Abstract
Prior work has suggested that loss of expression of one or more of the many C/D box small nucleolar RNAs (snoRNAs) encoded within the complex, paternally expressed SNRPN (small nuclear ribonuclear protein N) locus may result in the phenotype of Prader-Willi syndrome (PWS). We suggest that the minimal critical region for PWS is ∼121 kb within the >460-kb SNRPN locus, bordered by a breakpoint cluster region identified in three individuals with PWS who have balanced reciprocal translocations and by the proximal deletion breakpoint of a familial deletion found in an unaffected mother, her three children with Angelman syndrome, and her father. The subset of SNRPN-encoded snoRNAs within this region comprises the PWCR1/HBII-85 cluster of snoRNAs and the single HBII-438A snoRNA. These are the only known genes within this region, which suggests that loss of their expression may be responsible for much or all of the phenotype of PWS. This hypothesis is challenged by findings in two individuals with PWS who have balanced translocations with breakpoints upstream of the proposed minimal critical region but whose cells were reported to express transcripts within it, adjacent to these snoRNAs. By use of real-time quantitative reverse-transcriptase polymerase chain reaction, we reassessed expression of these transcripts and of the snoRNAs themselves in fibroblasts of one of these patients. We find that the transcripts reported to be expressed in lymphoblast–somatic cell hybrids are not expressed in fibroblasts, and we suggest that the original results were misinterpreted. Most important, we show that the PWCR1/HBII-85 snoRNAs are not expressed in fibroblasts of this individual. These results are consistent with the hypothesis that loss of expression of the snoRNAs in the proposed minimal critical region confers much or all of the phenotype of PWS.
Prader-Willi syndrome (PWS [MIM 176270]) is a neurodevelopmental disorder characterized by neonatal hypotonia and failure to thrive, hyperphagia leading to obesity, mental retardation, hypogonadotropic hypogonadism, characteristic facies, short stature, small hands and feet, and behavioral abnormalities. Most individuals with PWS have a 4–4.5-Mb deletion of their paternal chromosome 15. PWS must derive from loss of function of one or more paternally expressed genes within this region. The same deletion confers Angelman syndrome (AS [MIM 105830]) when inherited from the mother. Although a small number of patients with AS have mutations in the preferentially maternally expressed gene UBE3A, indicating that the gene is critical for the AS phenotype, no analogous coding-region mutations have been found in individuals with PWS. This has led to the suggestion that PWS is due to loss of function of multiple genes (Nicholls and Knepper 2001).
There are other causes of PWS that abrogate normal paternal expression of genes in this region, such as maternal uniparental disomy (UPD) 15 and imprinting center (IC) mutations (Nicholls and Knepper 2001). These affect all paternally expressed genes in the region and therefore are not helpful in determining the minimal critical region for PWS. Important clues to the subset of genes responsible for PWS derive from patients with chromosomal translocations that disrupt the region but maintain its imprinted expression pattern. Balanced reciprocal translocations with breakpoints distal to the IC are rare causes of PWS (Nicholls and Knepper 2001). In each of the five cases reported, the paternally derived chromosome 15 contains a translocation breakpoint within the paternally expressed SNRPN (small nuclear ribonuclear protein N) locus (Schulze et al. 1996; Sun et al. 1996; Conroy et al. 1997; Kuslich et al. 1999; Wirth et al. 2001).
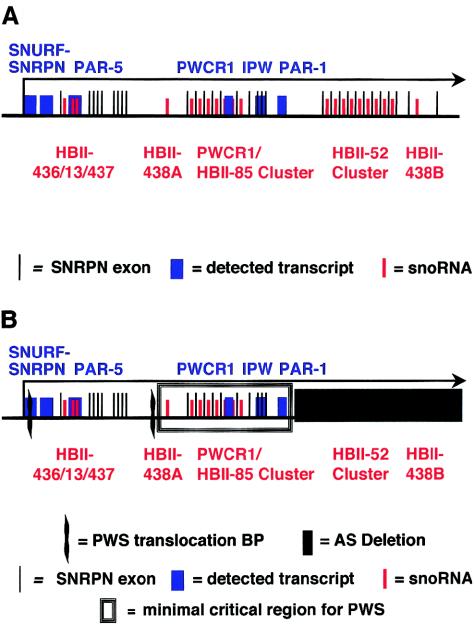
Multiple alternatively spliced and alternatively polyadenylated transcripts originate at the SNRPN promoter (Gray et al. 1999; Runte et al. 2001; Wirth et al. 2001). Overlapping cDNA clones and exon connection RT-PCR suggest that the largest transcripts are >460 kb and consist of at least 148 exons (Runte et al. 2001). Almost all transcripts encode two mRNAs at the 5′ end (Gray et al. 1999). Exons 1–3 encode a protein product of unknown function termed “SNURF” (SNRPN upstream reading frame) (Gray et al. 1999). Exons 4–10 encode SNRPN (SmN), a homolog of the SmB/B′ protein that binds small nuclear RNAs (snRNAs) involved in mRNA splicing. SmN replaces SmB/B′ in the brain (reviewed in Nicholls and Knepper 2001). An unusual feature of the large SNRPN transcripts is that multiple introns downstream of the protein-coding regions contain C/D box small nucleolar RNA (snoRNA) genes. There are two large multi–snoRNA gene clusters, three single-copy snoRNA genes, and one snoRNA gene present in two copies separated by 240 kb (Runte et al. 2001). Most introns contain only one snoRNA gene, and most snoRNAs are within introns (Runte et al. 2001). A simplified schematic of the SNRPN transcript, with approximate locations of snoRNA genes (Cavaille et al. 2000; de los Santos et al. 2000; Meguro et al. 2001; Runte et al. 2001), is shown in figure 1A. A similar organization has been reported for the maternally expressed Meg8 noncoding RNA gene at 14q32, which also encodes multiple C/D box snoRNAs within its introns; there are two snoRNA gene clusters, and there is one individual snoRNA gene (Cavaille et al. 2002).
