Abstract
There are many cyclic peptides with diverse biological activities, such as antibacterial activity, immunosuppressive activity, and anti-tumor activity, and so on. Encouraged by natural cyclic peptides with biological activity, efforts have been made to develop cyclic peptides with both genetic and synthetic methods. The genetic methods include phage display, intein-based cyclic peptides, and mRNA display. The synthetic methods involve individual synthesis, parallel synthesis, as well as split-and-pool synthesis. Recent development of cyclic peptide library based on split-and-pool synthesis allows on-bead screening, in-solution screening, and microarray screening of cyclic peptides for biological activity. Cyclic peptides will be useful as receptor agonist/antagonist, RNA binding molecule, enzyme inhibitor and so on, and more cyclic peptides will emerge as therapeutic agents and biochemical tools.
Keywords: Cyclic peptides, Split-and-pool synthesis, On-bead screening, Drug lead
INTRODUCTION
Cyclic peptides and peptide drugs
Cyclic peptides are polypeptide chains taking cyclic ring structure. The ring structure can be formed by linking one end of the peptide and the other with an amide bond, or other chemically stable bonds such as lactone, ether, thioether, disulfide, and so on. N-to-C (or head-to-tail) cyclization is amide bond formation between amino and carboxyl termini, and many biologically active cyclic peptides are formed this way. Several cyclic peptides found in nature are used in clinic. The examples are gramicidin and tyrocidine with bactericidal activity, cyclosporin A with immunosuppressive activity, and vancomycin with antibacterial activity, and so on. While peptides have been generally considered to be poor drug molecules, there are some advantages of peptide drugs too. Followings are the weakness of peptides and the strength will be discussed afterward. First, per oral absorption is poor for peptide drugs. In most cases, the route of administration is injection as peptides are not well absorbed in the gastrointestinal tract. Second, peptides are rapidly metabolized, even after successful absorption, by proteolytic enzymes. Third, peptides usually do not cross cell membrane as some small molecules do. If the target of a peptide drug is in the cytoplasm, the peptide may not even reach the target. In spite of these limitations, peptides can be good alternatives to small synthetic molecules because of following advantages. Compared to small synthetic molecules, peptides possess less toxicity and they would not accumulate in organs. Even the fact that peptides get degraded rapidly can be a good thing. Peptide drugs can be less harmful, after acting on target molecules, as they will disappear rapidly by proteolytic degradation. The degradation products are simply amino acids and would not have toxicity (Loffet, 2002). Peptides can work on their targets very selectively, as the interaction with the targets is very specific compared to small molecules (Hummel et al., 2006). Considering these strengths, it is not surprising that there are many peptide drugs available in the market. These peptide drugs include receptor agonists and antagonists, peptide hormones and analogs, HIV protease inhibitors, and so on (Vlieghe et al., 2010). In addition to the merits of peptides as drug molecules, cyclic peptides could make even better peptide drugs.
Usually, cyclic peptides show better biological activity compared to their linear counterparts due to the conformational rigidity (Edman, 1959; Horton et al., 2002). The rigidity of cyclic peptides decreases the entropy term of the Gibbs free energy, therefore allowing the enhanced binding toward target molecules, or receptor selectivity. Another benefit from cyclic structure is the resistance to hydrolysis by exopeptidases due to the lack of both amino and carboxyl termini. Cyclic peptides can be resistant even to endopeptidases, as the structure is less flexible than linear peptides. Some cyclic peptides, though not all, can cross the cell membrane. Cyclosporin A is a good example of the membrane permeable cyclic peptides. Until recently, it has been suggested that cyclic peptides cross the membrane better than the linear counterparts (Rezaiet al., 2006).
To test this, a group of peptides has been synthesized, and their cell permeability was compared between cyclic and linear peptides. The results indicated that a peptide does not cross the membrane better simply because it is cyclized (Kwon and Kodadek, 2007). If a certain cyclic peptide is membrane permeable, it is because there are structural features allowing the molecule to cross the cell membrane.For example, cyclosporin A has several intra-molecular hydrogen bonds keeping hydrophilic groups from the surface of the molecule. Overall, structural rigidity, receptor selectivity, biochemical stability are general features of cyclic peptides and some cyclic peptides can be membrane permeable. These features allow cyclic peptides to be good therapeutic agents or biochemical tools, and efforts have been made to develop synthetic cyclic peptide with biological activity. In this review, the role of cyclic peptides in therapeutics and biochemistry will be described, as well as the approaches to develop cyclic peptide compounds for such purposes.
