Abstract
The DNA-damage–signaling pathway has been implicated in all human cancers. However, the genetic defects and the mechanisms of this pathway in prostate carcinogenesis remain poorly understood. In this study, we analyzed CHEK2, the upstream regulator of p53 in the DNA-damage–signaling pathway, in several groups of patients with prostate cancer. A total of 28 (4.8%) germline CHEK2 mutations (16 of which were unique) were found among 578 patients. Additional screening for CHEK2 mutations in 149 families with familial prostate cancer revealed 11 mutations (5 unique) in nine families. These mutations included two frameshift and three missense mutations. Importantly, 16 of 18 unique CHEK2 mutations identified in both sporadic and familial cases were not detected among 423 unaffected men, suggesting a pathological effect of CHEK2 mutations in prostate cancer development. Analyses of the two frameshift mutations in Epstein Barr virus–transformed cell lines, using reverse-transcriptase polymerase chain reaction and western blot analysis, revealed abnormal splicing for one mutation and dramatic reduction of CHEK2 protein levels in both cases. Overall, our data suggest that mutations in CHEK2 may contribute to prostate cancer risk and that the DNA-damage–signaling pathway may play an important role in the development of prostate cancer.
Introduction
Genetic components contributing to prostate cancer (MIM 300200) have been difficult to identify, largely because of the complexity of this disease and the presence of phenocopies in high-risk families. Current genetic studies, using linkage analysis of “high-risk families” followed by positional cloning approaches, have identified more than six susceptibility loci (Ostrander and Stanford 2000). Only two studies have shown any success with the cloning of candidate susceptibility genes from these regions: HPC1 (MIM 601518) and HPC2/ELAC2 (MIM 605367), localized to chromosomes 1q and 17p, respectively (Tavtigian et al. 2001; Carpten et al. 2002). However, follow-up studies for HPC2/ELAC2 have failed to replicate the original findings (Wang et al. 2001; Xu et al. 2001) or have suggested only a limited role in hereditary prostate cancer (Rebbeck et al. 2000; Wang et al. 2001). Since prostate cancer is heterogeneous in nature, and because of the difficulty in identifying highly penetrant susceptibility genes, it may be that the pathogenesis of the disease is related, at least in part, to genomic mutations in multiple low-penetrance genes. Although less penetrant, such genes might play an important role at a population level.
Genomic instability is a common feature of many human cancers (Hoeijmakers 2001). The DNA-damage–signaling pathway plays a critical role in maintaining genomic stability in response to a variety of DNA-damaging events (Khanna and Jackson 2001). Disruption of this pathway has been shown to be pivotal in cancer development, since several proteins involved in this pathway (such as BRCA1 [MIM 113705], TP53 [MIM 191170], and ATM [MIM 208900]) are frequently mutated in human cancers and in several heritable cancer-prone syndromes, such as Li-Fraumeni syndrome (LFS [MIM 151623]) and ataxia telangiectasia (MIM 208900) (Malkin et al. 1990; Miki et al. 1994; Savitsky et al. 1995). Evidence that the DNA-damage–signaling pathway is also important in prostate cancer development comes from several studies. Adenovirus-mediated antisense ATM gene transfer has been shown to sensitize prostate cancer cells to radiation (Fan et al. 2000), and mutation in p53 is associated with amplification of the androgen receptor (MIM 313700) gene in prostate cancer (Koivisto and Rantala 1999). In addition, a low frequency of germline mutations in the breast cancer predisposition genes BRCA1 and BRCA2 (MIM 600185) has been identified in familial prostate cancer (Gayther et al. 2000). Moreover, the male mutation carriers in these families had been shown to have a 3.3-fold increased risk for prostate cancer, relative to the general population (Ford et al. 1994). Cumulatively, these data support the notion that the integrity of the DNA-damage–signaling pathway is essential for the prevention of prostate cancer. Since mutations in TP53, the key regulator of the DNA-damage–signaling pathway, are infrequent in prostate cancer but common in all other cancer types, we hypothesized that other components in this pathway could be mutation targets in prostate cancer.