Figure 1.
The SNRPN locus, starting at the major promoter.A, The large SNRPN transcript (black arrow). Several of the stable transcripts are in blue: SNURF, exons 1–3; SNRPN, exons 4–10; PAR-5; PWCR1; IPW and PAR-1. A subset of SNRPN exon sequences is illustrated by thin black lines. SnoRNA sequences are indicated by red lines and are identified by red text. The PWCR1/HBII-85 snoRNAs are a cluster of 27 sequences, the HBII-52 snoRNAs are a cluster of 47 sequences. Most are located within SNRPN introns, and most introns contain only one snoRNA. Not all are depicted, and the drawing is not to scale. B, Derivation of the PWS minimal critical region (box). Its borders are defined proximally by the wavy line that indicates the distal breakpoint cluster region within the alternatively spliced intron 20a/exon 21 (three individuals with PWS). The proximal breakpoint cluster region is in intron 2, within the SNURF gene (two individuals with PWS, including the individual studied here). The distal border of the minimal region is defined by the AS submicroscopic deletion that did not confer PWS when paternally inherited (blackened rectangle). The known genes and detected transcripts found within its borders are depicted; these are the transcripts PWCR1, IPW, and PAR-1 (blue) and the genes for snoRNAs HBII-438A and the PWCR1/HBII-85 cluster (red).
Among the five individuals who have features of PWS together with translocations, the breakpoints cluster into two regions: within SNRPN intron 2 (two individuals) (Kuslich et al. 1999; Sun et al. 1996) and within the untranslated, alternatively spliced, large (>30-kb) exon 20/intron 20a (three individuals) (Schulze et al. 1996; Conroy et al. 1997; Wirth et al. 2001). These define a promoter-proximal and a promoter-distal breakpoint cluster region within SNRPN (fig. 1B). We have chosen the promoter-distal translocation breakpoint cluster region as the upstream border of a proposed minimal critical region for PWS. A submicroscopic deletion that conferred AS when maternally inherited, but did not confer PWS when paternally inherited, defines the downstream border of the proposed minimal critical region (Hamabe et al. 1991). We have mapped the genomic locations of these borders in the Human Genome Working Draft, December 2001 version. The breakpoint cluster region is downstream of EST AI205506, which maps to nucleotides (nts) 22057217–22057639 (Wirth et al. 2001). The upstream breakpoint of the AS submicroscopic deletion was identified at nt 22178529, ∼4 kb downstream of PAR-1 and 28 kb upstream of the HBII-52 snoRNA cluster (Greger et al. 1993). The almost 121-kb region between these boundaries likely contains a gene (or genes) whose loss of expression confers, or contributes significantly to, the phenotype of PWS (fig. 1B).
The distal breakpoint cluster region can define an upstream border of a minimal critical region for PWS only if most of the individuals with this translocation breakpoint fulfill the diagnostic criteria for PWS (Holm et al. 1993). If they do not, it is important to identify features that consistently distinguish these individuals from those with PWS and proximal translocation breakpoints, so that potential contributions of genes in the intervening region to the full PWS phenotype may be determined (fig. 1B; SNRPN and snoRNAs HBII-436/13/437). The possibility that a clinical distinction exists between individuals with SNRPN promoter-proximal and promoter-distal balanced reciprocal translocation breakpoints is suggested by the original reports of these patients, but it must be critically assessed. The two individuals with proximal translocation breakpoints were described as having typical PWS (Sun et al. 1996; Kuslich et al. 1999), whereas two of the three individuals with distal translocation breakpoints were described as having atypical PWS phenotypes (Schulze et al. 1996; Conroy et al. 1997; Wirth et al. 2001).
Individual features of PWS are variably present in persons with the standard deletion or maternal UPD 15. Therefore, prior to the development of sensitive and specific molecular tests for PWS, a point system of 8 major criteria (neonatal hypotonia with poor suck, failure to thrive requiring special feeding techniques, hyperphagia, obesity after 1 year of age, hypogonadotropic hypogonadism, global developmental delay, characteristic facies, and chromosomal abnormality affecting the 15q11-13 region) and 11 minor criteria was established, to aid in diagnosis (Holm et al. 1993). Each major criterion fulfilled earns 1 point, and each minor criterion earns 1/2 point. A score of 8 points, with at least 4 from the major criteria, supports a diagnosis of PWS. Each of the five individuals with translocations within SNRPN was reported separately, by different authors, and was evaluated by different clinicians. In the original reports, the two individuals with proximal translocation breakpoints were reported to meet PWS clinical diagnostic criteria, and each had a score >8 points. The individual described by Kuslich et al. (1999) was reported to have a score of 10 but actually had a score of 11 (1 point added for characteristic facies). The individual reported by Sun et al. (1996) had a score of 10. Both of these individuals met all of the major clinical diagnostic criteria for PWS. Two of the three individuals with distal translocations also met criteria for PWS, with scores of 9 points (Conroy et al. 1997) and 9.5 points (Schulze et al. 1996) (reported as 8.5 points but with 1 point added for neonatal hypotonia). One of these individuals was reported to have atypical PWS, because he did not have hypogonadism or failure to thrive, although he did require special feeding techniques in the neonatal period (Conroy et al. 1997). The individual described by Wirth et al. (2001) was also reported to be atypical and had a clinical score of 7.0 points (reported as 6.5 points but with 1/2 point added for articulation defects). She did not meet the major criteria of neonatal hypotonia with poor suck, feeding problems with failure to thrive, or characteristic facies.