THE ROLE OF CYCLIC PEPTIDES IN THERAPEUTICS
Bactericidal activity of tyrocidine and gramicidin S
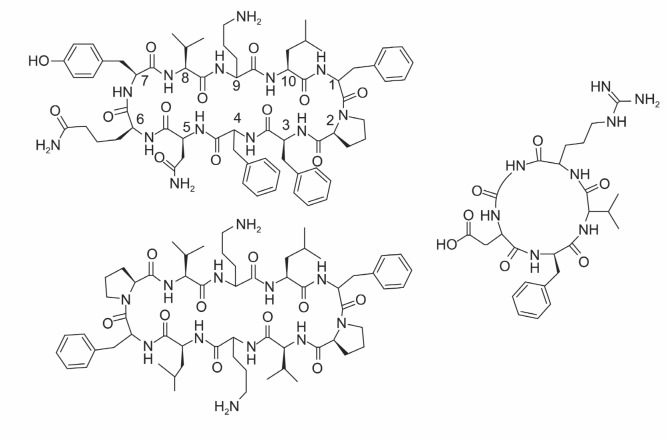
Tyrocidine is a cyclodecapeptide with antibacterial activity. It was found from a culture extract of a soil bacillus, Bacillus brevis, as bactericidal agent as early as 1939 (Dubos and Cattaneo, 1939) (Fig. 1 left top). Initially, this compound was characterized as a peptide lacking the free amino terminus, and therefore was proposed to have cyclic structure where amino terminus and carboxyl terminus are linked with an amide bond (Hotchkiss, 1941). While gramicidin S is a cyclic peptide, gramicidin refers to the mixture of linear pentadecapeptides with antibacterial activity (Fig. 1 left bottom). Tyrothricin (Hotchkiss and Dubos, 1941), the mixture of gramicidin and tyrocidine, was the first commercialized antibiotic and it is still used in clinic today (Tyrosur®). Gramicidin S, or Soviet gramicidin, is a cyclodecapeptide similar to tyrocidine, and was discovered in early 1940’s from the other side of the Earth for its antibacterial activity (Synge, 1945). Looking at the structure of these cyclic peptides, there can be two β-chains linked by proline residues and four intramolecular hydrogen bonds. This model was proposed in 1950’s (Hodgkin and Oughton, 1957) and later confirmed by X-ray crystallography (Hull et al., 1978). As there are four intramolecular hydrogen bonds, the peptide has a very rigid structure, a characteristic of cyclic peptides. In addition to the structural rigidity, these cyclic peptides are amphipathic (Zasloff, 2002). One side of molecule is hydrophobic while the other side is cationic. It appears the cationic face interacts with lipid head groups of cell membrane which is negatively charged (Matsuzaki, 1999). This initial interaction is followed by interaction between hydrophobic portions of cyclic peptide and membrane lipid, resulting in the rupture of bacterial cell membrane. The use of tyrocidine A is, however, limited to topical use as the membranolysis can happen even to the mammalian cells. There have been many approaches to reduce the toxicity toward mammalian cells to as will be discussed later in this review.
Fig. 1. Structure of Cyclic peptides used in clinic. Left top:Tyrocidine A, cyclo (Val-Orn-Leu-D-Phe-Pro-Phe-Phe-Asn-Gln-Tyr). Left bottom: Gramicidin S, cyclo (Val-Orn-Leu-D-Phe-Pro)2. Right:Cyclo-RGD Peptide EMD 66203, cyclo (Arg-Gly-Asp-D-Phe-Val).
Immunosuppressive activity of cyclosporin A
Cyclosporin A ([R-[R*,R*- (E)]]-cyclic (L-alanyl-D-alanyl-N-methyl-L-leucyl-N-methyl-L-leucyl-N-methyl-L-valyl-3-hy-droxy-N,4-dimethyl-L-2-amino-6-octenoyl-L-α-aminobutyryl-N-methylglycyl-N-methyl-L-leucyl-L-valyl-N-methyl-L-leucyl) is a cyclic peptide isolated from a fungus Tolypocladium inflatum. This compound is a well known immunosuppressant, clinically used worldwide to prevent graft rejection. The annual sale of cyclosporin A is more than US$ 1.0 billion in the USA only (Giroux, 2005). There are a couple of interesting structural features of cyclosporin A. First, there exist several Nα-methylated amino acids and non-standard amino acids. Second, cyclosporin A can freely pass the cell membrane, probably due to its structure. Cyclosporin A exerts its immuno-suppressive activity by inhibiting phosphatase activity of calcineurin. As Nα-methylated amino acids, found in cyclosporin A, are structurally rigid, it is technically challenging to obtain cyclosporin A from organic synthesis (Wenger, 1984). In contrast to the case of tyrocidine discussed above, there have been few approaches to improve cyclosporin A probably due to this challenge.
Anti-angiogenic activity of RGD peptide
For cells to interact with extracellular matrix, integrins play
an important role mediating signals from both in and out of cells. A functional integrin unit comprises of two subunits, α and β subunits in various combinations of isotypes. Many integrins recognize the tripeptide sequence -Arg-Gly-Asp- (RGD) for the interaction with the extracellular ligands. Interestingly, it was found that peptides containing RGD could inhibit tumor cell growth (Humphries et al., 1986; Gehlsen et al., 1988), and cyclic peptides with RGD sequence were more potent than the linear counterparts (Gurrath et al., 1992; Noiri et al., 1994). Cyclo-RGDfV (EMD 66203, f stands for D-Phe) itself can promote tumor regression by inducing apoptosis of angiogenic blood vessels (Fig. 1 right). In addition, cyclo-RGD compound can be used to deliver nanoparticles filled with anticancer drug to the area where tumor is growing (Murphy et al., 2008).
Cyclic peptides found in natural peptide hormones
We can find several cyclic peptides from natural peptide hormones such as calcitonin, oxytocin, somatostatin, vasopressin, and so on. These peptides form rigid structure by forming disulfide bond connecting two Cys residues in the peptide.
THE APPROACHES TO DEVELOP CYCLIC PEPTIDE COMPOUNDS
As described above, there are many cyclic peptides used in clinic, and most of them originate from the natural cyclic peptides. As several features make cyclic peptides attractive lead compounds for drug development as well as nice tools for biochemical research, scientists made diverse efforts to develop biologically active cyclic peptide compounds. Peptides can be prepared by either genetic or synthetic method. The genetic method, as described below, is usually limited to ribosomal 20 amino acids, whereas the sequence determination of hit compounds is straightforward. The synthetic method can provide more versatile cyclic peptide compounds as the repertoire of amino acids and the way of forming cyclic peptides is diverse. Solid-phase peptide synthesis combined with split-and-pool synthesis (Furka et al., 1991; Houghten et al., 1991; Lam et al., 1991) can prepare fairly large libraries. However, sequence determination is challenging after screening of these libraries. Conventional Edman degradation cannot be used for cyclic peptides once the free N-terminus disappears after cyclic peptide formation by N-to-C cyclization. While tandem mass spectrometry (MS) can be used to analyze peptide sesequences, the analysis of cyclic peptide sequence is more difficult than the analysis of linear peptide sequence (Eckert et al., 1985; Siegel et al., 1994; Ngoka and Gross, 1999; Schilling et al., 1999; Redman et al., 2003). The fragmentation pattern is very complex for cyclic peptides. For a hypothetical cyclic peptide containing only 3 amino acids, namely cyclo (ABC), the fragments formed from tandem MS would be ABC, BCA, CAB, AB, BC, CA, A, B, and C (total 9), while the linear peptide ABC would yield ABC, AB, BC, A, B, and C only (total 6). The fragmentation pattern gets more complex as the number of amino acid increases. Therefore, sequence analysis of cyclic peptides by tandem MS is not practical where the quantity of peptide from each microbead of split-and-pool synthesis library can be as little as about 100 pmol for -90 ㎛ beads.
