were able to show a significant relation between survivin expression and clinical outcome, the direction of this effect was not the same. When looking at these studies in detail it was noticed that they did not apply the same methods of analysis of the IHC results. This is of importance because survivin can be expressed in two cellular compartments: either in the cytoplasm or in the nucleus with different functions.98,105,106 In general, survivin is known to be involved in the regulation of cell viability as well as in the regulation of cell division. It is hypothesized that the nuclear subset is involved in controlling cell proliferation and the cytoplasmatic pool is more involved in regulating cell survival.107 Sarela et al94 found a relationship between survivin expression and a shorter DFS when scoring mainly the cytoplasm for survivin positivity. Ponnelle et al95 showed a positive influence of both cytoplasmatic and nuclear expression on survival in a very small patient population of only 46 patients. This only reached statistical significance for the cytoplasmic group. Fang et al108 showed a negative effect of survivin expression on OS, disease recurrence, and the development of liver metastasis. The same was true for the study by Sprenger et al99 in which pre-treatment biopsies of rectal cancer patients were analyzed for their survivin expression. In this study, low pre-treatment expression was related to a significantly better DFS. Unfortunately, neither of the groups elaborated on the specific location in the cell at which they scored survivin expression. The image of a tissue microarray (TMA) core that was immunohistochemically stained for survivin expression provided in the publication by Fang et al108 suggests that the staining pattern was predominantly cytosplasmatic. In conclusion, it appears that localization of survivin expression is in fact of great importance as it is likely related to the protein function and hence should be taken into account in future studies. This may be applicable to all of the other IAP family members since none have been studied widely with standardized scoring methods in large series.
Table 6.
Survivin as a clinical prognostic marker.
| Reference | Population size | Tumor type | Disease stage | Expression | Outcome parameter | Hazard ratio | P-value |
|---|---|---|---|---|---|---|---|
| Overview of the studies reporting on survivin as significant, prognostic marker within the pathway of apoptosis | |||||||
| Sarela94 | 49 | Colorectal | II | Up | OS | 9.1 | 0.03 |
| Ponnelle95 | 46 | Colorectal | I–IV | Up | OS | 0.35 | 0.045 |
| Fang108 | 630 | Colon | I–IV | Up | OS | 1.63 | 0.018 |
| Sprenger99 | 116 | Rectal | II–III | Down | DFS | – | 0.038 |
Notes:Table 6. This table provides the characteristics of all of the studies, identified by this review, that describe a significant prognostic effect of survivin expression determined by immunohistochemistry in colorectal cancer patients. For each marker, the important study characteristics are listed.
Abbreviations: OS, overall survival; Up, upregulation of marker expression; Down, downregulation of marker expression.
Discussion
This review gives an overview of the literature published on IHC-based prognostic biomarkers related to the pathway of apoptosis in colorectal cancer between January 1998 and June 2011. Particularly, we discussed those markers that were proven to be of independent, statistically significant, prognostic value by placing them in the context of their function within the apoptotic pathway. Based on this biological background information, we then analyzed the conclusions drawn by the authors of the studies included to determine whether their conclusions could provide valuable grounds to proceed investigating these markers for prognostic clinical application. The markers we discussed are all major regulatory players in this pathway such as p53, the Fas receptor, and the DR4 with their respective ligands, the Bcl-2 protein family, and survivin. In general, we concluded that none qualified as a single prognostic biomarker for colorectal cancer patients, despite the fact that it has been well-established that the outcome of the pathway of apoptosis is of prognostic value.29–37,99 Based on the information derived from all of the studies discussed, we postulate several explanations for this lack of sufficient clinical prognostic significance for individual markers of the apoptotic pathway. First, study characteristics of investigations of one specific biomarker varied widely. This sometimes provided a marker with prognostic significance, but only in a highly selected group of patients. For example, in the case of p53, some studies were able to reproduce the results of the review of Munro et al,71 but only when a selected patient cohort was studied which consisted of patients with better survival rates after surgery, as was the case in the study by Lim et al.75 In this study, the prognostic value of p53 was only present in the stage II/III cohort when both adjuvant and non-adjuvant treated patients were included, which were probably the patients with such good outcome perspectives after surgery that adjuvant treatment was deemed unnecessary. The prognostic value disappeared when only the adjuvant-treated cases were analyzed. This makes the applicability of p53 as a general prognostic marker at time of diagnosis and treatment allocation questionable for the entire colorectal cancer patient population. Second, well-standardized IHC protocols were applied for none of the markers. Possibly even more importantly, there seemed to be no standardized methods for quantifying expression level or a specific location in the cell at which the expression of a marker should be evaluated. This may greatly influence the interpretation of the results of these studies, as for instance the specific location of expression in a cell may be directly related to its biological function and thereby to its effect on the outcome of the apoptotic pathway and patient prognosis. Caution should be taken in the method of quantification, particularly for FasR and survivin expression. In the case of the FasR expression, a high level of expression found in the cytoplasm but not on the cells membrane can indicate the presence of decoy receptors.56 This may be an indication that the so-called Fas-counterattack hypothesis is indeed true, making it essential to evaluate the subcellular location of expression.55 In addition, in the case of survivin, staining location appeared to be significant since both nuclear and cytoplasmic expression showed a different relationship with outcome in the studies reviewed.94,95,99,108 We concluded that location of expression may implicate different biological functions of the same protein. Third, we showed that the pathway of apoptosis is strictly regulated at several levels by both stimulatory and inhibitory proteins that highly interact with each other. Using only one marker to describe the outcome of these interactions, therefore, seems inappropriate. To complicate matters even more, the function of a protein can differ depending on progression of the process of tumorigenesis as was shown for Bcl-2 expression.83 Intervention of key proteins of apoptosis in other pathways makes cellular outcome unpredictable when using expression of single proteins as prognostic biomarkers.