Our review of the reports of these patients and our reassessment of clinical scores indicate that it is difficult to reproducibly employ the diagnostic criteria. In addition, application of the diagnostic clinical criteria to individuals who have the standard deletion or maternal UPD 15 does not identify all of them as having PWS. A recent review of the clinical features of 90 individuals who had PWS, confirmed by standard molecular diagnostic tests and evaluated by the same clinician, found that 17% did not meet clinical diagnostic criteria for PWS (Gunay-Aygun et al. 2001). Given the small number of individuals reported to have balanced reciprocal translocations within the SNRPN locus—and given the fact that no feature of PWS was consistently different between the two groups—we find that we cannot distinguish between those with promoter-proximal and promoter-distal translocations. Because two of three individuals with SNRPN promoter-distal translocations meet clinical diagnostic criteria for PWS and because these criteria do not identify all individuals with the standard deletion as having PWS, we have chosen the promoter-distal translocation breakpoint cluster region as the upstream boundary of a proposed minimal critical region for PWS.
The assignment of the distal breakpoint cluster region as the upstream boundary of the proposed minimal critical region suggests that lack of expression of the PWCR1/HBII-85 snoRNA cluster and/or of HBII-438A may be responsible for many or all of the features of PWS. These snoRNAs appear to be the only putative functional molecules encoded within this region (fig. 1B). The PWCR1/HBII-85 snoRNAs are highly conserved between the human and mouse genomes (Cavaille et al. 2000; de los Santos et al. 2000; Meguro et al. 2001). Other stable transcripts from this region (PWCR1, IPW, and PAR-1) have limited coding potential and are minimally (PWCR1 and IPW) or not at all (PAR-1) conserved between human and mouse genomes (Sutcliffe et al. 1994; Wevrick et al. 1994; Wevrick and Francke 1997; de los Santos et al. 2000). Recent evidence suggests that these transcripts are portions of large transcripts that originate at the SNRPN promoter and may extend to UBE3A. It appears likely that PWCR1, IPW, and PAR-1 and other transcripts distal to the SmN coding region that lack coding potential are stable processing intermediates of these hypothetical large SNRPN transcripts (Runte et al. 2001). The only portions of the PWCR1 transcript that are conserved in the mouse ortholog Pwcr1 correspond to snoRNA sequences; this conservation first identified snoRNAs within the PWS region (de los Santos et al. 2000). The PWCR1 transcript may be stabilized by the presence of snoRNA sequences at the 5′ and 3′ ends that allow RNA duplex stem formation and/or bind snoRNP proteins (Kiss 2002). The putative SNRPN exon within PWCR1 is likely skipped because it has mutated splice sites, and, as a consequence, the two snoRNAs within it are likely not appropriately processed.
The hypothesis that loss of PWCR1/HBII-85 snoRNA expression is crucial for PWS is supported by mouse models of PWS, although the genomic characterization of the conserved syntenic region on mouse chromosome 7C is incomplete, and the upstream breakpoint of a crucial deletion mouse (Johnson et al. 1995; Tsai et al. 1999; Nicholls and Knepper 2001) has not been published. Therefore, a similarly detailed analysis is not possible. In addition, the mouse orthologs of HBII-438A, HBII-436, and HBII-437 have not been described to date. There are several mouse models of PWS, although the affected mice exhibit only one or two aspects of the human PWS phenotype. All mice with maternal UPD for the portion of chromosome 7 that is in conserved synteny with human chromosome 15q11-13 exhibited failure to thrive and died within 8 d of birth (Cattanach et al. 1992). This is believed to be similar to the failure-to-thrive phenotype of human newborns with PWS. In an attempt to identify the genes responsible for the mouse phenotype, mice have been engineered to have partial deletions of the region. Mice with paternal inheritance of a deletion in the IC also all died within 1 wk of birth (Yang et al. 1998). Mice with inherited paternal deletions of portions of Snrpn (or Snurf) that disrupt protein function had no detectable abnormal phenotype (Yang et al. 1998; Tsai et al. 1999); however, mice with a deletion from exon 2 of Snrpn to exon 2 of Ube3a exhibited 80% postnatal lethality, as well as growth retardation and hypotonia (Tsai et al. 1999). In contrast to findings in humans with PWS, mice that survived the neonatal period did not develop hyperphagia and obesity, and they were fertile (Tsai et al. 1999). Other aspects of the human phenotype are difficult to assess in mice. The results reported by Tsai et al. (1999) indicate that loss of expression of a gene or genes between exon 2 of Snrpn and Ube3a is responsible for the mouse phenotype. If the Snrpn locus is also the host gene for the orthologous snoRNAs in mice and if there are no other genes in this interval, these data would support a role for loss of expression of Snrpn-encoded snoRNAs for the mouse phenotype.
The hypothesis that absence of expression of the PWCR1/HBII-85 snoRNAs is crucial for the pathogenesis of PWS predicts that the expression of these snoRNAs should be absent in the individuals with PWS who have balanced reciprocal translocations. These snoRNAs are not expressed in brain of individuals with nontranslocation PWS (Cavaille et al. 2000; de los Santos et al. 2000). The PWCR1/HBII-85 snoRNAs were discovered after the reports of four of the five balanced translocation patients and were assayed directly only in one, in whom they were not expressed (Wirth et al. 2001). It is particularly important to determine PWCR1/HBII-85 snoRNA expression in the two patients with proximal translocation breakpoints, because cells from these patients have been reported to express IPW and PAR-1, which are encoded within the minimal critical region, IPW, and PAR-1 (Sun et al. 1996; Kuslich et al. 1999) (fig. 1). If these individuals with PWS express the PWCR1/HBII-85 snoRNAs appropriately, then loss of expression of these is unlikely to contribute significantly to the phenotype of PWS.
