James R. Lupski
I would like to thank the American Society of Human Genetics and my fellow human and medical geneticists for this great honor. The road to revelations of the concept of genomic disorders (Lupski 1998) did not always conform to my naïve preconceived notion of how it should be, but instead our molecular findings sometimes altered our view of genetic phenomena. The recognition of structural features of the human genome beyond primary DNA sequence, what I have referred to as “genome architecture,” has profound implications for how we as a species evolved and continue to evolve, as well as ramifications for common traits and human disease.
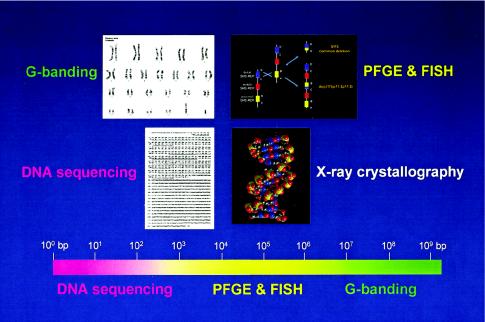
Figure 1 depicts different levels of genome architecture, each of which requires distinct methodological approaches to interrogate structure and alterations thereof. This relates to the ability to resolve the human genome at several levels, from single base-pair changes using DNA sequencing to the identification of chromosomal aberration through conventional G-banded karyotypes. Our ability to recognize genome alterations in the 104- to 106-bp range was only enabled through the development of techniques that could resolve genome changes of such magnitude. Pulsed-field gel electrophoresis (PFGE) (Schwartz and Cantor 1984) allowed us to separate molecules >104 bp in size, whereas fluorescence in situ hybridization (FISH) extended the reach of conventional cytogenetics (Pinkel et al. 1986, 1988). The latter technology, of course, was the subject of the inaugural Curt Stern Award last year, given to Dan Pinkel and Joe Gray. The genome architecture of special interest for our work consists of low-copy repeats that may be in a direct or inverted orientation. Nonallelic homologous recombination (NAHR) between direct repeats results in DNA duplication and deletion; many such rearrangements may be of a size that can only be assayed by FISH and/or PFGE. Armed with these powerful technologies and a little bit of luck, we recognized that the mechanisms for some genetic diseases are best understood at a genomic level when we identified a 1.5-Mb duplication associated with the common inherited peripheral neuropathy, Charcot-Marie-Tooth disease type 1A (CMT1A) (Lupski et al. 1991).
Figure 1.
Genome architecture and methods to resolve structure of varying DNA sizes. Above are shown four levels of genome architecture, from viewing the entire human genome, resolved by conventional G-banding, to the molecular double helical structure of DNA, revealed by x-ray crystallography. The focus of the Human Genome Project has been to determine the primary DNA sequence information. Below is shown a scale of the human genome from 1 bp (100 bp) to 3×109 bp and the size ranges (color coded) in which the different methods can physically resolve differences. Note that the genome architecture in the size range of ∼3×104 to 4×106 bp cannot be resolved either by DNA sequencing and agarose gel electrophoresis or by conventional G-banding. The techniques of PFGE and FISH extended the range of genetic technologies, enabling resolution of DNA rearrangements in the size range of >30 kb to <4 Mb. DNA rearrangements responsible for genomic disorders are often within the size range resolved only by PFGE and/or FISH.