Overall, we can conclude that studying outcome of the pathway of apoptosis, or deciding on a patient’s prognosis and treatment, using single markers seems inappropriate given the complexity of this pathway. We already highlighted the delicate balance that exists between expression and effects on apoptosis of the members of the Bcl-2 pathway and of the IAP’s and their inhibitors.78,83 To interpret the effect of expression of single proteins without the knowledge of the expression status of any of the others involved seems inappropriate. Therefore, we suggest improving the clinical applicability of these markers. Most importantly, we suggest that the apoptotic profile of a tumor should be determined rather than expression of single markers. This should include several markers that together represent the outcome of all regulatory thresholds within the pathway. An apoptotic profile would better represent the true function of the markers involved and provide insight on the outcome of the apoptotic pathway in individual tumors; thus, an apoptotic profile fulfills the need for prognostic biomarkers in colorectal cancer.22
In the literature studied for this review, several authors made an attempt to establish such a multi-marker apoptotic phenotype.59,60,83,99 However, in these studies, the selection of biomarkers was based on the markers that showed some prognostic relevance in their series as a single biomarker without any respect to the biology of the pathway. In addition, in most cases, the markers used in these ‘multi-marker phenotypes’ all belonged to just one sub-regulatory unit of the pathway. For example, Van Geelen et al59 and McLornan et al60 both studied multiple markers with respect to the DRs, but did not include any of the more downstream regulator proteins. Furthermore, in many of the studies the statistical methods used to analyze the results of these multi-marker phenotypes lacked power. A solution to this problem may be to approach the data as one would do in the case of gene expression array data by performing hierarchical clustering in order to develop a profile.109 Until then, the biomarkers that will eventually make up this apoptotic profile remain to be determined. In the meantime, we suggest that further studies focus on analyzing the clinical relevance of not only the outcome of (de)-regulation of the apoptotic pathway in colorectal cancer, but also on the outcome of the (de)-regulation of the pathway of proliferation. Under normal circumstances, a key factor in tissue homeostasis is the balance that exists between the level of cell death and the level of cell proliferation.110,111 Deregulation of either of these pathways can therefore cause disturbance of this balance, which may result in and maintain tumorigenesis. This hypothesis has been studied previously with success in a cohort of 100 colorectal cancer patients in which an AI:PI ratio was determined.112 This Apoptotic Index: Proliferation Index, based on M30 IHC for the level of apoptosis and Ki67 IHC for the proliferation index, was significantly related to patient outcome. It remains to be determined whether studies in larger patient populations will confirm these results.
In conclusion, to determine the prognostic relevance of biomarkers involved in apoptotic pathways in colorectal cancer using immunohistochemistry, multiple markers that together reflect the apoptotic status in individual tumors should be studied together. The introduction of such a multi-marker apoptotic phenotype or -profile into clinical practice demands standardization of technical assays and quantification methods. For future studies, therefore, we recommend considering the full pathway when beginning an exploratory phase towards the discovery of new biomarkers in colorectal cancer related to outcome of the apoptotic process. Moreover, we recommend applying hierarchical clustering-based statistical analysis and using knowledge regarding the biology of the pathway to identify promising markers. Furthermore, we recommend taking into account markers representing the pathway of proliferation when studying the prognostic effects of the apoptotic pathway in colorectal cancer. These measures will lead to multi-marker profiles that can then be validated in large retrospective studies and can ultimately be introduced into clinical practice.
Supplementary Data
List of abbreviations (in order of appearance)
- IHC
Immunohistochemistry
- CRC
Colorectal Cancer
- ASCO
American Association of Clinical Oncology
- EGTM
European Group on Tumor Markers
- Bcl-2
B-cell Lymphoma 2
- P53
Protein 53
- TRAILR1
Tumor necrosis factor Related Apoptosis Inducing Ligand Receptor 1
- UICC
International Union Against Cancer
- TNM
Tissue Node Metastasis
- ASCO-TEMP
American Association of Clinical Oncology Tumor Markers Expert Panel
- DNA
Deoxyribonucleic Acid
- UV
Ultraviolet
- IAP
Inhibitor of Apoptosis Protein
- DR
Death Receptor
- FasR
Fas Receptor
- TRAILR2/R3/R4
TRAIL Receptor 2/3/4 (see TRAILR1)
- FasL
Fas Ligand
- c-Flip
Cellular FADD Like Interleukin 1 beta-converting Enzyme Inhibitory Protein (see FADD)
- TNFR
Tumor Necrosis Factor Receptor
- DD
Death Domain
- FADD
Fas-Associated Death Domain
- DISC
Death Inducing Signalling Complexes
- DcR1/2
Decoy Receptor 1/2
- MHC
Major Histocompatibility Complex
- OS
Overall Survival
- DFS
Disease-Free Survival
- NF-κB
Nuclear Factor kappa-lightchain-enhancer of activated B cells
- CD8
Cluster of Differentiation 8
- CD95
Cluster of Differentiation 95
- T cell
Thymus cell
- CD95 L
CD 95 ligand (see CD 95)
- Bcl-2 XL
Bcl-2 Extra Large (see Bcl-2)
- MOM
Mitochondrial Outer Membrane
- TM
Transmembrane
- BAX
Bcl-2-Associated X protein (see Bcl-2)
- BAD
Bcl-2-Associated Death Promoter (see Bcl-2)
- Bim
Bcl-2 like interacting mediator of cell death (see Bcl-22)
- Bid
BH3 interacting domain death agonist (see BH3)
- Puma
P53 upregulated modulator of apoptosis
- TUCAN
Tumor-upregulated CARD containing antagonist of caspase 9 (see CARD)
- CARD
Caspase Recruitment Domain
- BH3
Bcl-2 Homology 3 (see Bcl-2)
- Apaf-1
Apoptosis-Activating factor 1
- S-phase
Synthesis phase
- XIAP
X-linked IAP (see IAP)
- BIR
Baculovirus IAP Repeat (see IAP)
- SMAC
Second Mitochondria-Derived Activator of Caspases
- DIABLO
Direct IAP Binding protein with Low pI (see IAP)
- HtrA2
High temperature requirement protein A2
- TMA
Tissue Micro Array
- N
Number of patients in study population
- Bag1
Bcl-2-associated Athanogene 1
- REC
Recurrence
- RFS
Recurrence-Free survival
Table 4.
P53 as a clinical prognostic marker.
| Reference | Population size | Tumor type | Disease stage | Outcome parameter | Hazard ratio | P-value |
|---|---|---|---|---|---|---|
| Overview of the literature on p53 expression in which p53 was shown to be an independent prognostic indicator of outcome in CRC patients | ||||||
| Noske76 | 116 | Colorectal | III | OS | – | 0.048 |
| Munro71,* | 12257 | Colorectal | I–IV | OS | 1.32 | <0.0001 |
| Torsello73 | 58 | Colorectal (<40 years age) | I–IV | OS | 2.48 | 0.046 |
| Lim75 | 213 | Colorectal | I–III | OS | 1.843 | 0.028 |
| Jurach74 | 83 | Rectal | II–III | OS | 2.32 | 0.01 |
| DFS | 2.45 | 0.04 | ||||
Notes:Table 4 provides the references of all the studies that were identified in this review to report on upregulation of p53 expression as a statistically significant, independent predictor of outcome in CRC patients.