Here, we report our studies of SNRPN transcripts in fibroblasts of the individual with t(4;15)(q27;q11.2). In the original study of this individual, Kuslich et al. (1999) used RT-PCR to show that interspecies somatic cell hybrids containing the der(4) chromosome did not express SNRPN exons 3–5, which are located downstream of the breakpoint, or the more distal PAR-5. The latter had been identified as a stable paternally imprinted transcript but is now known to be a portion of SNRPN intron 12 (fig. 1A). In contrast, they reported expression of the downstream transcripts IPW and PAR-1. These transcripts were also initially identified as independent imprinted transcripts, but IPW comprises sequences from intron 58 and exons 59–61 of SNRPN, and PAR-1 consists of sequences of exon 61 and intron 62 (Human Genome Browser). As mentioned above, these are likely to be stable processing intermediates of the hypothetical large SNRPN transcripts, and they are encoded within the proposed PWS minimal critical region (fig. 1B). Their expression in cells derived from this patient would imply the presence of an alternative or cryptic promoter downstream of PAR-5/intron 12 and upstream of IPW/intron 58. This would suggest that snoRNAs downstream of PAR-5, including the PWCR1/HBII-85 and HBII-438A snoRNAs within the proposed minimal critical region, might also be transcribed and produced (fig. 1A and 1B). To clarify the role of the snoRNAs in PWS, we determined expression of several of them in skin fibroblasts of the individual with t(4;15) by quantitative RT-PCR. We also assayed the expression of additional introns and exons of the SNRPN transcript, including IPW and PAR-1.
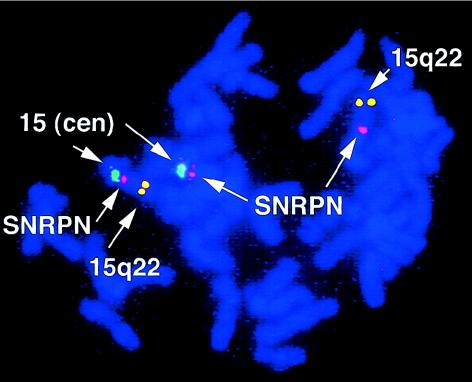
A karyotype confirmed the previously described t(4;15)(q27;q11.2) balanced translocation in the fibroblasts (data not shown). FISH demonstrated that both the der(15) and der(4) chromosomes contain sequences that hybridize to an SNRPN probe (Vysis), indicating that the translocation breakpoint on chromosome 15 is within SNRPN (fig. 2). The precise location of the breakpoint within SNRPN was confirmed by RT-PCR; SNRPN exon 2 is expressed, and SNRPN exon 3 is not, as reported elsewhere (Kuslich et al. 1999) (table 1).
Figure 2.
Results of FISH of fibroblast chromosomes with a t(4;15)(q27;q11.2) balanced reciprocal translocation. The chromosome 15 centromeric (cen) probe (green) identifies the normal chromosome 15 (left) and the der(15) (middle). The SNRPN probe (pink) hybridizes to the normal chromosome 15 (left), the der(15) (middle), and the der(4) (right), demonstrating that the breakpoint is within SNRPN. The distal chromosome 15q22 probe (yellow) identifies the normal chromosome 15 (left) and the der(4) (right).
Table 1.
Quantitative RT-PCR Data from Normal Control Fibroblasts, AS Lymphoblasts, PWS Fibroblasts, and t(4;15) Fibroblasts[Note]
| Transcripta | NormalFibroblasts | ASLymphoblasts | PWSFibroblasts | t(4;15)Fibroblasts |
| SNRPN exon 2 | .8 (.009) | .9 (.0004) | .0006 (.001) | 2.1 (.007) |
| SNRPN exon 3 | 2.8 (.03) | 2.7 (.001) | .002 (.002) | .02 (.008) |
| SNRPN exon 57/58 | 1.6 (.001) | 1.9 (.001) | .001 (.001) | .01 (.001) |
| IPW | 2.5 (.16) | 2.5 (.001) | .01 (.02) | .05 (.12) |
| PAR-1 | 3.1 (.11) | 2.4 (−.13) | .5 (−.3) | .3 (.005) |
| U21 | 1.5 (NC) | 1.4 (NC) | .6 (NC) | 1.4 (NC) |
| PWCR1/HBII-85 | 3.3 (.01) | 1.7 (.04) | .01 (.02) | .01 (.02) |
Note.— Data are relative number of nanograms of the transcript of interest (−RT results). CT PCR values from amplification plots, as in figure 3, were interpreted on the basis of a standard curve and were normalized to data for β-actin for each cell type. NC indicates that all −RT reactions gave no signal and so no value was calculated (CT >40). Primer sequences are as follows: SNRPN exon 2: 5′-ACGAACTACAGAACAGCACGTACC-3′ and 5′-CTGCGTTTGACTTGGACTTCC-3′; SNRPN exon 3, 5′-TTCTCAGCAGCAGCAAGTACCT-3′ and 5′-TGCCTCAGTTCAGCCTGGA-3′; SNRPN exon 57/58, 5′-CAGGAAAGATCAAAACGATGCA-3′ and 5′-GGCAGCTATCTGGACCAATCAC-3′; IPW, 5′-TGCATTCTTTTAGTGGATAGATGCA-3′ and 5′-TCCCCATAATGGCTTGTGTGT-3′; PAR-1, 5′-AGAGGCCAGCCATAACTAGCC-3′ and 5′-TGAAGAGCGTTCCCCTGTG-3′; U21, 5′-TGAGCAGTCAGTAGTTGGTCCTTT-3′ and 5′-GAAACAATTATCGCATCATATGCAA-3′; PWCR1/HBII-85, 5′-TCGATGATGAGTCCCCCATAA-3′ and 5′-CATTTTGTTCAGCTTTTCCAAGG-3′.