In the following paragraphs, both genetic and synthetic approaches to develop cyclic peptide compounds will be discussed.
Phage display technology
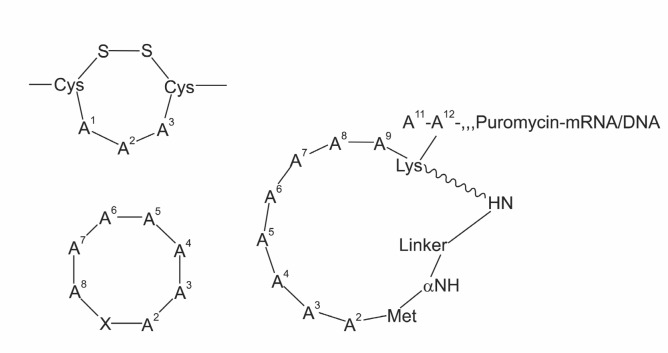
Phage display technology was not initially designed to develop cyclic peptide compounds when it was introduced first (Smith, 1985). In this technology, each phage particle displays unique peptide on its surface and the hit can be selected for binding toward a target molecule. Usually, peptides are displayed on the N-terminus, middle, or C-terminus of coat proteins, and the peptide sequence from each phage particle is directed by the DNA sequence of the same phage particle, allowing easy sequence determination. The screening can be repeated as long as encoding DNA molecules are preserved, and this repeated procedure, called bio-panning, is used to enrich the best binders. The diversity of phage display method can be typically up to billions (-109) of phages. As mentioned above, the peptide displayed on the surface of phage particle was not meant to be cyclic peptide. However, cyclic peptides formed from disulfide bond formation were sometimes obtained from phage display as exampled by RGD peptide (Koivunen et al., 1993) (Fig. 2 left top), and cyclic peptide can be prepared by designing the peptide sequences such as –Cys-X6-Cys for platelet glycoprotein binding (O'Neil et al., 1992). The phage particles are released to oxygen rich peri-plasmic space of bacteria, and two neighboring Cys residues would naturally form a disulfide bond to yield a cyclic peptide. One drawback of phage display is that the cyclic peptides are formed by disulfide formation, not head-to-tail cyclization. In addition, phage display is limited to natural, ribosomal amino acids. Many amino acids found in biologically active cyclic
Fig. 2. Structures of cyclic peptides from the genetic libraries. Left top: disulfide-containing cyclic peptide in phage display. Left bottom: intein-based cyclic peptide, X=Ser, or Cys. Right: cyclic peptide in mRNA display, a chemical linker is used to couple the N-terminal Met with the side chain of Lys at position 10.
peptides are nonribosomal, and they are not accessible with phage display.
Intein-based cyclic peptide
Inteins (Internal Proteins, or Intervening protein sequences) refer to the protein sequences that are spliced out during maturation (Perler et al., 1994; Perler, 2005). Scott and Benkovic reported Split-intein circuit ligation of peptides and proteins (SICLOPPS), utilizing a trans-intein DnaE to prepare cyclic peptide (Scott et al., 1999). The cyclic peptide formed from SICLOPPS takes a general structure cyclo (XA1A2A3…An), where X is either Cys or Ser (Fig. 2 left bottom). By constructing a SICLOPPS vector that allows the insertion of target peptide in the fusion protein, cyclic peptide libraries could be prepared in vivo, where each clone expresses a unique cyclic peptide. This method was used to screen for the inhibitors of protein-protein interactions (Horswill et al., 2004; Tavassoli and Benkovic, 2005; Tavassoli et al., 2008). SICLOPPS is a valuable tool to develop cyclic peptide compounds. The diversity is typically -108, and the cyclic peptides can be prepared in vivo. But, cyclic peptide synthesis in vivo can be a limitation as it prevents versatile in vitro screening. In addition, the method always requires Cys (or Ser) residue in the sequences, and the choice of amino acids is again limited to ribosomal amino acids.
Cyclic peptides from mRNA display
mRNA display is an in vitro method of displaying peptides/proteins coupled to the encoding mRNA. In this method, a library of peptides covalently linked to mRNA is screened for target binding. As the selected peptides from the screening are coupled to the encoding mRNA, hits can be amplified, and used for the next cycle of screening. This process is called bio-panning. This method was developed originally to prepare linear polypeptide libraries (Roberts and Szostak, 1997), and Roberts and co-workers recently came up with a method to prepare cyclic peptides based on mRNA display (Millward et al., 2005) (Fig. 2 right). They used a chemical crosslinker, dis-uccinimidyl glutarate (DSG) to couple the N-terminal end of peptides with the side chain of Lys residue at the C-terminal region of the peptides. They could select cyclic peptides binding the signaling molecule Gαi1 with high affinity (Millward et al., 2007). They claimed that Nα-methylphenylalanine (NMF) was used as one of the building blocks for in vitro transla-tion with the use of nonsense suppression. However, they did not show any cyclic peptides, among those selected from screening, that actually have this Nα-methylated amino acid. This implies at least two possibilities. One is that simply NMF was not favored for the binding of peptides toward their target molecule, and the other is that the steric hindrance prevented efficient coupling of NMF during in vitro translation. One drawback of mRNA display is that there is only one peptide produced from each mRNA template. If the poor efficiency of amino acid coupling results in the failure of peptide coupling, the mRNA encoding the corresponding peptide would be lost during the bio-panning regardless of actual binding affinity of the construct. Another limitation of this method is that they made cyclic peptides with the help of chemical crosslinker. In addition, when there are multiple Lys residues in the sequence, the cyclization reaction would not happen selectively to form a desired cyclic peptide. Also the dimer formation between two mRNA-peptide hybrid molecules mediated by crosslinker cannot be excluded.
