Abstract
The molecular mechanisms by which a variety of naturally-occurring dietary compounds exert chemopreventive effects have been a subject of intense scientific investigations. Induction of phase II detoxification and anti-oxidant enzymes through activation of Nrf2/ARE-dependent gene is recognized as one of the major cellular defense mechanisms against oxidative or xenobiotic stresses and currently represents a critical chemopreventive mechanism of action. In the present review, the functional significance of Keap1/Nrf2 protein module in regulating ARE-dependent phase II detoxification and anti-oxidant gene expression is discussed. The biochemical mechanisms underlying the phosphorylation and expression of Keap1/Nrf2 proteins that are controlled by the intracellular signaling kinases and ubiquitin-mediated E3 ligase system as well as control of nucleocytoplasmic translocation of Nrf2 by its innate nuclear export signal (NES) are described.
Keywords: Chemoprevention, NF-E2-related Factor-2 (Nrf2), Kelch-like ECH-associated protein 1 (Keap1), Antioxidant Response Element (ARE)
CONCEPT OF CARCINOGENESIS
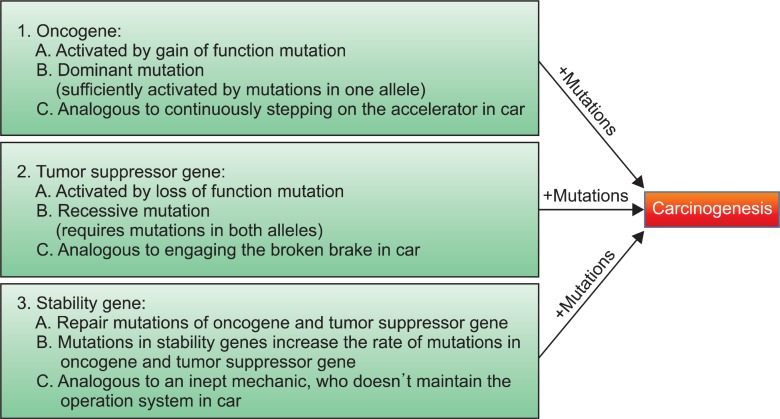
It is widely accepted that electrophiles and pro-oxidants promote carcinogenesis by causing damage to DNA. In fact, tumor cells are regarded as a collection of neoplastic cells that were transformed after a series of critical somatic mutations and then clonally expanded. Studies in the past decades have demonstrated that the genes whose mutations contributing to carcinogenesis can be categorized into three types: oncogene, tumor-suppressor gene and stability gene (Fig.1 ). Although the mode of genetic alterations in oncogene and tumor suppressor gene is different: (1) mutations in oncogene and tumor suppressor genes will lead to activation (gain of function) and inactivation (loss of function) of the gene product, respectively, and (2) activation of oncogene can be induced by mutations in either of maternal or paternal alleles (dominant mutation) but inactivation of tumor suppressor requires mutations in both alleles (recessive mutation), mutations in oncogene or tumor suppressor gene operate similarly at the physiological level: they increase the rate of carcinogenic process in normal cells by targeting key cellular processes. On the other hand, mutations in stability gene contribute to carcinogenesis in a slightly different manner that, if this class of gene is inactivated, mutation rates in oncogene and tumor suppressor gene will increase due to a lack of cellular systems that repair the genetic mistakes. In an analogy to automobile, Drs. Vogelstein and Kinzler have described that mutations in oncogene and tumor-suppressor gene are akin to stuck accelerator and dysfunctional brake that render the vehicle unable to stop when the driver attempts to engage them and that mutations in stability gene are akin to an inept mechanic who fails to oversee the operation systems in the car (Vogelstein and Kinzler, 2004).
Fig. 1. Three types of gene mutations responsible for tumor formation. Mutations in oncogene and tumor suppressor gene contribute to carcinogenesis. Mutations in stability gene, on the other hand, increase the frequency of mutations in oncogene and tumor suppressor gene due to a failure of cellular genetic repair system, facilitating the rate of carcinogenesis that is caused by mutations in oncogene and tumor suppressor gene.
In addition to somatic mutations and subsequent uncontrolled cell proliferation, other biological characteristics have been recognized as unequivocally important components in the development of human cancers. Drs. Hanahan and Weinberg initially classified these attributes of tumor cells and referred to them as six hallmarks of cancer: (1) self-sufficiency in growth signal, (2) insensitivity to anti-growth signals, (3) tumor invasion and metastasis, (4) limitless replicative potential, (5) sustained angiogenesis, and (6) evading apoptosis (Hanahan and Weinberg, 2000). With a remarkable progress in cancer research in the past decade, they have recently broadened the categories by adding several new hallmarks, including (1) reprogramming of energy metabolism, (2) evading immune destruction and (3) creation of tumor-prone microenvironment (Hanahan and Weinberg, 2011). This classification provides us with a clear idea that tumorigenesis in human is a complex and multistep process, in which the addition of individual hallmarks contributes to accumulation of genetic alterations that drive the progressive transformation of normal human cells into highly malignant ones. This concept is rendered more concrete by a large number of works, indicating that the genomes of human tumors are invariably altered at multiple sites and tumor formation in mice occurs through multiple rate-limiting steps (Sharpless and Depinho, 2006).