IPW and PAR-1 are stable transcripts from the SNRPN locus. U21 is a ubiquitously expressed nonimprinted C/D box snoRNA encoded on chromosome 1. PWCR1/HBII-85 primers recognize 4 of 27 PWCR1/HBII-85 snoRNA sequences.
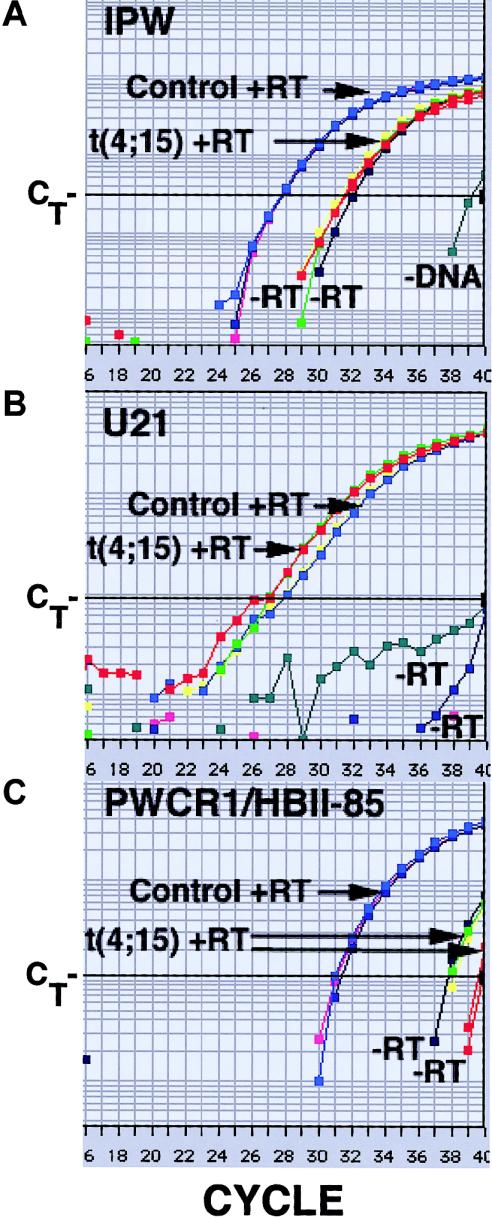
Our expression analysis of IPW and PAR-1 revealed that neither of these is expressed in fibroblasts of this individual. Conventional RT-PCR of IPW expression produced equivocal results. The primers used are located in the portion of IPW that corresponds to SNRPN intron 58, and they amplify genomic DNA. RNA was treated with DNase I prior to reverse transcription. The −RT control reaction amplified product, although the band was not as intense as that for the +RT reaction (data not shown). Therefore, we evaluated IPW expression by quantitative RT-PCR, using SYBR Green as a detection reagent (Applied Biosystems). SYBR Green fluoresces when bound to double-stranded DNA. Product amplification is detected by the change in fluorescence at each cycle of PCR. The PCR cycle at which product is significantly amplified above background is related to the amount of starting material and is called the threshold cycle (CT) (fig. 3) (Ginzinger 2002). Results reported here are relative, not absolute, and are interpreted by comparison to a standard curve produced by serial dilutions of pooled cDNA. To control for cell number, results are normalized to those of a housekeeping gene. This method allows relative quantification between samples of the amount of cDNA representing a transcript of interest. To facilitate reliable quantification, primers are designed to amplify small products, ∼50 nts. To ensure accurate quantification, reactions are performed in triplicate. To ensure that the correct product has been amplified, product size is confirmed by agarose gel electrophoresis. The IPW primers used were specified by the Applied Biosystems Primer Express program and do not span an intron. They are located within SNRPN exon 61. cDNA from fibroblasts of an unaffected control subject matched for age and sex (10-year-old boy) was prepared and assayed in parallel. The amplification plots demonstrate that fibroblasts of the individual with t(4;15) do not express IPW above the background identified in the −RT control (fig. 3A; compare the t(4;15) amplification plot to those of the two −RT controls). Similar results were obtained for PAR-1 and for SNRPN exons 57–58 (data not shown and table 1).
Figure 3.
Amplification plots of quantitative RT-PCR of cDNA from fibroblasts of the individual with t(4;15) and fibroblasts of a normal control for three transcripts: A, IPW; B, U21 snoRNA; C, PWCR1/HBII-85 snoRNAs. The Y-axis is the amount of fluorescence. The X-axis is the PCR cycle number. Each +RT reaction was done in triplicate, but not all individual plots can be distinguished, because they are superimposed. IPW and PWCR1/HBII-85: t(4;15) +RTs are shown in red, yellow, and green; −RTs are shown in red and dark blue. The −RT reactions, performed in parallel, gave similar results for both cell types and have not been labeled individually. In all cases, a no-DNA control was included. The CT values of these reactions were generally >40, but, with IPW primers, the value was 39. Samples with more starting cDNA of a transcript cross CT at a lower PCR cycle (e.g., the t(4;15) fibroblasts have more U21 cDNA than do the control fibroblasts).
In repeat experiments, we analyzed expression of these SNRPN derived transcripts using, as additional controls, lymphoblasts from an individual with AS and a maternal 15q11-q13 deletion confirmed by FISH and fibroblasts from an individual with PWS confirmed by methylation testing SNRPN exon 1 in parallel with the t(4;15) cDNA and the original unaffected control subjects (AS: 7-year-old boy; PWS: 38-year-old woman). This confirmed that the t(4;15) fibroblast results were similar to those of the PWS fibroblasts; the AS lymphoblast results were similar to those of the normal control fibroblasts (table 1). Although the t(4;15) sample appears to have some expression over the background of the −RT control (table 1), this does not appear to be significant, because, at high CT values, which are essentially negative reactions, there is considerable variability between replicas, (fig. 3C) for the t(4;15) +RT samples and the −RT samples. Therefore, −RT reactions were repeated in triplicate, to confirm the overlap between −RT and +RT amplification plots for the t(4;15) cells (SNPRN exons 3, 57, and 58; data not shown).