This article describes a review of multiple studies on the prognostic value of upregulated p53 expression determined with immunohistochemistry (IHC) in colorectal cancer patients.
Abbreviations: OS, overall survival; DFS, disease-free survival.
Footnotes
Author Contributions
Conceived and designed the experiments: EZ. Analyzed the data: EZ, PS. Wrote the first draft of the manuscript: EZ, PS. Contributed to the writing of the manuscript: EZ, AB, MR, PS. Agree with manuscript results and conclusions: EZ, AB, MR, PS, GL, CvdV, PK. Jointly developed the structure and arguments for the paper: EZ, GL, CvdV, PK. Made critical revisions and approved final version: GL, CvdV, PK, EZ, AB. All authors reviewed and approved of the final manuscript.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
Author(s) disclose no funding sources.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics. CA Cancer J Clin. 2003;53(1):5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2004;54(1):8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics. CA Cancer J Clin. 2005;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 6.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics. CA Cancer J Clin. 2006;56(2):106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics. CA Cancer J Clin. 2009;59(4):225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 11.Greene FL, Sobin LH. The staging of cancer: a retrospective and prospective appraisal. CA Cancer J Clin. 2008;58(3):180–90. doi: 10.3322/CA.2008.0001. [DOI] [PubMed] [Google Scholar]
- 12.Benson AB, III, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22(16):3408–19. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 13.Chun YS, Vauthey JN. Extending the frontiers of resectability in advanced colorectal cancer. Eur J Surg Oncol. 2007;33( Suppl 2):S52–8. doi: 10.1016/j.ejso.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Gunderson LL, Sargent DJ, Tepper JE, et al. Impact of T and N stage and treatment on survival and relapse in adjuvant rectal cancer: a pooled analysis. J Clin Oncol. 2004;22(10):1785–96. doi: 10.1200/JCO.2004.08.173. [DOI] [PubMed] [Google Scholar]
- 15.Kahlenberg MS, Sullivan JM, Witmer DD, Petrelli NJ. Molecular prognostics in colorectal cancer. Surg Oncol. 2003;12(3):173–86. doi: 10.1016/s0960-7404(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 16.Kozak KR, Moody JS. The impact of T and N stage on long-term survival of rectal cancer patients in the community. J Surg Oncol. 2008;98(3):161–6. doi: 10.1002/jso.21107. [DOI] [PubMed] [Google Scholar]
- 17.Poston GJ, Figueras J, Giuliante F, et al. Urgent need for a new staging system in advanced colorectal cancer. J Clin Oncol. 2008;26(29):4828–33. doi: 10.1200/JCO.2008.17.6453. [DOI] [PubMed] [Google Scholar]
- 18.Roukos DH, Murray S, Briasoulis E. Molecular genetic tools shape a roadmap towards a more accurate prognostic prediction and personalized management of cancer. Cancer Biol Ther. 2007;6(3):308–12. doi: 10.4161/cbt.6.3.3994. [DOI] [PubMed] [Google Scholar]
- 19.Locker GY, Hamilton S, Harris J, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24(33):5313–27. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 20.Duffy MJ, van Dalen A, Haglund C, et al. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43(9):1348–60. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Duffy MJ, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, et al. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. Eur J Cancer. 2003;39(6):718–27. doi: 10.1016/s0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 22.Pepe MS, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93(14):1054–61. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96(2):245–54. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 25.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–8. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 26.Philchenkov A, Zavelevich M, Kroczak TJ, Los M. Caspases and cancer: mechanisms of inactivation and new treatment modalities. Exp Oncol. 2004;26(2):82–97. [PubMed] [Google Scholar]
- 27.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108(2):153–64. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 28.Adell GC, Zhang H, Evertsson S, Sun XF, Stål OH, Nordenskjöld BA. Apoptosis in rectal carcinoma: prognosis and recurrence after preoperative radiotherapy. Cancer. 2001;91(10):1870–5. doi: 10.1002/1097-0142(20010515)91:10<1870::aid-cncr1208>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Bendardaf R, Ristamäki R, Kujari H, et al. Apoptotic index and bcl-2 expression as prognostic factors in colorectal carcinoma. Oncology. 2003;64(4):435–42. doi: 10.1159/000070304. [DOI] [PubMed] [Google Scholar]
- 30.de Bruin EC, van de Velde CJ, van de Pas S, et al. Prognostic value of apoptosis in rectal cancer patients of the dutch total mesorectal excision trial: radiotherapy is redundant in intrinsically high-apoptotic tumors. Clin Cancer Res. 2006;12(21):6432–6. doi: 10.1158/1078-0432.CCR-06-0231. [DOI] [PubMed] [Google Scholar]
- 31.Hilska M, Collan YU, Laine VJ, et al. The significance of tumor markers for proliferation and apoptosis in predicting survival in colorectal cancer. Dis Colon Rectum. 2005;48(12):2197–208. doi: 10.1007/s10350-005-0202-x. [DOI] [PubMed] [Google Scholar]
- 32.Jonges LE, Nagelkerke JF, Ensink NG, et al. Caspase-3 activity as a prognostic factor in colorectal carcinoma. Lab Invest. 2001;81(5):681–8. doi: 10.1038/labinvest.3780277. [DOI] [PubMed] [Google Scholar]
- 33.Langlois NE, Eremin O, Heys SD. Apoptosis and prognosis in cancer: rationale and relevance. J R Coll Surg Edinb. 2000;45(4):211–9. [PubMed] [Google Scholar]
- 34.Marijnen CA, Nagtegaal ID, Mulder-Stapel AA, et al. High intrinsic apoptosis, but not radiation-induced apoptosis, predicts better survival in rectal carcinoma patients. Int J Radiat Oncol Biol Phys. 2003;1;57(2):434–43. doi: 10.1016/s0360-3016(03)00580-7. [DOI] [PubMed] [Google Scholar]