CHEK2 (MIM 604373) is a mammalian homologue of the Saccharomyces cerevisiae Rad53 and Schizosaccharomyces pombe Cds1, both of which are involved in the DNA-damage–signaling pathway (Paulovich and Hartwell 1995; Sanchez et al. 1996; Boddy et al. 1998). CHEK2 is phosphorylated in response to various DNA-damage agents in an ATM-dependent fashion (Matsuoka et al. 1998). Activated CHEK2, along with other DNA-damage–activated protein kinases, stabilizes TP53 or enhances degradation of Cdc25A (MIM 116974) in the cell-cycle checkpoint control (Matsuoka et al. 1998; Hirao et al. 2000; Falck et al. 2001), through coordination of DNA repair, cell-cycle progression, and apoptosis (Caspari 2000; Bulavin et al. 2001). Recently, heterozygous germline mutations in the CHEK2 gene have been identified in patients with LFS, a highly penetrant familial cancer phenotype usually associated with inherited mutations in TP53 (Bell et al. 1999). One of the CHEK2 germline mutations (1100delC) identified in LFS was also identified in 5.1% of noncarriers of BRCA1 or BRCA2 mutations in families with breast cancer, suggesting its involvement in familial breast cancer, as well (Meijers-Heijboer et al. 2002). Subsequently, somatic CHEK2 mutations were also found in a subset of the primary tumors of LFS, such as sarcoma, breast cancer, and brain tumors, but were rare in other tumors (Allinen et al. 2001; Miller et al. 2002).
In this study, we examined DNA from patients with both sporadic and familial prostate cancers for mutations in CHEK2. We compared the frequency of the CHEK2 mutations in these two prostate-cancer groups with that in an unaffected control group, to determine whether defects in CHEK2 play a role in the development of prostate cancer.
Material and Methods
Ascertainment of Patients with Prostate Cancer
Tissue
Two separate sets of primary prostate tumor samples were collected at the Mayo Clinic and used in this study. The first set of tumor tissues (n=84) was unselected and was collected between 1997 and 1998. The second set (n=92) was selected for young age at diagnosis (<59 years) and was collected between 2000 and 2001. For these patients, neither family history information nor blood was available.
Blood
For a third group, blood was collected from patients with prostate cancer (n=400) with no family history of prostate cancer. These patients with sporadic prostate cancer were collected at the Mayo Clinic and were selected from respondents to a family history survey who reported no family history of prostate cancer (Wang et al. 2001). They were matched by year of diagnosis, age at diagnosis, and number of brothers in the familial group, which is described below. All but 11 of these men were treated surgically for their prostate cancer.
Familial Prostate Cancer Ascertainment
Families with familial clustering of prostate cancer were ascertained as described elsewhere (Berry et al. 2000). These families have occurrence of prostate cancer in a minimum of three men over at least two generations. Blood was collected from as many family members as possible. All men with prostate cancer who contributed a blood specimen had their cancers verified by review of medical records and pathologic confirmation. One family had Hispanic ancestry, and the remainder were white. Two affected members (the proband and one randomly selected affected man from the family) from each of 149 families were initially selected for mutation analysis. When mutations were identified, the other available family members were also screened for the specific mutation.
Unaffected Control Individuals
From a sampling frame of the local population, provided by the Rochester Epidemiology Project (Melton 1996), 475 men were randomly selected for a clinical urologic examination (Oesterling et al. 1993). This exam included digital rectal examination (DRE) and transrectal ultrasound (TRUS) of the prostate, abdominal ultrasound for post-void residual urine volume, serum prostate-specific antigen (PSA) and creatinine measurement, focused urologic physical examination, and cryopreservation of serum for subsequent sex-hormone assays. Any patient with an abnormal DRE, elevated serum PSA level, or suspicious lesion on TRUS was evaluated for prostatic malignancy. If the DRE and TRUS were unremarkable and the serum PSA level was elevated (>4.0 ng/ml), a sextant biopsy (three cores from each side) of the prostate was performed. An abnormal DRE or TRUS result, regardless of the serum PSA level, prompted a biopsy of the area in question. In addition, a sextant biopsy of the remaining prostate was performed. Those men who were found to be without prostate cancer on the basis of this extensive work-up, at baseline or at any of the follow-up exams through 1994, were used for the control population (n=372). To make up for study attrition, the sample was augmented with random samples from individuals in the population who were subjected to an identical workup (n=138), resulting in a total sample of 510 men without evidence of prostate cancer (Roberts et al. 2000). Three hundred and thirty-one of these individuals gave informed consent to participate in this particular study. The second group of normal control DNA samples was obtained from 92 men participating in an ongoing NCI prostate cancer chemo-prevention trial, all of whom were free of evidence of prostate cancer at the time blood was collected, on the basis of DRE and PSA (PSA level <3 ng/ml). The mean age for the 86 men with age data available was 65 years (range 57.6–75.9 years). These two groups of control individuals were combined for the analysis. This study was approved by the Mayo Clinic institutional review board.