REGULATION OF PHASE II DETOXIFICATION AND ANTI-OXIDANT GENE EXPRESSION BY Nrf2/Keap1 MODULE THROUGH REGULATORY PROTEINS AND INTRACELLULAR KINASE SIGNALING CASCADES
As mutagenesis plays a significant role in tumor development, it is likely that cancer prevention can be accomplished by two ways: (1) minimizing the exposure of DNA to endogenous and exogenous carcinogenic factors or (2) increasing the rate of their detoxification in our body. Considering that avoiding the putative chemical or viral carcinogens can be achieved by maintaining a healthful lifestyle, finding out how carcinogen detoxification system in our body can be increased seems to be a feasible theme of cancer prevention research. In order to combat against oxidative stress and electrophiles, normal cells have developed the elaborate defense enzyme systems during evolution, e.g. phase II detoxification and anti-oxidant enzymes, such as heme oxygenase-1 (HO-1), NAD[P] H:quinone oxidoreductase-1 (NQO1), superoxide dismutase (SOD), glutathione S-transferase (GST), and γ-glutamyl cysteine ligase (γ-GCL), etc. Transcriptional regulation of these enzymes is coordinated, largely in part, by the antioxidant response element (ARE), a nucleotide motif sequence that exists in 5’-upstream promoter region of these genes (Kensler et al., 2007). Now it is widely accepted that a transcriptional factor, e.g. NF-E2-related factor-2 (Nrf2), is responsible for ARE-dependent gene activation. Under a basal condition, Nrf2 is sequestered in the cytosol but, in response to a variety of cellular stresses, it translocates into the nucleus and activates ARE-dependent gene expression by binding to ARE sequence in the genome in association with small Maf proteins
and/or other coactivator proteins (Itoh et al., 1997).
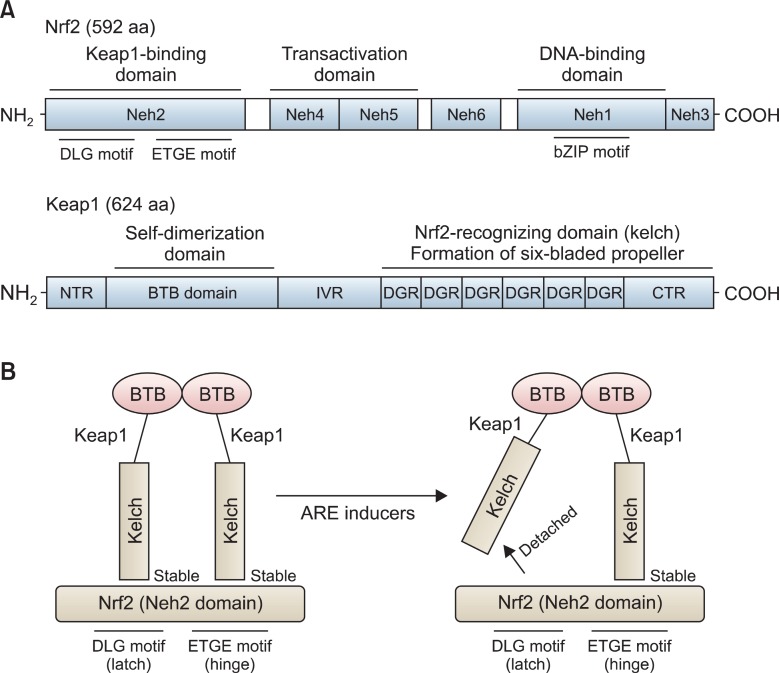
Nrf2 is a basic leucine zipper transcription factor that contains a Cap’n’Collar (CNC) structure. CNC is typically defined by the presence of a conserved 43-amino acid domain, and located in the N-terminal to the DNA binding domain (Sykiotis and Bohmann, 2010). Most CNC transcription factors are transcriptional activators, but they can act as transcriptional suppressors when naturally-truncated or Caspase-cleaved (Ohtsubo et al., 1999; Motohashi et al., 2002). Nrf2 protein is composed of 6 highly conserved Nrf2-ECH homology (Neh) domains (Fig.2 A, upper panel). The Neh1 domain contains a basic leucine zipper (bZIP) motif and behaves as a platform for ARE sequence binding. The Neh2 domain is located in the most N-terminal region and acts as a negative regulatory domain by binding to a cellular repressive regulator, Keap1(Kelch-like ECH-associated protein 1). The tandem Neh4 and Neh5 domains are essential for Nrf2 transactivation and the Neh3 domain, located in the most C-terminal region, is known to play a permissive role of Nrf2 transactivation (Nioi et al., 2005). The Neh6 domain, located between the transactivation domain (the Neh4 and Neh5) and the DNA binding domain (the Neh1), is known to be necessary for degradation of Nrf2 protein (McMahon et al., 2004).