- 35.Rödel F, Hoffmann J, Grabenbauer GG, et al. High survivin expression is associated with reduced apoptosis in rectal cancer and may predict disease-free survival after preoperative radiochemotherapy and surgical resection. Strahlenther Onkol. 2002;178(8):426–35. doi: 10.1007/s00066-002-1003-y. [DOI] [PubMed] [Google Scholar]
- 36.Rupa JD, de Bruïne AP, Gerbers AJ, et al. Simultaneous detection of apoptosis and proliferation in colorectal carcinoma by multiparameter flow cytometry allows separation of high and low-turnover tumors with distinct clinical outcome. Cancer. 2003;97(10):2404–11. doi: 10.1002/cncr.11366. [DOI] [PubMed] [Google Scholar]
- 37.de Heer P, de Bruin EC, Klein-Kranenbarg E, et al. Caspase-3 activity predicts local recurrence in rectal cancer. Clin Cancer Res. 2007;13(19):5810–15. doi: 10.1158/1078-0432.CCR-07-0343. [DOI] [PubMed] [Google Scholar]
- 38.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–12. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 39.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25(34):4798–811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 40.Winoto A. Cell death in the regulation of immune responses. Curr Opin Immunol. 1997;9(3):365–70. doi: 10.1016/s0952-7915(97)80083-0. [DOI] [PubMed] [Google Scholar]
- 41.Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22(53):8543–67. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 42.Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12(9):1543–68. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 43.Walczak H, Krammer PH. The CD95 (APO-1/Fas) and the TRAIL (APO-2 L) apoptosis systems. Exp Cell Res. 2000;256(1):58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 44.Mahmood Z, Shukla Y. Death receptors: targets for cancer therapy. Exp Cell Res. 2010;316(6):887–99. doi: 10.1016/j.yexcr.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Fulda S, Debatin KM. Exploiting death receptor signaling pathways for tumor therapy. Biochim Biophys Acta. 2004;1705(1):27–41. doi: 10.1016/j.bbcan.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Hu S, Vincenz C, Ni J, Gentz R, Dixit VM. I-FLICE, a novel inhibitor of tumor necrosis factor receptor-1-and CD-95-induced apoptosis. J Biol Chem. 1997;272(28):17255–7. doi: 10.1074/jbc.272.28.17255. [DOI] [PubMed] [Google Scholar]
- 47.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10(1):26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 48.Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2 L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12(6):611–20. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 49.Medema JP, Scaffidi C, Kischkel FC, et al. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC) EMBO J. 1997;16(10):2794–804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sprick MR, Rieser E, Stahl H, Grosse-Wilde A, Weigand MA, Walczak H. Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J. 2002;21(17):4520–30. doi: 10.1093/emboj/cdf441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 52.Mellier G, Huang S, Shenoy K, Pervaiz S. TRAILing death in cancer. Mol Aspects Med. 2010;31(1):93–112. doi: 10.1016/j.mam.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Scaffidi C, Fulda S, Srinivasan A, et al. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17(6):1675–87. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willis S, Day CL, Hinds MG, Huang DC. The Bcl-2-regulated apoptotic pathway. J Cell Sci. 2003;116(Pt 20):4053–6. doi: 10.1242/jcs.00754. [DOI] [PubMed] [Google Scholar]
- 55.Strand S, Hofmann WJ, Hug H, et al. Lymphocyte apoptosis induced by CD95 (APO-1/Fas) ligand-expressing tumor cells—a mechanism of immune evasion? Nat Med. 1996;2(12):1361–6. doi: 10.1038/nm1296-1361. [DOI] [PubMed] [Google Scholar]
- 56.Korkolopoulou P, Saetta AA, Levidou G, et al. c-FLIP expression in colorectal carcinomas: association with Fas/FasL expression and prognostic implications. Histopathology. 2007;51(2):150–6. doi: 10.1111/j.1365-2559.2007.02723.x. [DOI] [PubMed] [Google Scholar]
- 57.Strater J, Hinz U, Walczak H, et al. Expression of TRAIL and TRAIL receptors in colon carcinoma: TRAIL-R1 is an independent prognostic parameter. Clin Cancer Res. 2002;8(12):3734–40. [PubMed] [Google Scholar]
- 58.Sträter J, Hinz U, Hasel C, et al. Impaired CD95 expression predisposes for recurrence in curatively resected colon carcinoma: clinical evidence for immunoselection and CD95 L mediated control of minimal residual disease. Gut. 2005;54(5):661–5. doi: 10.1136/gut.2004.052696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Geelen CM, Westra JL, de Vries EG, et al. Prognostic significance of tumor necrosis factor-related apoptosis-inducing ligand and its receptors in adjuvantly treated stage III colon cancer patients. J Clin Oncol. 2006;24(31):4998–5004. doi: 10.1200/JCO.2006.06.8809. [DOI] [PubMed] [Google Scholar]
- 60.McLornan DP, Barrett HL, Cummins R, et al. Prognostic significance of TRAIL signaling molecules in stage II and III colorectal cancer. Clin Cancer Res. 2010;16(13):3442–51. doi: 10.1158/1078-0432.CCR-10-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tran SE, Holmstrom TH, Ahonen M, Kahari VM, Eriksson JE. MAPK/ERK overrides the apoptotic signaling from Fas, TNF, and TRAIL receptors. J Biol Chem. 2001;276(19):16484–90. doi: 10.1074/jbc.M010384200. [DOI] [PubMed] [Google Scholar]
- 62.Baader E, Toloczko A, Fuchs U, et al. Tumor necrosis factor-related apoptosis-inducing ligand-mediated proliferation of tumor cells with receptor-proximal apoptosis defects. Cancer Res. 2005;65(17):7888–95. doi: 10.1158/0008-5472.CAN-04-4278. [DOI] [PubMed] [Google Scholar]
- 63.Ishimura N, Isomoto H, Bronk SF, Gores GJ. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2006;290(1):G129–36. doi: 10.1152/ajpgi.00242.2005. [DOI] [PubMed] [Google Scholar]