Our quantitative RT-PCR results clearly demonstrate that IPW and PAR-1 are not expressed in fibroblasts of the individual with t(4;15) and PWS. Although Kuslich et al. (1999) used lymphoblast-derived interspecies somatic hybrid cells, we do not believe that the difference in cell type accounts for the apparent difference in results. The IPW primers used by Kuslich et al. also do not span an intron and are located within exon 61. Recommendations for RT-PCR are that PCR primers span introns so that contaminating genomic DNA is not detectably amplified. This is not always possible, as in the case of the intronless transcript PAR-1. Because of the possibility that genomic DNA may be amplified, a −RT control should always be included in the analysis. In the absence of this control, RT-PCR data are not conclusive, particularly when the PCR primers used do not span introns. Kuslich et al. did not perform a −RT reaction in parallel but used different primer sets that spanned introns to demonstrate a lack of genomic DNA amplification in their +RT samples; however, different primer pairs may give different amounts of background amplification from the same DNase I–treated samples (fig. 3; compare −RT controls in panels A–C). Therefore, because of the absence of a −RT control in their expression analysis, the data of Kuslich et al. do not demonstrate conclusively that IPW and PAR-1 are expressed in the cell type of the individual with t(4;15) PWS that they assayed. Of the seven transcripts downstream of the translocation breakpoint assayed in their work, only IPW and PAR-1 were apparently expressed. Because these data are not conclusive, we suspect that no SNRPN locus sequences downstream of the translocation breakpoint are expressed in the cells assayed by Kuslich et al. It was not possible to obtain these cells to test this.
Our results indicate that SNRPN locus sequences downstream of the translocation breakpoint are not expressed in fibroblasts of the individual with t(4;15) PWS. This finding predicts that the snoRNAs encoded within SNRPN are not expressed either, because these are all derived from SNRPN introns downstream of the translocation breakpoint. Because we have shown that IPW and PAR-1 are not expressed in the t(4;15) fibroblasts there is no evidence for a cryptic promoter or other activation of expression of sequences downstream of PAR-5, and there is no reason to expect that the snoRNAs are transcribed and produced. To confirm this, we used quantitative RT-PCR to determine expression of several of the SNRPN encoded snoRNAs. The ubiquitously expressed, nonimprinted, U21 snoRNA encoded on chromosome 1 served as a control. Amplification plots for U21 demonstrate that U21-containing sequences are detected, in approximately equivalent amounts, in both the t(4;15) fibroblasts and the control fibroblasts (normalized to β-actin; data not shown) (fig. 3B). The analysis was repeated with the additional controls described above (table 1). Sequence comparison reveals that the 27 PWCR1/HBII-85 snoRNAs can be divided into three classes (Runte et al. 2001). Class I is very similar to the more homogeneous mouse MBII-85 snoRNAs (Cavaille et al. 2000; B.P., unpublished results). One primer pair was designed that should amplify four of nine PWCR1/HBII-85 snoRNAs in class I. This primer pair gave no amplification above background from t(4;15) fibroblast cDNA or from cDNA of the PWS control fibroblasts (fig. 3C and table 1). Amplification above background was detected from cDNA of normal control fibroblasts and from cDNA of AS lymphoblasts (fig. 3C and table 1). Thus, the PWCR1/HBII-85 snoRNAs are detected in fibroblasts of a normal control individual and in lymphoblasts of an individual with AS, but they are not detected in control PWS fibroblasts or in fibroblasts of the individual with t(4;15) PWS.
There are multiple additional snoRNAs encoded within the SNRPN transcript (fig. 1A). We evaluated the expression of snoRNAs HBII-436 and HBII-437. We detected amplification product above background in control but not in t(4;15) fibroblasts with primers for both of these (data not shown). The sequence of HBII-437 has degenerate D and D′ box sequence elements and was not detected by northern blot analysis (Runte et al. 2001; it may not be a stable snoRNA. We also attempted to assay HBII-52 snoRNA expression. There are 47 HBII-52 snoRNAs in two classes (Cavaille et al. 2000; Runte et al. 2001). Only three (HBII-52 snoRNAs 17–19) are identical in sequence, and these constitute class II (Runte et al. 2001). We designed a primer pair to detect the HBII-52 snoRNA 17/18/19 sequence. These primers amplified product from cDNA of a normal control brain but not from that of a brain of an individual with PWS (data not shown). No product was amplified from either the control fibroblasts or the AS lymphoblasts or from the t(4;15) or PWS fibroblasts (data not shown). These results are consistent with the demonstration, described elsewhere by Cavaille et al. (2000), that the HBII-52 snoRNAs are detected only in the brain.