Genomic PCR and Mutation Analyses
DNA and RNA isolation from blood, tumor tissues, and cell lines were performed following the manufacturer’s protocol (QIAGEN). Thirteen pairs of intronic primers covering 14 exons of the CHEK2 gene (GenBank accession number XM_009898) were designed (available upon request). Primers used for amplification of exons 10–14 were particularly designed so that either one or both primers for each set of primers had a base mismatch in the most 3′ nucleotide, compared with sequences from nonfunctional copies of CHEK2. The primers thus preferentially amplified the functional CHEK2 on chromosome 22 rather than nonfunctional copies elsewhere in the genome. PCR amplification was performed in a volume of 12.5 μl containing 25 ng of genomic DNA, each primer at 0.2 μM, each dNTP at 0.2 mM, 2.0 mM MgCl2, 0.5 U of Taq polymerase (AmpliTaq Gold, Perkin Elmer), and 1× buffer provided by the manufacturer. Denaturing high-performance liquid chromatography (DHPLC) analyses and direct sequencing of the PCR products were performed as described elsewhere (Liu et al. 1997).
RT-PCR
Lymphoblastoid cell lines from the proband of each family were established on the basis of standard procedures. Lymphocytes from peripheral blood were transformed with Epstein-Barr virus (EBV) and were cultured in RPMI-1640 medium containing 10% fetal bovine serum. All transformed cells were frozen in liquid nitrogen for future use. CHEK2 germline mutations in these cell lines were confirmed by direct sequencing of genomic DNA. The two pairs of primers used for RT-PCR analysis of the mutations are as follows: CHK2F2 (5′-AAAAGAACAGATAAATACCGAACAT-3′) and CHKR2 (5′-TCTGCCTCTCTTGCTGAACC-3′), covering the mutations T470C, G715A, and A751T; and CHK2F3 (5′-AATTGATGGAAGGGGGAGAGCTGT-3′) and CHK2R3 (5′-TAGGTGGGGGTTCCACATAAGGT-3′), covering the 1100delC mutation. For RT-PCR analysis of the abnormal splicing products in the IVS2+1G→A mutant, one pair of exonic primers covering nucleotides 367 (in exon 2) to 564 (in exon 3) were designed (forward, 5′-TATTGCTTTGATGAACCACTGC-3′; reverse, 5′-TTCAGAATTGTTATTCAAAGGAC-3′). RT-PCR products were cloned into pGEM-T easy vector, according to the manufacturer’s protocol (Promega). The abnormal splicing products were detected by DHPLC and were then directly sequenced.
Western Blot Analysis
CHEK2 proteins in the cell lines with CHEK2 mutations were analyzed by western blot analysis. In brief, total protein from each cell line was harvested, denatured in Laemnli buffer (Bio-Rad), and separated on 8% polyacrylamide gels with prestained protein Benchmark (Gibco/BRL). After being transferred onto Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech), CHEK2 protein was visualized with rabbit polyclonal anti-CHEK2 antibody raised against N-terminal residues of human CHEK2 (kindly provided by Dr. J. Chen) by the ECL Western Blotting System (Amersham Pharmacia Biotech). The mouse monoclonal anti-β-actin antibody (clone AC-15, Sigma) was used as an internal control.
Statistical Methods
The frequencies of mutation carriers were compared among different groups, through use of Armitage’s test for trend. For statistical comparisons of patients with familial disease versus control subjects, a test for trend in the number of variant alleles, analogous to Armitage’s test for trend in proportions (Sasieni 1997) but with the appropriate variance to account for the correlated family data, was used (Slager and Schaid 2001).