- 64.Zeestraten EC, Van Hoesel AQ, Speetjens FM, et al. FoxP3-and CD8-positive infiltrating immune cells together determine clinical outcome in colorectal cancer. Cancer Microenviron. 2011;6(1):31–9. doi: 10.1007/s12307-011-0071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222(4):350–66. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krammer PH. CD95’s deadly mission in the immune system. Nature. 2000;407(6805):789–95. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- 67.Mills AA. p53: link to the past, bridge to the future. Genes Dev. 2005;19(18):2091–9. doi: 10.1101/gad.1362905. [DOI] [PubMed] [Google Scholar]
- 68.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 69.Bell HS, Dufes C, O’Prey J, et al. A p53-derived apoptotic peptide derepresses p73 to cause tumor regression in vivo. J Clin Invest. 2007;117(4):1008–18. doi: 10.1172/JCI28920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351(6326):453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 71.Munro AJ, Lain S, Lane DP. p53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;92(3):434–44. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N TP53-CRC Collaborative Study Group. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23(30):7518–28. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 73.Torsello A, Garufi C, Cosimelli M, et al. p53 and Bcl-2 in colorectal cancer arising in patients under 40 years of age: distribution and prognostic relevance. Eur J Cancer. 2008;44(9):1217–22. doi: 10.1016/j.ejca.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 74.Jurach MT, Meurer L, Moreira LF. Expression of the p53 protein and clinical and pathologic correlation in adenocarcinoma of the rectum. Arq Gastroenterol. 2006;43(1):14–9. doi: 10.1590/s0004-28032006000100006. [DOI] [PubMed] [Google Scholar]
- 75.Lim SC, Lee TB, Choi CH, Ryu SY, Min YD, Kim KJ. Prognostic significance of cyclooxygenase-2 expression and nuclear p53 accumulation in patients with colorectal cancer. J Surg Oncol. 2008;97(1):51–6. doi: 10.1002/jso.20907. [DOI] [PubMed] [Google Scholar]
- 76.Noske A, Lipka S, Budczies J, et al. Combination of p53 expression and p21 loss has an independent prognostic impact on sporadic colorectal cancer. Oncol Rep. 2009;22(1):3–9. doi: 10.3892/or_00000398. [DOI] [PubMed] [Google Scholar]
- 77.Baas IO, Mulder JW, Offerhaus GJ, Vogelstein B, Hamilton SR. An evaluation of six antibodies for immunohistochemistry of mutant p53 gene product in archival colorectal neoplasms. J Pathol. 1994;172(1):5–12. doi: 10.1002/path.1711720104. [DOI] [PubMed] [Google Scholar]
- 78.Thomadaki H, Scorilas A. BCL2 family of apoptosis-related genes: functions and clinical implications in cancer. Crit Rev Clin Lab Sci. 2006;43(1):1–67. doi: 10.1080/10408360500295626. [DOI] [PubMed] [Google Scholar]
- 79.Saelens X, Festjens N, Vande Walle L, van Gurp M, van Loo G, Vandenabeele P. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23(16):2861–74. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen M, Millar DG, Yong VW, Korsmeyer SJ, Shore GC. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J Biol Chem. 1993;268(34):25265–8. [PubMed] [Google Scholar]
- 81.Yang J, Liu X, Bhalla K, et al. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275(5303):1129–32. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 82.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14(1):56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 83.Buglioni S, D’Agnano I, Cosimelli M, et al. Evaluation of multiple bio-pathological factors in colorectal adenocarcinomas: independent prognostic role of p53 and bcl-2. Int J Cancer. 1999;84(6):545–52. doi: 10.1002/(sici)1097-0215(19991222)84:6<545::aid-ijc1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 84.Schwandner O, Schiedeck TH, Bruch HP, Duchrow M, Windhoevel U, Broll R. Apoptosis in rectal cancer: prognostic significance in comparison with clinical histopathologic, and immunohistochemical variables. Dis Colon Rectum. 2000;43(9):1227–36. doi: 10.1007/BF02237426. [DOI] [PubMed] [Google Scholar]
- 85.Chatla C, Jhala NC, Katkoori VR, et al. Recurrence and survival predictive value of phenotypic expression of Bcl-2 varies with tumor stage of colorectal adenocarcinoma. Cancer Biomark. 2005;1(4–5):241–50. doi: 10.3233/cbm-2005-14-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leahy DT, Mulcahy HE, O’Donoghue DP, Parfrey NA. Bcl-2 protein expression is associated with better prognosis in colorectal cancer. Histopathology. 1999;35(4):360–7. doi: 10.1046/j.1365-2559.1999.00743.x. [DOI] [PubMed] [Google Scholar]
- 87.Ilyas M, Hao XP, Wilkinson K, et al. Loss of Bcl-2 expression correlates with tumour recurrence in colorectal cancer. Gut. 1998;43(3):383–7. doi: 10.1136/gut.43.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elkablawy MA, Maxwell P, Williamson K, Anderson N, Hamilton PW. Apoptosis and cell-cycle regulatory proteins in colorectal carcinoma: relationship to tumour stage and patient survival. J Pathol. 2001;194(4):436–43. doi: 10.1002/path.894. [DOI] [PubMed] [Google Scholar]
- 89.Sinicrope FA, Hart J, Hsu HA, Lemoine M, Michelassi F, Stephens LC. Apoptotic and mitotic indices predict survival rates in lymph node-negative colon carcinomas. Clin Cancer Res. 1999;5(7):1793–804. [PubMed] [Google Scholar]
- 90.Krajewska M, Kim H, Kim C, et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res. 2005;11(15):5451–61. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 91.Bergmann A, Yang AY, Srivastava M. Regulators of IAP function: coming to grips with the grim reaper. Curr Opin Cell Biol. 2003;15(6):717–24. doi: 10.1016/j.ceb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 92.Deveraux QL, Stennicke HR, Salvesen GS, Reed JC. Endogenous inhibitors of caspases. J Clin Immunol. 1999;19(6):388–98. doi: 10.1023/a:1020502800208. [DOI] [PubMed] [Google Scholar]