Fig. 2. Molecular architecture of Nrf2 and Keap1 proteins and two-site substrate recognition model (Hinge and Latch Model) for the Keap1/Nrf2 System. Nrf2 protein consists of 592 amino acids (592 aa) and is composed of six Neh domains (Neh1-Neh6). Neh2 is the interacting domain with Keap1 and Neh4 and Neh5 are transactivation domain. Neh1 domain contains basic leucine zipper (bZIP) motif and is the binding domain for ARE. On the other hand Keap1 consists of NTR (N-terminal Region) BTB (a Broad complex, Tramtrack and Bric a brac domain), IVR (intervening region), six DGRs (dougle glycine repeats, also called as Kelch), and CTR (carboxyl terminal region) (A). Keap1proteins homo-dimerize each other by utilizing the BTB domains in cells. The Keap1 protein homodimer recognizes the DLG (weak interaction) and ETGE (strong interaction) motifs in the Neh2 domain of Nrf2. After stress, detachment of the weak-binding DLG motif from Keap1(latch) occurs, but the strong binding ETGE motif from Keap1 (hinge), however, remains attached (B).
Keap1 is a negative regulatory protein of Nrf2 by binding to the Neh2 domain of Nrf2. In fact, Keap1 was initially identified by yeast 2-hybrid assay, using the Neh2 domain of Nrf2 as bait (Itoh et al., 1999). Keap1 consists of 5 different domains:an amino-terminal region (NTR), a Broad complex, Tramtrack and Bric a brac domain (BTB), an intervening region (IVR), six Kelch/dougle glycin repeats (DGRs), and a carboxyl terminal region (CTR) (Fig.2 A, bottom panel). Keap1 is a cytosolic protein whose subcellular location can be explained, at least in part, by binding to a cytoplasmic actin or myosin VIIa through the DGR domain (Kang et al., 2004). Therefore, it is conceivable that Keap1 acts as a cytosolic anchor of Nrf2, sequestering Nrf2 in the cytoplasm during basal conditions and this fact can be easily confirmed by overexpression of GFP-tagged Keap1 in cells. By conducting intensive biophysical analyses, Yamamoto and colleagues have demonstrated that Keap1 protein employs the DGR regions by forming six-bladed propeller to recognize two primary sequences, e.g. the ETGE and DLG motifs, existing in the Neh2 domain of Nrf2 protein, in which Keap1 homodimerizes via the BTB domain to bind to Nrf2 protein at a ratio of 2:1 (Tong et al., 2006). The overlapping ETGE and DLG motifs in Nrf2 protein seem to bind to two Keap1 proteins with a highly differential affinity. Isothermal calorimetry (ITC) measurement showed that the Keap1 binding affinity to ETGE motif (Ka=20×107 M−1) is much stronger than DLG motif (Ka=0.1×107 M−1) (Tong et al., 2007). Based on these observations, they have proposed, so called “hinge and latch” model to explain the regulation of Nrf2 by Keap1 in the cytoplasm, in which the “hinge” mediates a high-affinity interaction between the Nrf2 ETGE motif and Keap1 and this interaction is unaffected by stress inducers, whereas the “latch” mediates displacement of the Nrf2 DLG motif from Keap1 in response to stress inducers (Fig.2 B). While this model is currently considered as a prime mechanism of action that explains the mode of Nrf2/Keap1 interaction in the cytoplasm, there are some disputes against this model (Li and Kong, 2009).
After identification of Keap1 as a binding partner of Nrf2, scientists have identified a number of putative Nrf2 or Keap1-regulatory proteins. For example, He et al. have identified activating transcription factor 4 (ATF4) as an Nrf2-interacting protein in which ATF4 heterodimerizes with Nrf2 and potentiates ARE activation by Nrf2 (He et al., 2001). Karapetian et al. have identified prothymosin-α as a novel binding partner of human
Keap1 protein that competes with Nrf2 protein for binding to the same domain of Keap1 (Karapetian et al., 2005). Sun et al. have demonstrated that Bach1 protein inhibits transcription of HO-1 in a basal condition by making the enhancer regions of HO-1 (E1 and E2) inaccessible to Nrf2 protein or the other unknown activator proteins (Sun et al., 2002). Clements et al.have demonstrated that DJ-1/PARK7 is an indispensable protein for Nrf2-mediated transcriptional activation, although its direct interaction with Nrf2 or Keap1 protein was unclear (Clements et al., 2006). More recently, it was shown that the selective autophagy substrate, p62 interacts with the Nrf2-binding site on Keap1. As such, overproduction of p62 or a deficiency in autophagy interferes with the interaction between Nrf2 and Keap1, resulting in the stabilization of Nrf2 (Komatsu et al., 2010). Collectively, these studies point out that Nrf2-mediated regulation of ARE-dependent gene expression is a complicated process that is orchestrated by many intracellular cofactor proteins.