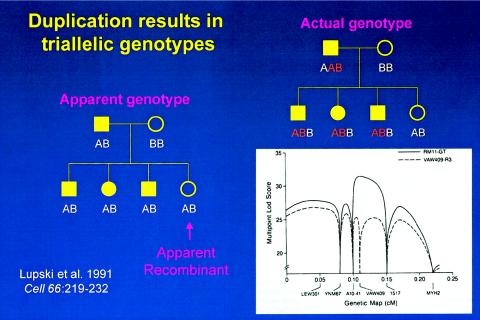
However, we stumbled along the way, recognizing that some CMT1A-linked probes gave dosage differences of cross hybridizing bands but not initially recognizing that this presumed artifact segregated in a Mendelian fashion. The duplication forced us to rethink the interpretation of marker allele segregation. When scoring as a simple biallelic system (fig. 2), the apparent genotype of this simple pedigree reveals an apparent recombinant. However, duplication results in triallelic genotypes. Rescoring the actual genotypes as a triallelic system, after the molecular recognition of the duplication, clearly documents that the unaffected individual does not, in fact, represent a recombinant (fig. 2). This had profound consequences for linkage analysis, with the triallelic scoring enabling the peak LOD score to now coincide in map position with the marker revealing molecular duplication (fig. 2).
Figure 2.
The effects of molecular duplication on the interpretation of marker genotypes and linkage mapping. Standard pedigree symbols are used: females depicted by circles and males by squares. Filled-in symbols denote affected individuals. On the left is a simple pedigree with marker genotypes scored as a usual biallelic system with one of the two alleles inherited from each parent. One unaffected daughter is an apparent recombinant, since she has the same apparent genotype as her three affected siblings. To the right is shown the actual genotypes scored as a triallelic system, accounting for the molecular duplication. The lower right shows how the different scoring biallelic (dashed line) versus triallelic (bold line) affects the multipoint LOD score. Note the differences in peak LOD scores and the fact that the failure to account for three alleles (or dosage differences in heterozygous RFLPs) results in an erroneous map position.
The CMT1A duplication was actually visualized by multiple molecular methods (Patel and Lupski 1994), including FISH, PFGE, and dosage differences of heterozygous alleles for restriction-fragment–length polymorphisms (RFLPs), but it was the three alleles observed in affected individuals by short-tandem repeat (STR) or microsatellite analysis that first illuminated the molecular duplication for us. The cosegregation of the disease with a junction fragment measuring 500 kb and resolved by PFGE suggested a stable mutation and a precise recombination mechanism (Lupski et al. 1991). These findings, in combination with observations from Vincent Timmerman in Christine van Broeckhoven’s group that marker genotypes which revealed the de novo duplication were accompanied by unequal crossing-over, suggested that there might be repeated sequences flanking the genomic segment that was duplicated. Indeed, Pentao Liu in my laboratory identified the repeat, which was >20 kb in size and highly homologous, that we termed “CMT1A-REP.” As predicted by the unequal crossing-over model, CMT1A-REP was found to be present in three copies on the CMT1A duplication–bearing chromosome (Pentao et al. 1992) and one copy in the reciprocal deletion responsible for a clinically milder episodic neuropathy known as hereditary neuropathy with liability to pressure palsies (HNPP) (Chance et al. 1994; Reiter et al. 1996). However, what was completely surprising was that the CMT1A-REP repeat, as well as the later-identified SMS-REP repeat responsible for the Smith-Magenis syndrome (SMS) (Chen et al. 1997) and its duplication reciprocal (Potocki et al. 2000), were not present in lower mammals, including mouse and hamster.
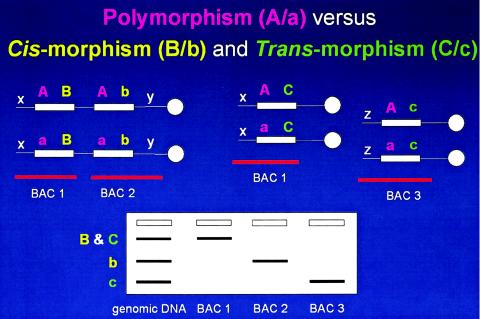
Another interesting observation from Southern analyses was that the two identified different size cross-hybridizing fragments do not represent polymorphic alleles, as both are present in the monochromosomal 17 hybrid MH22-6 (Pentao et al. 1992). I must digress now to differentiate variation of allelic sequences on different chromosome homologues, or polymorphisms, from variation in low-copy repeat (LCR) sequences on the same chromosomes—a phenomenon we have termed cis-morphisms (fig. 3) (Boerkoel et al. 1999). This is of particular interest, given the recent excitement about single-nucleotide polymorphisms (SNPs). If 10% or more of the human genome consists of LCRs, are 10% of SNPs actually cis-morphisms? What are the implications for mapping complex traits using SNPs? I draw your attention to an excellent recent paper by Xavier Estivill, Lap-Chee Tsui, and colleagues that appeared in Human Molecular Genetics. This paper documents that a significant proportion of the SNPs in the NCBI database correspond to paralogous sequence variants (PSVs) that could represent either cis-morphisms or trans-morphisms (fig. 3) within segmental duplications (LCRs) of the human genome sequence (Estivill et al. 2002).