Although our analysis of gene expression was performed in fibroblasts and, thus, may not reflect tissue-specific and cell type–specific expression within the brain (the critical affected organ in PWS), peripheral tissues are the only ones to which we have access for these rare, living patients with translocations. All gene expression studies of these individuals have, of necessity, been performed in fibroblasts or lymphoblasts, and all conclusions derived from these studies are limited by this fact. Our data indicate that the previously published expression analysis, which suggested that some of the SNRPN-encoded snoRNAs might be expressed in the individual with PWS and t(4;15) was misinterpreted and that SNRPN locus sequences downstream of the breakpoint are not expressed in peripheral cells of this individual. Although we cannot assay SNRPN expression in the brain of this individual, we suggest that our results remove one challenge to the hypothesis that loss of PWCR1/HBII-85 snoRNA expression contributes significantly to the phenotype of PWS (Kuslich et al. 1999; Nicholls and Knepper 2001; Wirth et al. 2001). There is yet another challenge to this hypothesis. In fibroblasts of the individual with PWS and a t(19;15) (the chromosome 15 breakpoint is also within SNRPN intron 2), SNRPN exons immediately downstream of the breakpoint, as well as PAR-5, IPW, and PAR-1 were reported to be expressed (Sun et al. 1996) (fig. 1). The IPW primers used in this work do span an intron, and the −RT control had no detectable product. (The PAR-1 and PAR-5 primers did not span introns; the PAR-1 −RT control had diffuse product amplification; and the PAR-5 −RT control had no amplification.) Two primer pairs were designed to detect SNRPN exons 1–2 (first pair) and exons 3–9 (second pair). Exons 1 –2 were expressed, as was expected, but exons 3–9, downstream of the breakpoint, were also expressed. Expression of snoRNAs was not evaluated, because these had not yet been identified, but it is likely that the intron-encoded snoRNAs are also detectably produced in these fibroblasts, because the exons appear to be transcribed. We suggest that translocation to a gene-rich and/or transcriptionally active region of chromosome 19 has resulted in a transcript that extends into SNRPN exons on der(19) in the fibroblasts studied, as was suggested originally by Sun et al. (1996). Processing of this transcript could lead to the generation of the SNRPN intron-encoded snoRNAs. Because this individual has PWS but may express the SNRPN-encoded snoRNAs at some level in some tissues, we suggest that expression of the fusion transcript is either too low to confer normal function, is not present in the appropriate cell type(s), and/or lacks the correct developmental regulation. To distinguish between these possibilities, it will be necessary to identify the gene on chromosome 19 to which the SNRPN transcript has been fused, to determine the level of expression quantitatively, and/or to determine the expression of the putative chromosome 19 gene/SNRPN fusion transcript in additional tissues.
The function of the RNAs whose loss may be crucial for the development of PWS is unknown. C/D box snoRNAs are characterized by short sequence elements (the C, C′, D, and D′ boxes) (Kiss 2002). These elements bind proteins that stabilize these nontranslated RNAs and are important for their nucleolar localization (Samarsky et al. 1998; Kiss 2002). The nucleolus is the site of ribosome biogenesis; most C/D box snoRNAs direct 2′-O-ribose methylation of ribosomal RNA (rRNA) (Kiss 2002). The role of this methylation is not known. The sequence upstream of the D or D′ box of these snoRNAs is complementary to the region of rRNA in which the modified nt resides. There are >100 such modifications in vertebrate rRNA, and corresponding snoRNAs have been identified for many of these sites (Huttenhofer et al. 2001). Some C/D box snoRNAs methylate snRNAs involved in splicing, others modify tRNA. Finally, a growing number lack complementarity to rRNA, snRNA, or tRNA (Huttenhofer et al. 2001; Jady and Kiss 2000; Kiss 2002). These are termed “orphan snoRNAs,” because their targets, which are presumed to be RNAs, have not yet been identified. The snoRNAs encoded within the SNRPN introns fall into this category (Cavaille et al. 2000; de los Santos et al. 2000; Meguro et al. 2001). Their targets, the modification(s) they direct, and the molecular and cellular function(s) they affect are unknown. It has been suggested that some orphan snoRNAs may target mRNA. The HBII-52 snoRNAs have a striking 18-nt complementarity to mRNA of serotonin receptor 2c, in a region that is both edited and alternatively spliced (Cavaille et al. 2000); however, no alteration of serotonin receptor structure or function in cells lacking HBII-52 has yet been reported. In addition, these snoRNAs appear not to be crucial for the pathogenesis of PWS, because they are absent in an apparently normal woman who inherited the submicroscopic deletion from her father (Hamabe et al. 1991; Greger et al. 1993).
Analysis of the available data has led us to propose that the minimal critical region for PWS is ∼121 kb and that it contains the PWCR1/HBII-85 snoRNA cluster and the HBII-438A snoRNA as the only putative functional genes. We recognize that this proposal is limited by the fact that it is based on analysis of individuals who have PWS and a translocation breakpoint within SNRPN, rather than deletions of the minimal critical region. Gene expression has been assayed only in peripheral tissues of these individuals, not in cells of the critical organ affected in PWS, the brain. We cannot confirm that SNRPN sequences upstream of the translocation breakpoint are expressed and that SNRPN sequences downstream of the translocation breakpoints are not expressed in the crucial cell types within the brain. Testing of our hypothesis will require further work in both mice and humans regarding the role of the products of the SNRPN/Snrpn locus in brain function. That the snoRNAs encoded within the proposed critical region are functional has not yet been demonstrated; however, they have canonical C and D boxes (Kiss 2002). PWCR1/HBII-85 snoRNAs are highly conserved between human and mouse and have been identified as stable molecules, by northern blot analysis (Cavaille et al. 2000; de los Santos et al. 2000; Runte et al. 2001).
The demonstration that IPW, PAR-1, and, most important, four of the PWCR1/HBII-85 snoRNAs encoded by SNRPN are not expressed in fibroblasts of the individual with t(4;15) and PWS removes an important barrier to the conclusion that loss of expression of the snoRNAs in the proposed minimal critical region contributes to, or is responsible for, the phenotype of PWS. To further evaluate this hypothesis, expression of the PWCR1/HBII-85 snoRNAs should be assayed in those rare patients whose phenotype suggests PWS but whose molecular test results are unusual (Dupont et al. 1999; Gillessen-Kaesbach et al. 1999). Loss of expression of additional genes encoded by the large SNRPN transcripts may also contribute to the phenotype of PWS. A true understanding of the pathogenesis of PWS will be possible only when the targets of the many snoRNAs have been identified, the nature of the modification(s) the snoRNAs direct clarified, and the cellular function(s) of these modifications brought to light.