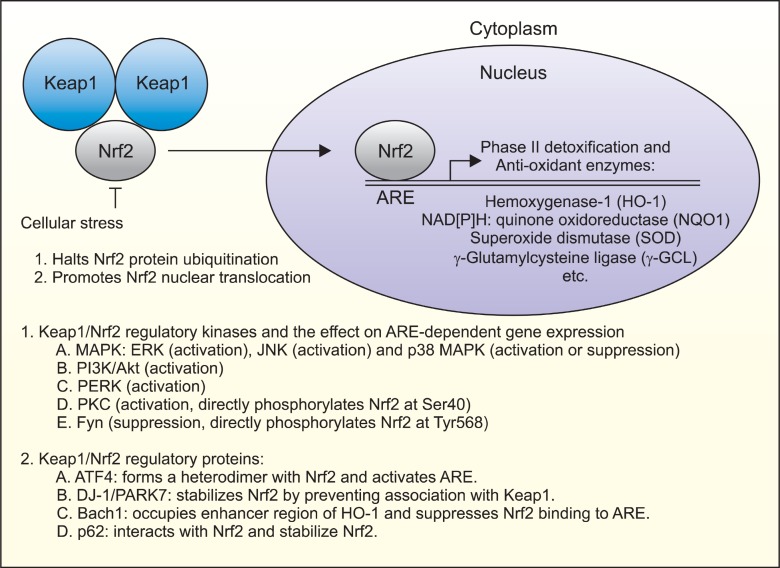
Natural chemopreventive agents are reported to be strong chemical inducers of ARE-dependent gene expression as well as phase II detoxification and antioxidant enzymes (Jeong et al., 2005). Accumulating evidence indicates that ARE activation by chemopreventive compounds is mediated by modulating the activities of intracellular signaling kinase pathways. Kong and colleagues have shown that mitogen-activated protein kinases (MAPKs) are implicated in the regulation of ARE-dependent gene expression, in which ERK and JNK are positive regulators while p38 MAPK is a negative regulator (Keum et al., 2004). On the other hand, there exist a number of other kinases involved in ARE-dependent gene regulation, such as phosphatidylinositol 3’-kinase (PI3K), PKR-like endoplasmic reticulum kinase (PERK), protein kinase C (PKC), Fyn kinase, and glycogen synthase kinase-3β (GSK3β). Due to their ability to phosphorylate many proteins, the possibility that these kinases directly phosphorylate Nrf2 protein have been raised and tested. Huang et al. have reported that a direct phosphorylation of Nrf2 at Ser40 by PKC play a positive role in Nrf2-mediated ARE activation by interfering with the interaction of Nrf2 with Keap1 (Huang et al., 2000). Cullinan et al. also showed that Nrf2 could be directly phosphorylated by PERK, although its target sites remain to be identified (Cullinan et al., 2003). In both cases, Nrf2 phosphorylation contributes to the activation of ARE-dependent gene expression. On the contrary, Jain et al. have reported an intriguing observation that Fyn kinase can phosphorylate Tyr568 of Nrf2 and causes a nuclear exclusion of Nrf2 protein (Jain and Jaiswal, 2006). Although the detailed mechanisms how phosphorylation of Nrf2 protein differentially contributes to ARE-dependent gene expression are still unclear, these observations suggest that phosphorylation of Nrf2 protein can dictate the activation or inhibition of ARE-dependent gene expression, probably depending on the amino residue(s) that are phosphorylated. In summary, Fig. 3 depicts the mode of Nrf2-mediated ARE gene regulation and its modulation by Keap1, intracellular signaling kinases and Keap1/Nrf2/ARE-binding proteins.
Fig. 3. Regulation of Nrf2-mediated are activity by Nrf2 phosphorylation and Keap1/Nrf2/ARE-interacting proteins. Cellular Nrf2 protein is constantly synthesized and degraded by Keap1-mediated polyubiquitination in the cytoplasm. In response to endogenous or exogenous stresses degradation of Nrf2 protein halts, which causes Nrf2 to translocate into the nucleus and bind to ARE, contributing to the synthesisn of phase II detoxification and anti-oxidant enzymes. In addition, Nrf2-mediated ARE gene activation is placed under a tight control of Nrf2 phosphorylation directly or indirectly by numerous intracellular kinases or can be modulated by interactions with other Nrf2/Keap1/ARE-binding proteins in cells.