Synthetic methods for cyclic peptides
As mentioned already, cyclic peptides can be synthesized by solid-phase synthesis in addition to typical organic synthesis. Until recently, the modification or improvement of the cyclic peptides involved individual chemical synthesis. This process is very time consuming to extract the structure and activity relationship. Compared to the theoretical diversity for a cyclodecapeptide (over 1013 from 2010, assuming 20 ribosomal amino acids are used), the practical diversity from individual synthesis is very little. Typically, scientists have tried to modify a couple of positions in a cyclic peptide to obtain better, improved compounds with the diversity not exceeding hundreds. When individual compounds are synthesized separately from the beginning to the end, the process is called sequential synthesis. When the synthetic intermediates are split during the synthesis and separate vessels are used for later steps, the process is called the parallel synthesis. In either case, it is difficult to prepare a large size cyclic peptide library. One nice example of synthetic approach can be seen in the development of cyclic RGD peptide. As described above, RGD peptide has affinity toward many integrins which play an important role in angiogenesis. Kessler (Mas-Moruno et al., 2010) and coworkers started the optimization of RGD peptide in late 1980’s. Beginning with a peptide sequence RGD, they restricted the conformation of peptide by cyclization, and did spatial screening to optimize the ligand structure. This led to the synthesis of cyclo (RGDfV). Later, Cilengitide, cyclo (RGDf(NMe)V) was derived from N-methyl scan; N-methylation is known to improve biological activity and stability of peptide drugs. Cilengitide is currently under phase III clinical trial for treating glioblastoma.
While Kessler and coworkers relied on individual synthesis, there are other efforts with higher throughput. Walsh and co-workers employed an enzyme domain from non-ribosomal peptide synthetase for cyclization of precursor synthesized on solid phase (Kohli et al., 2002). They fed the terminal thioesterase domain the peptide precursor bound to the resin support and the enzyme could catalyze the cyclization of precursor peptide, releasing the cyclic peptide tyrocidine A. By replacing amino acids at position 1 and 4 (D-Phe1 and D-Phe4) with different amino acids, natural and unnatural (8 and 24 each), they synthesized 192 different tyrocidine A analogs and optimized the selectivity. They obtained one tyrocidine A analog with almost the same antibacterial activity but reduced hemolytic effect.
Another remarkable approach was the one taken by Guo and co-workers (Qin et al., 2003; Qin et al., 2004), in which they utilized the propensity of tyrocidine A precursor to form a conformation highly favorable for head-to-tail cyclization (Bu et al., 2002). They prepared tyrocidine A precursor peptides with amino acids at positions 3, 4, 5, and 6 replaced by different amino acids (4, 2, 4, and 6 different amino acids each position). The parallel synthesis allowed fairly large number of diversity (4×2×4×6=192), and they could obtain more potent and less toxic compound.
Cyclic peptides from split-and-pool synthesis
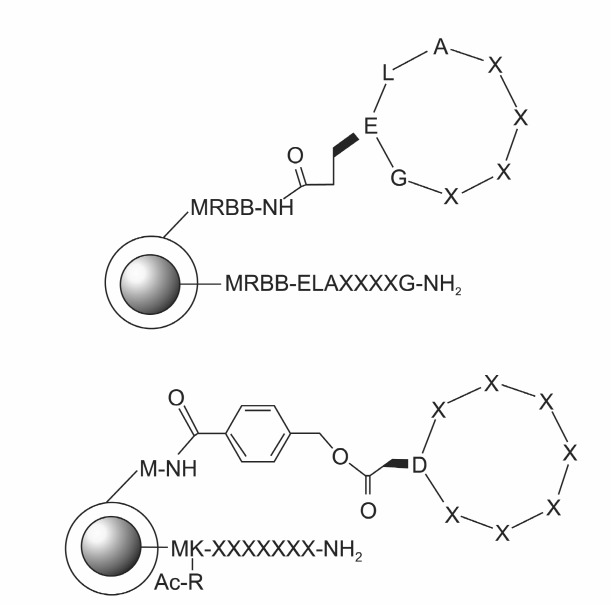
As described above, synthetic approach allows the incorporation of non-ribosomal amino acids in the cyclic peptide. However, the diversity that could be obtainable from the ge-netic approach cannot be reached with the sequential parallel synthesis. Split-and-pool synthesis allows the preparation of peptide libraries in large scales, and many linear peptide libraries have been prepared and screened for biological applications. Preparation of cyclic peptide libraries by split-and-pool synthesis is not difficult in terms of synthesis. The bottle-neck is the sequence identification after the hit is selected. Recent advancement in high-throughput sequence determination allows the peptide sequence determination for hundreds micro-beads in a short time, compared to automated Edman analysis which analyzes each bead individually. During partial Edman degradation process, the portion of peptides is kept intact with the help of capping reagents while the peptides are degraded by PITC. At the end of the degradation, each bead yields a ladder of peptides for mass analysis (Sweeney and Pei, 2003; Thakkar et al., 2006). While this technology was not applicable to cyclic peptides, spatial segregation of cyclic peptides on the bead surface allowed both high-throughput screening and high-throughput sequence determination of cyclic peptides for binding to the target molecules (Joo et al., 2006). The cyclic peptides are displayed on the surface of the microbeads while the corresponding linear peptide is inside the beads for sequence determination (Fig. 3 top). The cyclic peptides displayed on the outer layer can be used for screening, and the linear peptide can be used for sequence determination by Edman degradation. With this approach, cyclic peptides binding to streptavidin and prolactin receptor have been selected (Joo et al., 2006; Liu et al., 2009), and cyclic peptides inhibiting enzymes were optimized (Liu et al., 2010). Split-and-pool synthesis combined with spatial segregation allows high throughput screening and sequence determination of cyclic peptides for biological activity, and insertion of non-ribosomal amino