Figure 3.
Paralagous sequence variations (PSVs). Above are depicted homologous chromosomes (thin horizontal lines), with centromeres (white circles) to the right. LCR sequences (white rectangles) are shown with sequence variations depicted as colored letters. The white x, y, and z denote unique flanking sequences in the genome used to anchor BAC clones (horizontal red lines). Below is shown a hypothetical Southern analysis with lanes containing genomic DNA and DNA from BAC clones. Cis-morphisms refer to paralogous sequence variations on the same chromosome B/b (yellow). Trans-morphisms (C/c) refer to PSVs on different chromosomes. Both cis-morphisms and trans-morphisms are revealed by BAC-specific cross-hybridizing restriction endonuclease fragment bands when using an LCR-specific probe. These PSVs are distinguished from polymorphisms (A/a; purple), the latter referring to allelic variation of the same sequence on different chromosome homologues. It is not clear what percentage of SNPs are actually paralogous sequence variants. Nor is it immediately obvious how the lack of recognition of this distinction (i.e., SNP vs. PSV) may effect the interpretation of mapping and other genetic studies. Potentially, the lack of this distinction in mapping studies could have one or more of three effects (since the PSVs can be several megabases apart or even on different chromosomes): erroneous map position, loss of linkage, or false positive linkage.
Let’s return to the evolution of low-copy repeats. Since no CMT1A-REP copies were identified in mouse and hamster, after determining that CMT1A-REP represents segmental duplication of a portion of the COX10 gene encoding hemeA:farnesyltransferase (Murakami et al. 1997; Reiter et al. 1997), we and others performed genomic Southern and cis-morphism analyses in closely related primate species (Reiter et al. 1997; Boerkoel et al. 1999). Remarkably, CMT1A-REP is duplicated in human and chimpanzee but not in gorilla, orangutan, or another 1 dozen closely related primate species (Kiyosawa and Chance 1996; Reiter et al. 1997; Keller et al. 1999)! Of even greater interest, by comparing human and mouse genomic and EST databases, the segmental duplication of CMT1A-REP resulted in the creation of two new genes with completely different expression profiles (Inoue et al. 2001; Inoue and Lupski 2002). Studies of the derivation of a number of LCRs reveal that each of them evolved relatively recently and predominately during primate speciation (reviewed in Samonte and Eichler 2002; Stankiewicz and Lupski 2002b). Thus, as the human genome evolves, we appear to accumulate LCRs through segmental duplication (Bailey et al. 2002). These LCRs create a genome architecture likely important to ongoing genome evolution and make us particularly vulnerable to genomic disorders (Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002a). LCRs may also be involved in karyotypic evolution during primate speciation (Stankiewicz et al. 2001). Recently, utilizing a human genome diversity cell-line panel (Cann et al. 2002) and CMT1A-REP probes, we have obtained evidence suggesting that these LCRs are continuing to evolve and may vary among selected world populations.