REGULATION OF Nrf2 PROTEOSOMAL DEGRADATION AND NUCLEOCYTOPLASMIC TRAFFICKING
Nrf2 is a redox-sensitive transcriptional factor and such sensitiveness will involve at least two key steps: (1) the modulation of protein phosphorylation and/or cellular expression level and (2) the regulation of nuclear import and export. Considering that many intracellular signaling kinase cascades are involved in the regulation of ARE-dependent gene expression, it is possible to assume that the activation of selected kinase pathways might be linked to the proteosomal degradation of Nrf2 protein. This conjecture is based on the facts that (1) phosphorylation of a protein can serve as a precursor signal to elicit or inhibit ubiquitin-mediated proteosomal degradation (Pickart, 2004) and (2) the regulation of cellular Nrf2 protein level is largely mediated by the ubiquitin-mediated protein degradation (Furukawa and Xiong, 2005). Ubiquitin is a small conserved regulatory protein whose main function is to mark the proteins for proteolysis. Ubiquitin-mediated proteolysis requires a cascade of three enzymes: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating) and E3 (ubiquitin-ligase) enzymes. Unlike circumscribed functions of E1 and E2 enzymes, E3 enzymes are loosely defined with at least two distinct functions: catalyzing isopeptide formation and recruiting specific substrates to this catalytic activity. At present, hundreds of E3 enzymes have been identified and many of these E3 ligases contain either the homologous to E6-associated protein (E6-AP) COOH-terminus (HECT) domain or the really interesting new gene (RING) finger domain, in which RING finger-type E3 promotes the ubiquitination of substrates by positioning the substrates in a close proximity to the activated E2, while HECT-type E3 ligases display a catalytic activity by itself (Chen et al., 2006).
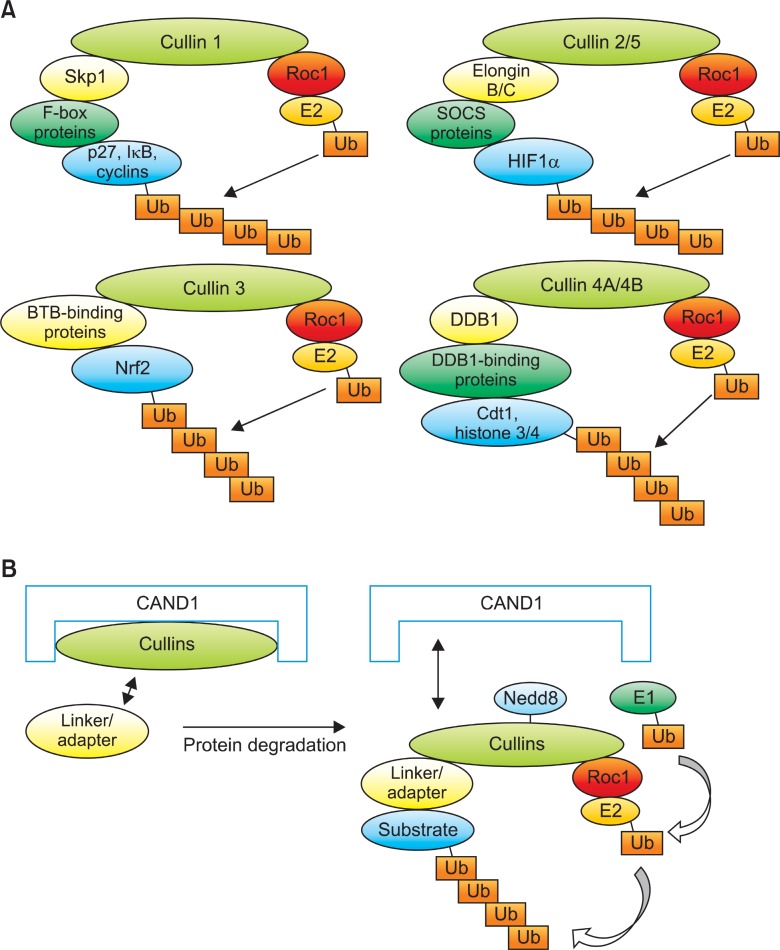
Cullins (Culs) are a group of evolutionary conserved proteins consisting seven different isotypes in human (Cul1, 2, 3, 4A, 4B, 5 and 7) and serve as a scaffold to assemble Cullin-RING E3 Ligases (CRLs) by its C-terminal interaction with the small RING finger-containing protein, ROC1 or ROC2 and its N-terminal interaction with a number of substrate adaptor proteins that recognize and bind to specific domains in the substrates (Fig.4A). The SCF complex, one of a classical RING finger-type E3 ligase, consists of 4 distinct modular proteins (Skp1, Cullin 1, F-box and ROC1) and serves as the prototype of other cullin-based E3 ligases, including the well-defined ECS complex (ElonginB/C-Cullin 2-SOCS-ROC1), Cul3-
Fig. 4. Modular composition and CAND1-mediated regulation of cullin-based E3 ligase activity. Cullin 1 (Cul1) uses F-box proteins to degrade the substrates as adaptors that recognize a variety of substrates through Skp1 linker protein. Representative substrates of Cul1-dependent E3 ligase include p27, IkB and Cyclins. Likewise, Cul2/5 recruits SOCS proteins as adaptor to recognize a variety of substrates, including HIF1α using Elongin B/C as linker proteins. Without a linker protein, Cul3 utilize BTB proteins (for example, Keap1) as an adaptor and recruits many protein substrates, including Nrf2. Cul4 recruits DDB1 as a linker and proteins with WD-40 motif as an adaptor protein to recognize and degrade the substrates, including Cdt1 and Histone 3/4 (H3/4) (A). CAND1 protein inhibits the activity of Cullins by enabling linker or adaptor proteins inaccessible to Cullins at the N-terminus and Roc1 and E2 inaccessible to Cullins at the C-terminus. When a substrate is signaled for degradation, CAND1 is removed from Cullins and Nedd8 is conjugated to Cullins (Neddylation) by ubiquitin-conjugating enzymes (Ubcs), which helps to recruit the E1 and E2 ligase components to E3 enzymes and subject substrates to poly-ubiquitin (Ub) chain formation and proteolysis (B).