acids are possible. This method utilizes on-bead screening for binding to the target molecules and the amount of peptides on microbeads can be as little as 100 pmol. Millions of unique cyclic peptides can be screened at a time with this method in a fairly short time, while the throughput with individual synthesis was hundreds of compound at a time with a lot of time and effort. In addition to on-bead screening, cyclic peptides could be released from the beads and used for in-solution screening (Fig. 3 bottom). In that case, peptides released from microbeads are not sufficient for the screening and macrobeads have to be used instead of microbeads. Although the use of macrobeads reduces the throughput, this still provides fast and high throughput screening compared to individual synthesis (Xiao and Pei, 2007). Separate from the spatial segregation approach, “two compounds, one bead” approach was developed by Kodadek and coworkers (Kwon and Kodadek, 2008). In each bead of their library, unique cyclic peptoid containing Cys residue is synthesized; Cys is included for the coupling on the maleimide-activated glass slide for microarray screening. On the same bead is synthesized the corresponding linear peptoid not containing Cys residue for sequence analysis by MS. This approach is useful for microarray screening of cyclic peptoid as well as cyclic peptide. As shown so far, split-and-pool synthesis allows the screening of cyclic peptides with on-bead screening, in-solution screening, and microarray screening for biological activity with high throughput.
Fig. 3. Structures of cyclic peptide libraries based on split-and-pool synthesis. Top: Cyclic peptide for on-bead screening. Bottom: Cyclic peptide for in-solution screening. Cyclic peptides can be released from the bead by hydrolysis of ester linkage, and then the remaining linear peptides are used for sequence determination.
CYCLIC PEPTIDES AS BIOCHEMICAL TOOLS
In the previous paragraphs, cyclic peptides with clinical applications, the genetic and synthetic approaches to develop cyclic peptide compounds were discussed. Now, potential applications of cyclic peptides as biochemical tools will be discussed.
Cyclic peptides as receptor agonists/antagonists
Structural rigidity combined with diverse peptide sequence can provide a binding motif toward target molecules. Compared to small molecules, they can be more selective while the size of molecule can be smaller than protein molecules such as antibodies and growth factors. The RGD peptide shown above can be a good example of cyclic peptide as receptor binding molecule. We can find more examples in which cyclic peptides work on receptors. One nice approach from Park and coworkers was to synthesize a mimetic of a monoclonal antibody specific for the p185HER2/neu growth factor receptor (Park et al., 2000). A cyclic peptide formed with a disulfide linkage with the sequence FCDGFYACYMDV could inhibit p185HER2/neu tyrosine kinases both in vitro and in vivo. Cell surface receptors can be good targets for cyclic peptide compounds as the peptide would work on the cell surface and it is not necessary for the peptide to cross the cell membrane. Another example is the antagonist against CXCR4 chemokine G protein-coupled receptor (GPCR) (Lalonde et al., 2011). CXCR4 chemokine GPCR has been recognized to be involved in the metastasis of cancer, and the entry of virus into host cells. T140 analogs are disulfide containing cyclic peptides modified from tachyplesin, a self-defense peptide of horseshoe crabs (Tamamura et al., 1998; Tamamura et al., 2006). These peptides can effectively bind to the receptor as antagonist. Even though diverse libraries were not screened, these compounds show reasonable binding affinities toward target molecules with biochemical activity. Cyclic peptides screened for prolactin receptor binding by Pei and coworkers are good example of cyclic peptide synthesized de novo (Liu et al., 2009). Millions of cyclic peptides were screened using on-bead screening of peptide library synthesized with split-and-pool synthesis described above, two rounds of screening yielded peptides with dissociation constants in micromolar range.
RNA binding cyclic peptides
Gene expression can be regulated in multiple levels, and RNA can be a target of regulation as seen in RNA interference (RNAi). The use of cyclic peptides can be one way to modulate RNA activity or stability as shown by Varani and co-workers (Athanassiou et al., 2004; Athanassiou et al., 2007). Initially, they developed BIV2 and related molecules that bind to transactivator response element (TAR) RNA of bovine immunodeficiency virus (BIV). BIV2 works as an inhibitor of Tat-TAR interaction, which is essential for the viral replication. They developed this compound by coupling a D-Pro-L-Pro dipeptide to the peptide sequence from Tat protein, mimicking β-hairpin structure found in the Tat-BIV TAR complex. They could synthesize a compound with nanomolar affinity, and their compound was later modified to work on HIV virus with the same principle (Lalonde et al., 2011). Interestingly, their compounds happened to be cell permeable and could work on the target inside the cell.
While the above example was limited to certain type of RNA, it would be nice to have cyclic peptide molecules binding to the RNA of interest. This would allow us temporal regulation of gene expression with stable peptide compounds.