Acknowledgments
Prader-Willi control fibroblasts were provided by the University of Miami Brain and Tissue Bank for Developmental Disorders (BTB number 1889), and PWS and control brain samples were provided by the University of Maryland Brain and Tissue Bank (UMB numbers 1290 and 1156, respectively) through National Institute of Child Health and Development contract N01-HD83284. We thank Jeff Traynor, for help with quantitative RT-PCR, and members of the Francke laboratory and Ross Metzger, for critical review of this manuscript. This research was supported by a Charles E. Culpeper Biomendical Pilot Initiative grant from the Rockefeller Brothers Fund and by National Institutes of Health postdoctoral training grant T32-GM08748.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PWS [MIM 176270] and AS [MIM 105830])
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the Human Genome Browser)
References
- Cattanach BM, Barr JA, Evans EP, Burtenshaw M, Beechey CV, Leff SE, Brannan CI, Copeland NG, Jenkins NA, Jones J (1992) A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet 2:270–274 [DOI] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhoffer A (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA 97:14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Seitz H, Paulsen M, Ferguson-Smith AC, Bachellerie JP (2002) Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum Mol Genet 11:1527–1538 [DOI] [PubMed] [Google Scholar]
- Conroy JM, Grebe TA, Becker LA, Tsuchiya K, Nicholls RD, Buiting K, Horsthemke B, Cassidy SB, Schwartz S (1997) Balanced translocation 46,XY,t(2;15)(q37.2;q11.2) associated with atypical Prader-Willi syndrome. Am J Hum Genet 61:388–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Schweizer J, Rees CA, Francke U (2000) Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am J Hum Genet 67:1067–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont JM, Le Tessier D, Rabineau D, Cuisset L, Vasseur C, Jeanpierre M, Delpech M, Pinton F, Ponsot G, Denavit MF (1999) Unexpected Angelman syndrome molecular defect in a girl displaying clinical features of Prader-Willi syndrome. J Med Genet 36:652–654 [PMC free article] [PubMed] [Google Scholar]
- Gillessen-Kaesbach G, Demuth S, Thiele H, Theile U, Lich C, Horsthemke B (1999) A previously unrecognised phenotype characterised by obesity, muscular hypotonia, and ability to speak in patients with Angelman syndrome caused by an imprinting defect. Eur J Hum Genet 7:638–644 [DOI] [PubMed] [Google Scholar]
- Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–312 [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD (1999) An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96:5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger V, Woolf E, Lalande M (1993) Cloning of the breakpoints of a submicroscopic deletion in an Angelman syndrome patient. Hum Mol Genet 2:921–924 [DOI] [PubMed] [Google Scholar]
- Gunay-Aygun M, Schwartz S, Heeger S, O'Riordan MA, Cassidy SB (2001) The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics 108:E92 [DOI] [PubMed] [Google Scholar]
- Hamabe J, Kuroki Y, Imaizumi K, Sugimoto T, Fukushima Y, Yamaguchi A, Izumikawa Y, Niikawa N (1991) DNA deletion and its parental origin in Angelman syndrome patients. Am J Med Genet 41:64–68 [DOI] [PubMed] [Google Scholar]
- Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, Greenberg F (1993) Prader-Willi syndrome: consensus diagnostic criteria. Pediatrics 91:398–402 [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 20:2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Kiss T (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res 28:1348–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DK, Stubbs LJ, Culiat CT, Montgomery CS, Russell LB, Rinchik EM (1995) Molecular analysis of 36 mutations at the mouse pink-eyed dilution (p) locus. Genetics 141:1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145–148 [DOI] [PubMed] [Google Scholar]
- Kuslich CD, Kobori JA, Mohapatra G, Gregorio-King C, Donlon TA (1999) Prader-Willi syndrome is caused by disruption of the SNRPN gene. Am J Hum Genet 64:70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro M, Mitsuya K, Nomura N, Kohda M, Kashiwagi A, Nishigaki R, Yoshioka H, Nakao M, Oishi M, Oshimura M (2001) Large-scale evaluation of imprinting status in the Prader-Willi syndrome region: an imprinted direct repeat cluster resembling small nucleolar RNA genes. Hum Mol Genet 10:383–394 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL (2001) Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2:153–175 [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K (2001) The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10:2687–2700 [DOI] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J 17:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Hansen C, Skakkebaek NE, Brondum-Nielsen K, Ledbeter DH, Tommerup N (1996) Exclusion of SNRPN as a major determinant of Prader-Willi syndrome by a translocation breakpoint. Nat Genet 12:452–454 [DOI] [PubMed] [Google Scholar]
- Sun Y, Nicholls RD, Butler MG, Saitoh S, Hainline BE, Palmer CG (1996) Breakage in the SNRPN locus in a balanced 46,XY,t(15;19) Prader-Willi syndrome patient. Hum Mol Genet 5:517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL (1994) Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet 8:52–58 [DOI] [PubMed] [Google Scholar]
- Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL (1999) Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet 8:1357–1364 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Francke U (1997) An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet 6:325–232 [DOI] [PubMed] [Google Scholar]
- Wevrick R, Kerns JA, Francke U (1994) Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet 3:1877–1882 [DOI] [PubMed] [Google Scholar]
- Wirth J, Back E, Huttenhofer A, Nothwang HG, Lich C, Gross S, Menzel C, Schinzel A, Kioschis P, Tommerup N, Ropers HH, Horsthemke B, Buiting K (2001) A translocation breakpoint cluster disrupts the newly defined 3′ end of the SNURF-SNRPN transcription unit on chromosome 15. Hum Mol Genet 10:201–210 [DOI] [PubMed] [Google Scholar]
- Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, Francke U, Jenkins NA, Copeland NG, Brannan CI (1998) A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19:25–31 [DOI] [PubMed] [Google Scholar]