based (BTB-Cul3-ROC1) and Cul4-based E3 ligase structures (DDB1-Cul4A/B-ROC1, respectively) (Petroski and Deshaies, 2005). It is known that Cul1 uses Skp1 and F-box proteins to recruit and degrade the substrates to Cul1-dependent E3 ligase (Nakayama and Nakayama, 2006). Likewise, Cul2 and Cul5 utilizes elongins B and C and SOCS box proteins (Kaelin, 2002). Cul3 and Cul4 utilize the BTB and DDB1 motif proteins to recognize and ubiquitinate the substrates, respectively (Furukawa et al., 2003; Higa and Zhang, 2007).
Because Keap1 possesses BTB domain, this protein can act as an adaptor protein for the Cul3-mediated Nrf2 ubiquitination (Cullinan et al., 2004; Kobayashi and Yamamoto, 2005) and that two cysteine residues in Keap1, e.g. Cys273 and Cys288, appear to be critical for the degradation of Nrf2 protein (Zhang and Hannink, 2003). The activity of Nrf2 proteosomal degradation is regulated, at least in part, by the assembly of Cul3 with its adaptor protein, Keap1. Cullin-associated and neddylation-dissociated 1 (CAND1) is an inhibitory protein of all cullins that form a ternary complex with cullins and ROC1 by binding to both N-terminal and C-terminal sequences of cullins (Fig.4B). When the cells perceive a degradation signal of proteins, CAND1 protein is removed from Culs by the COP9-signalosome (CSN), which in turn leads to NEDD8 modification of Culs, e.g. Neddylation, leading to the decreased affinity of CAND1 for the cullin protein that enables another substrate adaptor protein (presumably with its bound substrate) to displace CAND1 and initiate another cycle of substrate ubiquitination and degradation (Fig.4B) (Liu et al., 2002). Lo and Hannink have previously observed that endogenous expression of Nrf2 was decreased by an ectopic expression of CAND1, but increased by anti-CAND1 siRNA and that (2) Neddylation of Cul3 at Lys712 was required for an efficient Keap1-dependent Nrf2 ubiquitiation (Lo and Hannink, 2006). This fact suggests that CAND1 is an important component in regulating Nrf2 protein stability.
Treatment of chemopreventive compounds (for example, β-NF, curcumin, and sulforaphane) induces the expression of Nrf2 protein by attenuating its proteosomal degradation without affecting Nrf2 mRNA level (Nguyen et al., 2003). Considering the critical role of CAND1, Cul3, Keap1 and Nrf2 proteins in regulating ARE-dependent gene expression, it can be presumed that chemopreventive agents will induce Nrf2 protein expression by affecting the expression and/or assembly of CAND1, Cul3 and Keap1 proteins. However, CAND1, Cul3, and Keap1 proteins are very stable proteins (data not shown). Therefore, modulation of CRL assembly by chemopreventive agents seems an appropriate mechanism responsible for Nrf2 stability and a couple of putative biochemical mechanisms can be proposed to explain how chemopreventive agents attenuate Nrf2 proteosomal degradation. First, it is possible to speculate that chemopreventive compounds affect CAND1 binding to Cul3 possibly by modulating Neddylation status of Cul3 to accomplish Nrf2 protein stabilization. Second, chemopreventive compounds might suppress Nrf2 ubiquitination by disrupting the association of Keap1/Nrf2 proteins. To the best of my knowledge, however, no clear-cut studies have been performed to address these issues yet. Third, the possibility that unknown protein factor(s) other than Keap1 might be involved in Nrf2 protein stabilization by chemopreventive agents can be raised. This view is supported by findings of Hayes and colleagues that deletion of the ETGE from Nrf2-Gal4 fusion protein could undergo proteosomal degradation without the need to interact with Keap1 (McMahon et al., 2003) and DIDLID element in Nrf2 might serve as a component for Keap1-independent degradation by recruiting an unidentified ubiquitin ligase (McMahon et al., 2004). If these observations truly hold, it is proposed that protein(s) possessing BTB domain other than Keap1 may take part in Keap1-independent proteosomal degradation of Nrf2 by binding to Cul3. In fact, there exist a large number of putative proteins with BTB domain that can potentially bind to Cul3 in cells (Sumara et al., 2008).