Cyclic peptides as enzyme inhibitors
As explained above, cyclosporin A inhibits phosphatase activity of calcineurin to exert its immunosuppressive action. Among naturally occurring cyclic peptides, there are protease inhibitors such as sunflower trypsin inhibitor-1 (sfti-1) comprising in 14 amino acids (Colgrave et al., 2010). In addition to these natural peptides, several cyclic peptides have been developed as enzyme inhibitors. Peptidylprolyl isomerase Pin1 is an enzyme altering the conformation of Pro residue in specific peptide sequences containing phosphorylated Thr/Ser followed by Pro. Cis-trans isomerization mediated by Pin1 is important in cell cycle regulation, and Pin1 can be a target for regulating cell cycle progress. While there were many inhibitors developed against Pin1 enzyme, small molecules were not selective enough, and peptide-based inhibitors were liable to proteolyic degradation. Pei and co-workers synthesized and screened cyclic peptide library against the catalytic domain of Pin1 to derive an inhibitor with nanomolar affinity (Liu et al., 2010). Their screening was based on the binding of target protein to the cyclic peptide displayed on the microbead surface, and millions of unique cyclic peptides were tested during screening. Next example is the inhibitor against E. coli dam methyltransferase. Using the SICLOPPS technology, Benkovic and coworkers screened for cyclic peptides inhibiting the dam methyltransferase (Naumann et al., 2008). In addition to the examples shown here, we can find cyclic peptide inhibitors against proteasome (Stauch et al., 2010), HIV integrase (Hayouka et al., 2010), phosphatase (Hayashi et al., 2011), and the list is getting longer and longer.
CONCLUSION
Cyclic peptides have several structural features making them good drug leads, and there are several naturally occurring cyclic peptides in clinical use. In addition, biologically active cyclic peptides have been developed with genetic and synthetic approaches and they are useful as therapeutics and biochemical tools. With the introduction of new high throughput screening methods, there will be more cyclic peptides working as receptor agonists/antagonists, RNA binding molecules, and enzyme inhibitors.
Acknowledgments
This study was supported by National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST, 2010-0003008), and by the Catholic University of Daegu Research Grant in 2010.
References
- 1.Athanassiou Z. Dias R. L. Moehle K. Dobson N. Varani G. Robinson J. A. Structural mimicry of retroviral tat proteins by constrained beta-hairpin peptidomimetics: ligands with high affinity and selectivity for viral TAR RNA regulatory elements. J. Am. Chem. Soc. (2004);126:6906–6913. doi: 10.1021/ja0497680. [DOI] [PubMed] [Google Scholar]
- 2.Athanassiou Z. Patora K. Dias R. L. Moehle K. Robinson J. A. Varani G. Structure-guided peptidomimetic design leads to nanomolar beta-hairpin inhibitors of the Tat-TAR interaction of bovine immunodeficiency virus. Biochemistry. (2007);46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 3.Bu X. Wu X. Xie G. Guo Z. Synthesis of tyrocidine A and its analogues by spontaneous cyclization in aqueous solution. Org. Lett. (2002);4:2893–2895. doi: 10.1021/ol0263191. [DOI] [PubMed] [Google Scholar]
- 4.Colgrave M. L. Korsinczky M. J. Clark R. J. Foley F. Craik D. J. Sunflower trypsin inhibitor-1 proteolytic studies on a trypsin inhibitor peptide and its analogs. Biopolymers. (2010);94:665–672. doi: 10.1002/bip.21415. [DOI] [PubMed] [Google Scholar]
- 5.Dubos R. J. Cattaneo C. Studies on a bactericidal agent extracted from a soil bacillus : Iii. preparation and activity of a protein-free fraction. J. Exp. Med. (1939);70:249–256. doi: 10.1084/jem.70.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert K. Schwarz H. Tomer K. B. Gross M. L. Tandem mass spectrometry methodology for the sequence determination of cyclic peptides. J. Am. Chem. Soc. (1985);107:6765–6769. doi: 10.1021/ja00310a003. [DOI] [Google Scholar]
- 7.Edman P. Chemistry of amino acids and peptides. Annu. Rev.Biochem. (1959);28:69–96. doi: 10.1146/annurev.bi.28.070159.000441. [DOI] [PubMed] [Google Scholar]
- 8.Furka A. Sebestyén F. Asgedom M. Dibó G. General method for rapid synthesis of multicomponent peptide mixtures. Int. J. Pept. Protein. Res. (1991);37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 9.Gehlsen K. R. Argraves W. S. Pierschbacher M. D. Ruoslahti E. Inhibition of in vitro tumor cell invasion by Arg-Gly-Asp-containing synthetic peptides. J. Cell. Biol. (1988);106:925–930. doi: 10.1083/jcb.106.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giroux R. Cyclosporine. Chem. Eng. News. (2005);83:56. [Google Scholar]
- 11.Gurrath M. Müller G. Kessler H. Aumailley M. Timpl R. Conformation/activity studies of rationally designed potent anti-adhesive RGD peptides. Eur. J. Biochem. (1992);210:911–921. doi: 10.1111/j.1432-1033.1992.tb17495.x. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi R. Tanoue K. Durell S. R. Chatterjee D. K. Jenkins L. M. Appella D. H. Appella E. Optimization of a cyclic peptide inhibitor of Ser/Thr phosphatase PPM1D (Wip1). Biochemistry. (2011);50:4537–4549. doi: 10.1021/bi101949t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayouka Z. Hurevich M. Levin A. Benyamini H. Iosub A. Maes M. Shalev D. E. Loyter A. Gilon C. Friedler A. Cyclic peptide inhibitors of HIV-1 integrase derived from the LEDGF/p75 protein. Bioorg. Med. Chem. (2010);18:8388–8395. doi: 10.1016/j.bmc.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 14.Hodgkin D. C. Oughton B. M. Possible molecular models for gramicidin S and their relationship to present ideas of protein structure. Biochem. J. (1957);65:752–756. doi: 10.1042/bj0650752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horswill A. R. Savinov S. N. Benkovic S. J. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc. Natl. Acad. Sci. USA. (2004);101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton D. A. Bourne G. T. Smythe M. L. Exploring privileged structures: the combinatorial synthesis of cyclic peptides. J. Comput. Aided. Mol. Des. (2002);16:415–430. doi: 10.1023/A:1020863921840. [DOI] [PubMed] [Google Scholar]