- 93.Martinez-Ruiz G, Maldonado V, Ceballos-Cancino G, Grajeda JP, Melendez-Zajgla J. Role of Smac/DIABLO in cancer progression. J Exp Clin Cancer Res. 2008;27:48. doi: 10.1186/1756-9966-27-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sarela AI, Scott N, Ramsdale J, Markham AF, Guillou PJ. Immunohistochemical detection of the anti-apoptosis protein, survivin, predicts survival after curative resection of stage II colorectal carcinomas. Ann Surg Oncol. 2001;8(4):305–10. doi: 10.1007/s10434-001-0305-0. [DOI] [PubMed] [Google Scholar]
- 95.Ponnelle T, Chapusot C, Martin L, et al. Cellular localisation of survivin: impact on the prognosis in colorectal cancer. J Cancer Res Clin Oncol. 2005;131(8):504–10. doi: 10.1007/s00432-005-0682-z. [DOI] [PubMed] [Google Scholar]
- 96.Terzi C, Canda AE, Sagol O, et al. Survivin, p53, and Ki-67 as predictors of histopathologic response in locally advanced rectal cancer treated with preoperative chemoradiotherapy. Int J Colorectal Dis. 2008;23(1):37–45. doi: 10.1007/s00384-007-0376-x. [DOI] [PubMed] [Google Scholar]
- 97.Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58(22):5071–4. [PubMed] [Google Scholar]
- 98.Li F, Yang J, Ramnath N, Javle MM, Tan D. Nuclear or cytoplasmic expression of survivin: what is the significance? Int J Cancer. 2005;114(4):509–12. doi: 10.1002/ijc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sprenger T, Rödel F, Beissbarth T, et al. Failure of downregulation of survivin following neoadjuvant radiochemotherapy in rectal cancer is associated with distant metastases and shortened survival. Clin Cancer Res. 2011;17(6):1623–31. doi: 10.1158/1078-0432.CCR-10-2592. [DOI] [PubMed] [Google Scholar]
- 100.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22(53):8581–9. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 101.Marusawa H, Matsuzawa S, Welsh K, et al. HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 2003;22(11):2729–40. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dohi T, Okada K, Xia F, et al. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279(33):34087–90. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 103.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14(16):5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 104.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 105.Li F. Survivin study: what is the next wave? J Cell Physiol. 2003;197(1):8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- 106.Fortugno P, Wall NR, Giodini A, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115(Pt 3):575–85. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 107.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. SurvivindeltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59(24):6097–102. [PubMed] [Google Scholar]
- 108.Fang YJ, Lu ZH, Wang GQ, et al. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24(8):875–84. doi: 10.1007/s00384-009-0725-z. [DOI] [PubMed] [Google Scholar]
- 109.Knosel T, Emde A, Schluns K, et al. Immunoprofiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia. 2005;7(8):741–7. doi: 10.1593/neo.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bedi A, Pasricha PJ, Akhtar AJ, et al. Inhibition of apoptosis during development of colorectal cancer. Cancer Res. 1995;55(9):1811–6. [PubMed] [Google Scholar]
- 111.Takano Y, Saegusa M, Ikenaga M, Mitomi H, Okayasu I. Apoptosis of colon cancer: comparison with Ki-67 proliferative activity and expression of p53. J Cancer Res Clin Oncol. 1996;122(3):166–70. doi: 10.1007/BF01366957. [DOI] [PubMed] [Google Scholar]
- 112.Michael-Robinson JM, Reid LE, Purdie DM, et al. Proliferation, apoptosis, and survival in high-level microsatellite instability sporadic colorectal cancer. Clin Cancer Res. 2001;7(8):2347–56. [PubMed] [Google Scholar]
- 113.Zlobec I, Terracciano L, Tornillo L, et al. Role of RHAMM within the hierarchy of well-established prognostic factors in colorectal cancer. Gut. 2008;57(10):1413–9. doi: 10.1136/gut.2007.141192. [DOI] [PubMed] [Google Scholar]
- 114.Tornillo L, Lugli A, Zlobec I, et al. Prognostic value of cell cycle and apoptosis regulatory proteins in mismatch repair-proficient colorectal cancer: a tissue microarray-based approach. Am J Clin Pathol. 2007;127(1):114–23. doi: 10.1309/6RT941W1G6GDEHUE. [DOI] [PubMed] [Google Scholar]
- 115.Popat S, Chen Z, Zhao D, et al. A prospective, blinded analysis of thymidylate synthase and p53 expression as prognostic markers in the adjuvant treatment of colorectal cancer. Ann Oncol. 2006;17(12):1810–7. doi: 10.1093/annonc/mdl301. [DOI] [PubMed] [Google Scholar]
- 116.Watson NFS, Madjd Z, Scrimegour D, et al. Evidence that the p53 negative/Bcl-2 positive phenotype is an independent indicator of good prognosis in colorectal cancer: a tissue microarray study of 460 patients. World J Surg Oncol. 2005;3:47. doi: 10.1186/1477-7819-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sinicrope FA, Rego RL, Halling KC, et al. Thymidylate synthase expression in colon carcinomas with microsatellite instability. Clin Cancer Res. 2006;12(9):2738–44. doi: 10.1158/1078-0432.CCR-06-0178. [DOI] [PubMed] [Google Scholar]
- 118.Zlobec I, Terracciano LM, Lugli A. Local recurrence in mismatch repair-proficient colon cancer predicted by an infiltrative tumor border and lack of CD8(+) tumor-infiltrating lymphocytes. Clin Cancer Res. 2008;14(12):3792–7. doi: 10.1158/1078-0432.CCR-08-0048. [DOI] [PubMed] [Google Scholar]
- 119.Peng JJ, Cai SJ, Lu HF, et al. Predicting prognosis of rectal cancer patients with total mesorectal excision using molecular markers. World J Gastroenterol. 2007;13(21):3009–15. doi: 10.3748/wjg.v13.i21.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lan YT, Chang SC, Li AF, et al. p53 protein accumulation as a prognostic marker in sporadic colorectal cancer. Int J Colorectal Dis. 2007;22(5):499–506. doi: 10.1007/s00384-006-0194-6. [DOI] [PubMed] [Google Scholar]