While it is generally held that Keap1 protein behaves as a sensor protein to regulate the release of Nrf2 protein in the cytosol (Zhang, 2006), Kong and colleagues have identified a nuclear export signal (NES, 537LKKQLSTLYL546) that overlaps with the basic leucine zipper (bZIP) domain in Nrf2 protein (Li et al., 2005). This sequence appears to be redox-insensitive, since cytoplasmic distribution of overexpressed Nrf2 zip-domain truncated protein was unaltered by treatments of oxidants (sulforaphane and diethyl maleate) or reducing agents (N-acetyl cysteine or reduced glutathione). Also, they have identified another nuclear export signal (NES, 173LLSIPELQ-CLNI186), located in the transactivation (TA) domain of Nrf2 protein (Li et al., 2006) and, in contrast to NES located in bZIP domain, this NES seemed redox-sensitive, as reflected by an observation that treatments of sulforaphane, tert-butylhydroquinone and H2O2 effectively induced nuclear translocation of overexpressed GFP-NES protein (amino acids 162-295). Together, these study open up an interesting possibility that, in addition to Keap1, Nrf2 protein by itself can act as a sensor protein to dictate transcription of phase II detoxification and anti-oxidant enzymes by modulating its own subcellular location in response to oxidative stress or electrophiles.
CONCLUDING REMARKS
Chemoprevention research has emerged as an important research discipline in recent years. A number of population-based clinical trials are currently in progress to affirm the chemopreventive efficacy of natural or synthetic compounds against various types of tumors in human (Lippman and Hawk, 2009). However, the number of chemopreventive agents proven to be effective in human is still few and several naturally-occurring ingredients, observed to possess chemopreventive effects in preclinical studies were ineffective or even harmful against selected types of tumors in the clinical settings. Such examples include selenium and vitamin E against prostate cancer (Lippman et al., 2009) and alpha-tocopherol and beta-carotene against lung cancer (Omenn et al., 1996). The possible reason for these surprising results probably stemmed, at least in part, from our ignorance of their exact molecular targets in our body. Therefore, understanding the in-depth regulatory mechanisms of chemopreventive targets, including Keap1/Nrf2 module will increase the chance to develop more effective chemopreventive and/or chemotherapeutic agents in the future.
Acknowledgments
This work was supported by the GRRC program of Gyeong-gi province [(GRRC-DONGGUK2011-A01), Study of control of viral diseases].
References
- 1.Chen C. Seth A. K. Aplin A. E. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol. Cancer Res. (2006);4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 2.Clements C. M. McNally R. S. Conti B. J. Mak T. W. Ting J. P. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA. (2006);103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullinan S. B. Gordan J. D. Jin J. Harper J. W. Diehl J. A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. (2004);24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullinan S. B. Zhang D. Hannink M. Arvisais E. Kaufman R. J. Diehl J. A. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell Biol. (2003);23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furukawa M. He Y. J. Borchers C. Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. (2003);5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 6.Furukawa M. Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell Biol. (2005);25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D. Weinberg R. A. Hallmarks of cancer: the next generation. Cell. (2011);144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D. Weinberg R. A. The hallmarks of cancer. Cell. (2000);100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 9.He C. H. Gong P. Hu B. Stewart D. Choi M. E. Choi A. M. Alam J. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J. Biol. Chem. (2001);276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 10.Higa L. A. Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell. Div. (2007);2:5. doi: 10.1186/1747-1028-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H. C. Nguyen T. Pickett C. B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA. (2000);97:12475–12480. doi: 10.1073/pnas.220418997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh K. Chiba T. Takahashi S. Ishii T. Igarashi K. Katoh Y. Oyake T. Hayashi N. Satoh K. Hatayama I. Yamamoto M. Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. (1997);236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 13.Itoh K. Wakabayashi N. Katoh Y. Ishii T. Igarashi K. Engel J. D. Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. (1999);13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain A. K. Jaiswal A. K. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. (2006);281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 15.Jeong W. S. Keum Y. S. Chen C. Jain M. R. Shen G. Kim J. H. Li W. Kong A. N. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J. Biochem. Mol. Biol. (2005);38:167–176. doi: 10.5483/BMBRep.2005.38.2.167. [DOI] [PubMed] [Google Scholar]