We also identified similar experimental findings of a common, recurrent rearrangement, as evidenced by a specific junction fragment detected by PFGE, in the Smith-Magenis contiguous gene syndrome (SMS) (Chen et al. 1997). A hint for a recurrent rearrangement was obtained several years earlier, when the same genetic markers were shown to be deleted for the majority of SMS patients, suggesting clustering of breakpoints (Greenberg et al. 1991). The SMS deletion and reciprocal duplication rearrangements are mediated through a large LCR, termed “SMS-REP,” that represents a repeat gene cluster (Chen et al. 1997). We used cis-morphisms within SMS-REPs to position large insert BAC clones for genomic sequencing. The nucleotide-sequence analysis revealed complex structure sharing >160 kb of 98% sequence identity among the three SMS-REP copies (Park et al. 2002). These SMS-REP sequences are not present in the mouse, as documented by direct genomic sequence comparisons (Bi et al. 2002). Interestingly, portions of SMS-REPs are also repeated elsewhere on chromosome 17. FISH analysis of metaphase chromosome 17 reveals the multiple cross-hybridizing signals and higher order architecture of SMS-REP–like sequences throughout chromosome 17 (Park et al. 2002).
For the medical geneticist, a FISH test using only two probes can detect multiple rearrangements within proximal 17p responsible for genomic disorders. These include the CMT1A duplication, the HNPP deletion, and the SMS del(17)(p11.2p11.2) and its reciprocal dup(17)(p11.2p11.2). Incredibly, using this assay, we identified a patient with DNA rearrangements on both homologues of chromosome 17 whose phenotype consisted of mild delay and a family history of autosomal dominant carpal tunnel syndrome (Potocki et al. 1999). The occurrence of both the 17p11.2 duplication and the HNPP deletion in this patient likely reflects the relatively high frequency at which these abnormalities arise and the underlying molecular characteristics of the genome in this region.
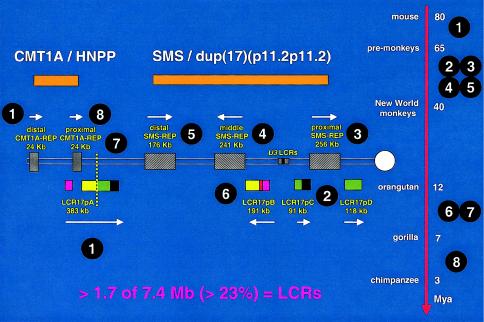
Our recent efforts have focused on studying the breakpoints of patients with uncommon rearrangements in proximal 17p. The majority of these breakpoints (∼60%) also occurred in LCRs and actually enabled the identification of yet more LCRs (Park et al. 2002). In fact, at least 23% of genome sequences within proximal 17p are contained within LCRs. FISH studies using primate cell lines in conjunction with molecular clock analysis enabled a working model that most parsimoniously explains how higher-order genomic architecture in proximal 17p evolved through a series of consecutive segmental duplications during primate speciation (fig. 4). Note that the most recent event is the segmental duplication of distal CMT1A-REP to result in proximal CMT1A-REP.
Figure 4.
Evolution of proximal 17p LCR during primate speciation. Proximal chromosome 17p is depicted by two thin horizontal lines with the centromere (white circle) shown to the right. LCRs are shown as horizontal rectangles with the same color or black-and-white graphic, representing highly homologous (identity usually >97%) sequence. Above are shown the genomic segments (orange), either duplicated in CMT1A and dup(17)(p11.2p11.2) or deleted in HNPP and SMS, respectively. To the right is shown a time line of mammalian, mainly primate, speciation, with the millions of years (Mya) as indicated. The white numbers circled in black indicate the approximate time at which the segmental duplication occurred and the given LCR appeared. The depicted sequence of events represents a working model that most parsimoniously explains present experimental observations. Note that one of the oldest LCRs, LCR17pA (1), was split by the most recent segmental duplication (8) that resulted in the proximal CMT1A-REP, which is present in humans and chimpanzees. Likewise, the proximal SMS-REP (3) has split a more ancient LCR (2) into LCR17pC and LCR17pD. The present orientation of middle SMS-REP and LCR17pB in the human genome may result from an inversion event after a directly oriented copy of middle SMS-REP appeared during primate speciation through a segmental duplication of proximal SMS-REP (6). The vertical dotted yellow line demarcates the evolutionary chromosome translocation 4;19 in Gorilla gorilla (7). Incredibly, LCRs represent >23% of the 7.4-Mb genomic sequence analyzed in proximal 17p. The majority of these LCRs have been found at the breakpoint of at least one DNA rearrangement.