- 17.Hotchkiss R. D. The chemical nature of gramicidin and tyrocidine. J. Biol. Chem. (1941);141:171–185. [Google Scholar]
- 18.Hotchkiss R. D. Dubos R. J. The isolation of bactericidal substances from cultures of Bacillus brevis. J. Biol. Chem. (1941);141:155–162. [Google Scholar]
- 19.Houghten R. A. Pinilla C. Blondelle S. E. Appel J. R. Dooley C. T. Cuervo J. H. Generation and use of synthetic pep-tide combinatorial libraries for basic research and drug discovery. Nature. (1991);354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 20.Hull S. E. Karlsson R. Main P. Woolfson M. M. Dodson E. J. The crystal structure of a hydrated gramicidin S-urea complex. Nature. (1978);275:206–207. doi: 10.1038/275206a0. [DOI] [Google Scholar]
- 21.Hummel G. Reineke U. Reimer U. Translating peptides into small molecules. Mol. Biosyst. (2006);2:499–508. doi: 10.1039/b611791k. [DOI] [PubMed] [Google Scholar]
- 22.Humphries M. J. Olden K. Yamada K. M. A synthetic peptide from fibronectin inhibits experimental metastasis of murine melanoma cells. Science. (1986);233:467–470. doi: 10.1126/science.3726541. [DOI] [PubMed] [Google Scholar]
- 23.Joo S. H. Xiao Q. Ling Y. Gopishetty B. Pei D. High-throughput sequence determination of cyclic peptide library members by partial Edman degradation/mass spectrometry. J. Am. Chem. Soc. (2006);128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 24.Kohli R. M. Walsh C. T. Burkart M. D. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. (2002);418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 25.Koivunen E. Gay D. A. Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J. Biol. Chem. (1993);268:20205–20210. [PubMed] [Google Scholar]
- 26.Kwon Y. U. Kodadek T. Quantitative comparison of the relative cell permeability of cyclic and linear peptides. Chem. Biol. (2007);14:671–677. doi: 10.1016/j.chembiol.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Kwon Y. U. Kodadek T. Encoded combinatorial libraries for the construction of cyclic peptoid microarrays. Chem. Commun. (Camb). (2008);44:5704–5706. doi: 10.1039/b812735b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalonde M. S. Lobritz M. A. Ratcliff A. Chamanian M. Athanas-siou Z. Tyagi M. Wong J. Robinson J. A. Karn J. Varani G. Arts E. J. Inhibition of both HIV-1 reverse transcription and gene expression by a cyclic peptide that binds the Tat-transactivating response element (TAR) RNA. PLoS. Pathog. (2011);7:e1002038. doi: 10.1371/journal.ppat.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam K. S. Salmon S. E. Hersh E. M. Hruby V. J. Kazmierski W. M. Knapp R. J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. (1991);354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 30.Liu T. Joo S. H. Voorhees J. L. Brooks C. L. Pei D. Synthesis and screening of a cyclic peptide library: discovery of small-molecule ligands against human prolactin receptor. Bioorg. Med. Chem. (2009);17:1026–1033. doi: 10.1016/j.bmc.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu T. Liu Y. Kao H. Y. Pei D. Membrane permeable cyclic peptidyl inhibitors against human Peptidylprolyl Isomerase Pin1. J. Med. Chem. (2010);53:2494–2501. doi: 10.1021/jm901778v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loffet A. Peptides as drugs: is there a market? J. Pept. Sci. (2002);8:1–7. doi: 10.1002/psc.366. [DOI] [PubMed] [Google Scholar]
- 33.Mas-Moruno C. Rechenmacher F. Kessler H. Cilengitide: the first anti-angiogenic small molecule drug candidate design synthesis and clinical evaluation. Anticancer. Agents. Med. Chem. (2010);10:753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. (1999);1462:1–10. doi: 10.1016/S0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 35.Millward S. W. Fiacco S. Austin R. J. Roberts R. W. Design of cyclic peptides that bind protein surfaces with antibody-like affinity. ACS. Chem. Biol. (2007);2:625–634. doi: 10.1021/cb7001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millward S. W. Takahashi T. T. Roberts R. W. A general route for post-translational cyclization of mRNA display libraries. J. Am. Chem. Soc. (2005);127:14142–14143. doi: 10.1021/ja054373h. [DOI] [PubMed] [Google Scholar]
- 37.Murphy E. A. Majeti B. K. Barnes L. A. Makale M. Weis S. M. Lutu-Fuga K. Wrasidlo W. Cheresh D. A. Nanoparticle-mediated drug delivery to tumor vasculature suppresses metastasis. Proc. Natl. Acad. Sci. USA. (2008);105:9343–9348. doi: 10.1073/pnas.0803728105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naumann T. A. Tavassoli A. Benkovic S. J. Genetic selection of cyclic peptide Dam methyltransferase inhibitors. Chembiochem. (2008);9:194–197. doi: 10.1002/cbic.200700561. [DOI] [PubMed] [Google Scholar]
- 39.Ngoka L. C. Gross M. L. Multistep tandem mass spectrometry for sequencing cyclic peptides in an iontrap mass spectrometer. J. Am. Soc. Mass. Spectrom. (1999);10:732–746. doi: 10.1016/S1044-0305(99)00049-5. [DOI] [PubMed] [Google Scholar]
- 40.Noiri E. Gailit J. Sheth D. Magazine H. Gurrath M. Muller G. Kessler H. Goligorsky M. S. Cyclic RGD peptides ameliorate ischemic acute renal failure in rats. Kidney. Int. (1994);46:1050–1058. doi: 10.1038/ki.1994.366. [DOI] [PubMed] [Google Scholar]