- 16.Kaelin W. G. Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer. (2002);2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 17.Kang M. I. Kobayashi A. Wakabayashi N. Kim S. G. Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA. (2004);101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karapetian R. N. Evstafieva A. G. Abaeva I. S. Chichkova N. V. Filonov G. S. Rubtsov Y. P. Sukhacheva E. A. Melnikov S. V. Schneider U. Wanker E. E. Vartapetian A. B. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol. Cell Biol. (2005);25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kensler T. W. Wakabayashi N. Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. (2007);47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 20.Keum Y. S. Jeong W. S. Kong A. N. Chemoprevention by isothiocyanates and their underlying molecular signaling mechanisms. Mutat. Res. (2004);555:191–202. doi: 10.1016/j.mrfmmm.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M. Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox. Signal. (2005);7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M. Kurokawa H. Waguri S. Taguchi K. Kobayashi A. Ichimura Y. Sou Y. S. Ueno I. Sakamoto A. Tong K. I. Kim M. Nishito Y. Iemura S. Natsume T. Ueno T. Kominami E. Motohashi H. Tanaka K. Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. (2010);12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 23.Li W. Jain M. R. Chen C. Yue X. Hebbar V. Zhou R. Kong A. N. Nrf2 Possesses a redox-insensitive nuclear export signal overlapping with the leucine zipper motif. J. Biol. Chem. (2005);280:28430–28438. doi: 10.1074/jbc.M410601200. [DOI] [PubMed] [Google Scholar]
- 24.Li W. Kong A. N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. (2009);48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W. Yu S. W. Kong A. N. Nrf2 possesses a redox-sensitive nuclear exporting signal in the Neh5 transactivation domain. J. Biol. Chem. (2006);281:27251–27263. doi: 10.1074/jbc.M602746200. [DOI] [PubMed] [Google Scholar]
- 26.Lippman S. M. Hawk E. T. Cancer prevention: from 1727 to milestones of the past 100 years. Cancer Res. (2009);69:5269–5284. doi: 10.1158/0008-5472.CAN-09-1750. [DOI] [PubMed] [Google Scholar]
- 27.Lippman S. M. Klein E. A. Goodman P. J. Lucia M. S. Thompson I. M. Ford L. G. Parnes H. L. Minasian L. M. Gaziano J. M. Hartline J. A. Parsons J. K. Bearden J. D. 3rd. Crawford E. D. Goodman G. E. Claudio J. Winquist E. Cook E. D. Karp D. D. Walther P. Lieber M. M. Kristal A. R. Darke A. K. Arnold K. B. Ganz P. A. Santella R. M. Albanes D. Taylor P. R. Probstfield J. L. Jagpal T. J. Crowley J. J. Meyskens F. L. Jr. Baker L. H. Coltman C. A. Jr. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA. (2009);301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J. Furukawa M. Matsumoto T. Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell. (2002);10:1511–1518. doi: 10.1016/S1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 29.Lo S. C. Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol. Cell Biol. (2006);26:1235–1244. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon M. Itoh K. Yamamoto M. Hayes J. D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. (2003);278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 31.McMahon M. Thomas N. Itoh K. Yamamoto M. Hayes J. D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. (2004);279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 32.Motohashi H. O'Connor T. Katsuoka F. Engel J. D. Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. (2002);294:1–12. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama K. I. Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. (2006);6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen T. Sherratt P. J. Huang H. C. Yang C. S. Pickett C. B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. (2003);278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 35.Nioi P. Nguyen T. Sherratt P. J. Pickett C. B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell Biol. (2005);25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohtsubo T. Kamada S. Mikami T. Murakami H. Tsujimoto Y. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. (1999);6:865–872. doi: 10.1038/sj.cdd.4400566. [DOI] [PubMed] [Google Scholar]
- 37.Omenn G. S. Goodman G. E. Thornquist M. D. Balmes J. Cullen M. R. Glass A. Keogh J. P. Meyskens F. L. Valanis B. Williams J. H. Barnhart S. Hammar S. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. (1996);334:1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 38.Petroski M. D. Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nature Reviews Molecular Cell Biology. (2005);6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 39.Pickart C. M. Back to the future with ubiquitin. Cell. (2004);116:181–190. doi: 10.1016/S0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 40.Sharpless N. E. Depinho R. A. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. (2006);5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 41.Sumara I. Maerki S. Peter M. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. (2008);18:84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Sun J. Hoshino H. Takaku K. Nakajima O. Muto A. Suzuki H. Tashiro S. Takahashi S. Shibahara S. Alam J. Taketo M. M. Yamamoto M. Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO. J. (2002);21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sykiotis G. P. Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. (2010);3 (112):re3.. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong K. I. Kobayashi A. Katsuoka F. Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. (2006);387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 45.Tong K. I. Padmanabhan B. Kobayashi A. Shang C. Hirotsu Y. Yokoyama S. Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell Biol. (2007);27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogelstein B. Kinzler K. W. Cancer genes and the pathways they control. Nat. Med. (2004);10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D. D. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. (2006);38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D. D. Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell Biol. (2003);23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]