In summary, molecular studies of the CMT1A duplication, the SMS deletion, their reciprocal recombination products, and other rearrangements that cause genomic disorders have revealed general features regarding the mechanisms for these disorders. First, it is clear that these rearrangements are not random events but rather reflect genome architecture. This genome architecture consists of region-specific LCRs that contribute to the susceptibility to DNA rearrangement. Furthermore, LCRs provide substrates for NAHR or unequal crossing-over. Thus, genomic disorders are recombination-based diseases and not resultant from errors in DNA replication/repair. The LCRs appear to have arisen recently during primate speciation through segmental duplication and likely play a role in genome evolution. The human genome has evolved an architecture that may make us as a species more susceptible to rearrangements causing genomic disorders. Finally, genomic disorders contribute in a significant way to disease burden.
Acknowledgments
I would like to thank my colleagues, including students, postdoctoral fellows, and faculty members in the Department of Molecular and Human Genetics for an incredibly stimulating environment and Art Beaudet for the wisdom and leadership to enable science to thrive at Baylor College of Medicine. The work in my laboratory has been generously supported by the National Institute of Neurological Disorders and Stroke, the National Institute of Child Health and Human Development, the National Cancer Institute, the National Eye Institute, the Muscular Dystrophy Association, the Foundation Fighting Blindness, and the Baylor College of Medicine Mental Retardation Research and Child Health Research Centers. None of these findings would have been possible without the hard work of a number of gifted students, technicians, and postdocs whom I have had the pleasure to collaborate with, and have had fun doing science with, for several years.
Footnotes
Previously presented at the annual meeting of The American Society of Human Genetics, in Baltimore, on October 19, 2002.
References
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 [DOI] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJM, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR (1999) Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann NY Acad Sci 883:22–35 [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli-Sforza LL (2002) A human genome diversity cell line panel. Science 296:261–262 [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed] [Google Scholar]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC (2002) Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum Mol Genet 11:1987–1995 [DOI] [PubMed] [Google Scholar]
- Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith ACM, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR (1991) Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2). Am J Hum Genet 49:1207–1218 [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B (2001) The 1.4 Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 [DOI] [PubMed] [Google Scholar]
- Keller MP, Seifried BA, Chance PF (1999) Molecular evolution of the CMT1A-REP region: a human- and chimpanzee-specific repeat. Mol Biol Evol 16:1019–1026 [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Chance P (1996) Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet 5:745–753 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–420 [DOI] [PubMed] [Google Scholar]
- Lupski JR, Montes de Oca-Luna R, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI (1991) DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66:219–232 [DOI] [PubMed] [Google Scholar]
- Murakami T, Reiter LT, Lupski JR (1997) Genomic structure and expression of the human heme A:farnesyltransferase (COX10) gene. Genomics 42:161–184 [DOI] [PubMed] [Google Scholar]
- Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PI, Lupski JR (1994) Charcot-Marie-Tooth disease: a new paradigm for the mechanism of inherited disease. Trends Genet 10:128–133 [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J (1988) Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA 85:9138–9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA 83:2934–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, Shapira SK, Kashork CD, Spikes AS, Shaffer LG, Lupski JR (1999) DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet 64:471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Kashork CD, Shaffer LG, Lupski JR (2000) Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR (1997) The human COX10 gene is disrupted during homologous recombination between the 24 KB proximal and distal CMT1A-REPs. Hum Mol Genet 6:1595–1603 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny D, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 (correction: Nat Genet 19:303) [DOI] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Cantor CR (1984) Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 37:67–75 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74-82 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park SS, Inoue K, Lupski JR (2001) The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11:1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]