- 41.O'Neil K. T. Hoess R. H. Jackson S. A. Ramachandran N. S. Mousa S. A. DeGrado W. F. Identification of novel peptide antagonists for GPIIb/IIIa from a conformationally constrained phage peptide library. Proteins. (1992);14:509–515. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 42.Park B. W. Zhang H. T. Wu C. Berezov A. Zhang X. Dua R. Wang Q. Kao G. O'Rourke D. M. Greene M. I. Murali R. Rationally designed anti-HER2/neu peptide mimetic disables P185HER2/neu tyrosine kinases in vitro and in vivo. Nat. Biotechnol. (2000);18:194–198. doi: 10.1038/72651. [DOI] [PubMed] [Google Scholar]
- 43.Perler F. B. Protein splicing mechanisms and applications. IUBMB. Life. (2005);57:469–476. doi: 10.1080/15216540500163343. [DOI] [PubMed] [Google Scholar]
- 44.Perler F. B. Davis E. O. Dean G. E. Gimble F. S. Jack W. E. Neff N. Noren C. J. Thorner J. Belfort M. Protein splicing elements: inteins and exteins--a definition of terms and recommended nomenclature. Nucleic. Acids. Res. (1994);22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin C. Bu X. Zhong X. Ng N. L. Guo Z. Optimization of antibacterial cyclic decapeptides. J. Comb. Chem. (2004);6:398–406. doi: 10.1021/cc030117u. [DOI] [PubMed] [Google Scholar]
- 46.Qin C. Zhong X. Bu X. Ng N. L. Guo Z. Dissociation of antibacterial and hemolytic activities of an amphipathic peptide antibiotic. J. Med. Chem. (2003);46:4830–4833. doi: 10.1021/jm0341352. [DOI] [PubMed] [Google Scholar]
- 47.Redman J. E. Wilcoxen K. M. Ghadiri M. R. Automated mass spectrometric sequence determination of cyclic peptide library members. J. Comb. Chem. (2003);5:33–40. doi: 10.1021/cc0200639. [DOI] [PubMed] [Google Scholar]
- 48.Rezai T. Yu B. Millhauser G. L. Jacobson M. P. Lokey R. S. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. (2006);128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- 49.Roberts R. W. Szostak J. W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA. (1997);94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schilling B. Wang W. McMurray J.S. Medzihradszky K.F. Fragmentation and sequencing of cyclic peptides by matrix-assisted laser desorption/ionization post-source decay mass spectrometry. Rapid. Commun. Mass. Spectrom. (1999);13:2174–2179. doi: 10.1002/(SICI)1097-0231(19991115)13:21<2174::AID-RCM771>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 51.Scott C. P. Abel-Santos E. Wall M. Wahnon D. C. Benkovic S. J. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. USA. (1999);96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel M. M. Huang J. Lin B. Tsao R. Edmonds C. G. Structures of bacitracin A and isolated congeners: sequencing of cyclic peptides with blocked linear side chains by electrospray ionization mass spectrometry. Biol. Mass. Spectrom. (1994);23:186–204. doi: 10.1002/bms.1200230403. [DOI] [PubMed] [Google Scholar]
- 53.Smith G. P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. (1985);228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 54.Stauch B. Simon B. Basile T. Schneider G. Malek N. P. Kalesse M. Carlomagno T. Elucidation of the structure and intermolecular interactions of a reversible cyclic-peptide inhibitor of the proteasome by NMR spectroscopy and molecular modeling. Angew. Chem. Int. Ed. Engl. (2010);49:3934–3938. doi: 10.1002/anie.201000140. [DOI] [PubMed] [Google Scholar]
- 55.Sweeney M. C. Pei D. An improved method for rapid sequencing of support-bound peptides by partial edman degradation and mass spectrometry. J. Comb. Chem. (2003);5:218–222. doi: 10.1021/cc020113+. [DOI] [PubMed] [Google Scholar]
- 56.Synge R. L. 'Gramicidin S': overall chemical characteristics and amino-acid composition. Biochem. J. (1945);39:363–367. doi: 10.1042/bj0390363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamamura H. Tsutsumi H. Masuno H. Mizokami S. Hiramatsu K. Wang Z. Trent J. O. Nakashima H. Yamamoto N. Peiper S. C. Fujii N. Development of a linear type of low molecular weight CXCR4 antagonists based on T140 analogs. Org. Biomol. Chem. (2006);4:2354–2357. doi: 10.1039/b603818b. [DOI] [PubMed] [Google Scholar]
- 58.Tamamura H. Xu Y. Hattori T. Zhang X. Arakaki R. Kanbara K. Omagari A. Otaka A. Ibuka T. Yamamoto N. Nakashima H. Fujii N. A low-molecular-weight inhibitor against the chemokine receptor CXCR4: a strong anti-HIV peptide T140. Biochem. Biophys. Res. Commun. (1998);253:877–882. doi: 10.1006/bbrc.1998.9871. [DOI] [PubMed] [Google Scholar]
- 59.Tavassoli. A. Benkovic S. J. Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew. Chem. Int. Ed. Engl. (2005);44:2760–2763. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 60.Tavassoli A. Lu Q. Gam J. Pan H. Benkovic S. J. Cohen S. N. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS. Chem. Biol. (2008);3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 61.Thakkar. A. Wavreille. A.S. Pei D. Traceless capping agent for peptide sequencing by partial edman degradation and mass spectrometry. Anal. Chem. (2006);78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 62.Vlieghe P. Lisowski V. Martinez J. Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug. Discov. Today. (2010);15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Wenger R. M. Synthesis of cylosporine. Total syntheses of 'cyclosporin A' and 'cyclosporin H' two fungal metabolites isolated from the species. Tolypocladium inflatum GAMS. Helvetica. Chimica. Acta. (1984);67:502–525. doi: 10.1002/hlca.19840670220. [DOI] [Google Scholar]
- 64.Xiao Q. Pei D. High-throughput synthesis and screening of cyclic peptide antibiotics. J. Med. Chem. (2007);50:3132–3137. doi: 10.1021/jm070282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. (2002);415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]