- 121.Zlobec I, Baker K, Terracciano LM, Lugli A. RHAMM, p21 combined phenotype identifies microsatellite instability-high colorectal cancers with a highly adverse prognosis. Clin Cancer Res. 2008;14(12):3798–806. doi: 10.1158/1078-0432.CCR-07-5103. [DOI] [PubMed] [Google Scholar]
- 122.Lam AKY, Ong K, Ho YH. hTERT expression in colorectal adenocarcinoma: correlations with p21, p53 expressions and clinicopathological features. Int J Colorectal Dis. 2008;23(6):587–94. doi: 10.1007/s00384-008-0455-7. [DOI] [PubMed] [Google Scholar]
- 123.Nehls O, Okech T, Hsieh CJ, et al. Studies on p53, BAX and Bcl-2 protein expression and microsatellite instability in stage III (UICC) colon cancer treated by adjuvant chemotherapy: major prognostic impact of proapoptotic BAX. Br J Cancer. 2007;96(9):1409–18. doi: 10.1038/sj.bjc.6603728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Benevolo M, Mottolese M, Piperno G, et al. HLA-A, -B, -C expression in colon carcinoma mimics that of the normal colonic mucosa and is prognostically relevant. Am J Surg Pathol. 2007;31(1):76–84. doi: 10.1097/01.pas.0000213343.55605.b9. [DOI] [PubMed] [Google Scholar]
- 125.Fernández-Cebrián JM, Nevado SM, Vorwald KP, et al. Can the clinical outcome in stage II colon carcinomas be predicted by determination of molecular marker expression? Clin Transl Oncol. 2007;9(10):663–70. doi: 10.1007/s12094-007-0119-z. [DOI] [PubMed] [Google Scholar]
- 126.Koshiji M, Kumamoto K, Morimura K, et al. Correlation of N-myc downstream-regulated gene 1 expression with clinical outcomes of colorectal cancer patients of different race/ethnicity. World J Gastroenterol. 2007;13(20):2803–10. doi: 10.3748/wjg.v13.i20.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Broll R, Busch P, Duchrow M, et al. Influence of thymidylate synthase and p53 protein expression on clinical outcome in patients with colorectal cancer. Int J Colorectal Dis. 2005;20(2):94–102. doi: 10.1007/s00384-004-0621-5. [DOI] [PubMed] [Google Scholar]
- 128.Zhao DP, Ding XW, Peng JP, Zheng YX, Zhang SZ. Prognostic significance of bcl-2 and p53 expression in colorectal carcinoma. J Zhejiang Univ Sci B. 2005;6(12):1163–9. doi: 10.1631/jzus.2005.B1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Theodoropoulos GE, Lazaris AC, Theodoropoulos VE, et al. Hypoxia, angiogenesis and apoptosis markers in locally advanced rectal cancer. Int J Colorectal Dis. 2006;21(3):248–57. doi: 10.1007/s00384-005-0788-4. [DOI] [PubMed] [Google Scholar]
- 130.Bertolini F, Bengala C, Losi L, et al. Prognostic and predictive value of baseline and posttreatment molecular marker expression in locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2007;68(5):1455–61. doi: 10.1016/j.ijrobp.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 131.Lyall MS, Dundas SR, Curran S, Murray GI. Profiling markers of prognosis in colorectal cancer. Clin Cancer Res. 2006;12(4):1184–91. doi: 10.1158/1078-0432.CCR-05-1864. [DOI] [PubMed] [Google Scholar]
- 132.Lin LC, Lee HH, Hwang WS, et al. p53 and p27 as predictors of clinical outcome for rectal-cancer patients receiving neoadjuvant therapy. Surg Oncol. 2006;15(4):211–6. doi: 10.1016/j.suronc.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 133.Terzi C, Canda AE, Sagol O, Atila K, Sonmez D, Fuzun M, et al. Survivin, p53, and Ki-67 as predictors of histopathologic response in locally advanced rectal cancer treated with preoperative chemoradiotherapy. Int J Colorectal Dis. 2008;23(1):37–45. doi: 10.1007/s00384-007-0376-x. [DOI] [PubMed] [Google Scholar]
- 134.Tzouvala M, Lazaris AC, Papatheodoridis GV, et al. Potential role of apoptosis and apoptotic regulatory proteins in colorectal neoplasia: correlations with clinico-pathological parameters and survival. Dig Dis Sci. 2008;53(2):451–60. doi: 10.1007/s10620-007-9857-6. [DOI] [PubMed] [Google Scholar]
- 135.Lebe B, Sarioğlu S, Sokmen S, Ellidokuz H, Fuzun M, Kupelioglu A. The clinical significance of p53, p21, and p27 expressions in rectal carcinoma. Appl Immunohistochem Mol Morphol. 2005;13(1):38–44. doi: 10.1097/00129039-200503000-00007. [DOI] [PubMed] [Google Scholar]
- 136.King-Yin LA, Ong K, Ho YH. Colorectal mucinous adenocarcinoma: the clinicopathologic features and significance of p16 and p53 expression. Dis Colon Rectum. 2006;49(9):1275–83. doi: 10.1007/s10350-006-0650-y. [DOI] [PubMed] [Google Scholar]
- 137.Garrity MM, Burgart LJ, Mahoney MR, et al. North Central Cancer Treatment Group. Prognostic value of proliferation, apoptosis, defective DNA mismatch repair, and p53 overexpression in patients with resected Dukes’ B2 or C colon cancer: a North Central Cancer Treatment Group Study. J Clin Oncol. 2004;22(9):1572–82. doi: 10.1200/JCO.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 138.Fernebro E, Bendahl PO, Dictor M, Persson A, Ferno M, Nilbert M. Immunohistochemical patterns in rectal cancer: application of tissue microarray with prognostic correlations. Int J Cancer. 2004;111(6):921–8. doi: 10.1002/ijc.20229. [DOI] [PubMed] [Google Scholar]
- 139.Manne U, Weiss HL, Grizzle WE. Bcl-2 expression is associated with improved prognosis in patients with distal colorectal adenocarcinomas. Int J Cancer. 2000;89(5):423–30. doi: 10.1002/1097-0215(20000920)89:5<423::aid-ijc5>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 140.Schwandner O, Schiedeck TH, Bruch HP, Duchrow M, Windhoevel U, Broll R. p53 and Bcl-2 as significant predictors of recurrence and survival in rectal cancer. Eur J Cancer. 2000;36(3):348–56. doi: 10.1016/s0959-8049(99)00271-3. [DOI] [PubMed] [Google Scholar]
- 141.Kouraklis G, Kakisis J, Theoharis S, et al. Prognostic significance and correlation with survival of Bcl-2 and TGF-beta RII in colon cancer. Dig Dis Sci. 2003;48(12):2284–9. doi: 10.1023/b:ddas.0000007864.24866.77. [DOI] [PubMed] [Google Scholar]
- 142.Giatromanolaki A, Stathopoulos GP, Tsiobanou E, et al. Combined role of tumor angiogenesis, Bcl-2, and p53 expression in the prognosis of patients with colorectal carcinoma. Cancer. 1999;86(8):1421–30. doi: 10.1002/(sici)1097-0142(19991015)86:8<1421::aid-cncr6>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 143.Rosati G, Chiacchio R, Reggiardo G, De SD, Manzione L. Thymidylate synthase expression, p53, Bcl-2, Ki-67 and p27 in colorectal cancer: relationships with tumor recurrence and survival. Tumour Biol. 2004;25(5–6):258–63. doi: 10.1159/000081389. [DOI] [PubMed] [Google Scholar]
- 144.Hoos A, Nissan A, Stojadinovic A, et al. Tissue microarray molecular profiling of early, node-negative adenocarcinoma of the rectum: a comprehensive analysis. Clin Cancer Res. 2002;8(12):3841–9. [PubMed] [Google Scholar]
- 145.Bhatavdekar JM, Patel DD, Chikhlikar PR, et al. Molecular markers are predictors of recurrence and survival in patients with Dukes B and Dukes C colorectal adenocarcinoma. Dis Colon Rectum. 2001;44(4):523–33. doi: 10.1007/BF02234324. [DOI] [PubMed] [Google Scholar]
- 146.Bukholm IK, Nesland JM. Protein expression of p53, p21 (WAF1/CIP1), Bcl-2, Bax, cyclin D1 and pRb in human colon carcinomas. Virchows Arch. 2000;436(3):224–8. doi: 10.1007/s004280050034. [DOI] [PubMed] [Google Scholar]
- 147.Paradiso A, Simone G, Lena MD, et al. Expression of apoptosis-related markers and clinical outcome in patients with advanced colorectal cancer. Br J Cancer. 2001;84(5):651–8. doi: 10.1054/bjoc.2000.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meterissian SH, Kontogiannea M, Al-Sowaidi M, et al. Bcl-2 is a useful prognostic marker in Dukes’ B colon cancer. Ann Surg Oncol. 2001;8(6):533–7. doi: 10.1007/s10434-001-0533-3. [DOI] [PubMed] [Google Scholar]
- 149.Qiu H, Sirivongs P, Rothenberger M, Rothenberger DA, Garcia-Aguilar J. Molecular prognostic factors in rectal cancer treated by radiation and surgery. Dis Colon Rectum. 2000;43(4):451–9. doi: 10.1007/BF02237186. [DOI] [PubMed] [Google Scholar]
- 150.Ishijima N, Miki C, Ishida T, Kinoshita T, Suzuki H. The immunohistochemical expression of BCL-2 oncoprotein in colorectal adenocarcinoma. Surg Today. 1999;29(7):682–4. doi: 10.1007/BF02483002. [DOI] [PubMed] [Google Scholar]
- 151.Sun N, Meng Q, Tian A. Expressions of the anti-apoptotic genes Bag-1 and Bcl-2 in colon cancer and their relationship. Am J Surg. 2010;200(3):341–5. doi: 10.1016/j.amjsurg.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 152.Zlobec I, Minoo P, Baker K, et al. Loss of APAF-1 expression is associated with tumour progression and adverse prognosis in colorectal cancer. Eur J Cancer. 2007;43(6):1101–7. doi: 10.1016/j.ejca.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 153.Zlobec I, Minoo P, Baumhoer D, et al. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112(3):495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]
- 154.Paik SS, Jang KS, Song YS, et al. Reduced expression of Apaf-1 in colorectal adenocarcinoma correlates with tumor progression and aggressive phenotype. Ann Surg Oncol. 2007;14(12):3453–9. doi: 10.1245/s10434-007-9541-2. [DOI] [PubMed] [Google Scholar]
- 155.Strater J, Herter I, Merkel G, Hinz U, Weitz J, Moller P. Expression and prognostic significance of APAF-1, caspase-8 and caspase-9 in stage II/III colon carcinoma: caspase-8 and caspase-9 is associated with poor prognosis. Int J Cancer. 2010;127(4):873–80. doi: 10.1002/ijc.25111. [DOI] [PubMed] [Google Scholar]
- 156.Nehls O, Okech T, Hsieh CJ, et al. Low BAX protein expression correlates with disease recurrence in preoperatively irradiated rectal carcinoma. Int J Radiat Oncol Biol Phys. 2005;61(1):85–91. doi: 10.1016/j.ijrobp.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 157.Schelwies K, Sturm I, Grabowski P, et al. Analysis of p53/BAX in primary colorectal carcinoma: low BAX protein expression is a negative prognostic factor in UICC stage III tumors. Int J Cancer. 2002;99(4):589–96. doi: 10.1002/ijc.10380. [DOI] [PubMed] [Google Scholar]
- 158.Rau B, Sturm I, Lage H, et al. Dynamic expression profile of p21 WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003;21(18):3391–401. doi: 10.1200/JCO.2003.07.077. [DOI] [PubMed] [Google Scholar]
- 159.Cascinu S, Graziano F, Catalano V, et al. An analysis of p53, BAX and vascular endothelial growth factor expression in node-positive rectal cancer. Relationships with tumour recurrence and event-free survival of patients treated with adjuvant chemoradiation. Br J Cancer. 2002;86(5):744–9. doi: 10.1038/sj.bjc.6600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sinicrope FA, Rego RL, Foster NR, et al. Proapoptotic Bad and Bid protein expression predict survival in stages II and III colon cancers. Clin Cancer Res. 2008;14(13):4128–33. doi: 10.1158/1078-0432.CCR-07-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sinicrope FA, Rego RL, Okumura K, et al. Prognostic impact of bim, puma, and noxa expression in human colon carcinomas. Clin Cancer Res. 2008;14(18):5810–8. doi: 10.1158/1078-0432.CCR-07-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ponnelle T, Chapusot C, Martin L, et al. Subcellular expression of c-IAP1 and c-IAP2 in colorectal cancers: relationships with clinicopathological features and prognosis. Pathol Res Pract. 2003;199(11):723–31. doi: 10.1078/0344-0338-00488. [DOI] [PubMed] [Google Scholar]
- 163.Quirke P, Durdey P, Dixon MF, Williams NS. Local recurrence of rectal adenocarcinoma due to inadequate surgical resection. Histopathological study of lateral tumour spread and surgical excision. Lancet. 1986;2(8514):996–9. doi: 10.1016/s0140-6736(86)92612-7. [DOI] [PubMed] [Google Scholar]
- 164.De Oliveira LF, De Oliveira CH, Barrezueta LF, et al. Immunoexpression of inhibitors of apoptosis proteins and their antagonist SMAC/DIABLO in colorectal carcinoma: correlation with apoptotic index, cellular proliferation and prognosis. Oncol Rep. 2009;22(2):295–303. [PubMed] [Google Scholar]
- 165.Endo K, Kohnoe S, Watanabe A, et al. Clinical significance of Smac/DIABLO expression in colorectal cancer. Oncol Rep. 2009;21(2):351–5. [PubMed] [Google Scholar]