Results
CHEK2 Mutation Screening
In this study, we screened the CHEK2 gene for mutations in several groups of men with prostate cancer. For the first two groups, only tissue (tumor and matched normal) was available for study (clinic tumors 1 and 2 in table 1). In the 178 patients with available tissue, 13 CHEK2 mutations were identified (table 1; fig. 1). Nine (10.7%) were detected among the 84 unselected patients with prostate cancer, and 4 (4.3%) were detected among the 94 patients with early-onset cancer. These included eight different missense mutations and 1-bp deletion mutations at nucleotide 1100 (1100delC). All of the mutations altered evolutionarily conserved amino acids, with the exception of the Arg181Cys mutation in exon 3. These mutations were considered to most likely be germline mutations, since they were present in both tumor and matched normal prostate tissues. However, since DNA from blood was not available for analysis, there is a possibility that they may represent very early somatic events. In an effort to address this concern, DNA from blood leukocytes was obtained from an additional 400 patients with prostate cancer without a family history of prostate cancer. Fifteen CHEK2 mutations were identified in this third group (3.75%) (table 1). Although there are differences in the frequency of CHEK2 mutations among the three prostate cancer groups, the overall incidence of CHEK2 mutations present in these patients (28/578, 4.8%) suggest that CHEK2 germline mutations are likely to be associated with development of a subset of prostate cancer.
Table 1.
CHK2 Germline Mutations Identified in Men with Prostate Cancer and in Unaffected Control Individuals
| Mutation Number | Mutation | Amino AcidChange | Exon | Domain | ClinicTumors 1(n=84)a | ClinicTumors 2(n=94)b | Individualswith SporadicProstate Cancer(n=400)c | Individualswith FamilialProstate Cancer(n=298)d | Unaffected Men(n=423)e |
| 1 | G190A | Glu64Lys | 1 | S/TQ-rich | 1 | 0 | 1 | 0 | 0 |
| 2 | 245del15 bp | del DQEPE | 1 | S/TQ-rich | 0 | 0 | 1 | 0 | 0 |
| 3 | G434C | Arg145Pro | 2 | FHA | 0 | 0 | 1 | 0 | 0 |
| 4 | IVS2+1G→A | Frameshift | 2 | FHA | 0 | 0 | 0 | 1 | 0 |
| 5 | T470C | Ile157Thr | 3 | FHA | 1 | 0 | 6 | 7 | 5 |
| 6 | G499A | Gly167Arg | 3 | FHA | 0 | 0 | 1 | 0 | 0 |
| 7 | C538T | Arg180Cys | 3 | Unknown | 0 | 2 | 0 | 0 | 1 |
| 8 | G539A | Arg180His | 3 | Unknown | 0 | 0 | 1 | 0 | 0 |
| 9 | C541T | Arg181Cys | 3 | Unknown | 1 | 0 | 0 | 0 | 0 |
| 10 | G542A | Arg181His | 3 | Unknown | 0 | 0 | 1 | 0 | 0 |
| 11 | G715T | Glu239Stop | 5 | Kinase | 0 | 0 | 1 | 0 | 0 |
| 12 | G715A | Glu239Lys | 5 | Kinase | 1 | 0 | 0 | 1 | 0 |
| 13 | A751T | Ile251Phe | 5 | Kinase | 0 | 0 | 0 | 1 | 0 |
| 14 | G954A | Arg318His | 8 | Kinase | 0 | 1 | 0 | 0 | 0 |
| 15 | A967C | Thr323Pro | 8 | Kinase | 1 | 0 | 0 | 0 | 0 |
| 16 | A980G | Tyr327Cys | 8 | Kinase | 0 | 0 | 1 | 0 | 0 |
| 17 | 1100delC | Frameshift | 10 | Kinase | 3 | 1 | 1 | 1 | 0 |
| 18 | C1427A | Thr476Lys | 12 | Kinase | 1 |
0 |
0 |
0 |
0 |
| Total | 9 (10.7%) | 4 (4.3%) | 15 (3.75%) | 11 (3.7%) | 6 (1.4%) | ||||
| P value (Ile157Thr included)f | <.0001 | .07 | .03 | .08 | |||||
| P value (Ile157Thr excluded)g | <.0001 | .0003 | .008 | .11 |
Unselected prostate-cancer tumor samples collected in 1997 and 1998.
Prostate-cancer tumor samples, with a younger age at onset (age <59 years), collected in 2000 and 2001.
Blood samples from patients without a family history of prostate cancer.
Two affected men from each of 149 families were screened.
Population-based control group (n=331) with a mean age at diagnosis of 53.4 years (range 42–83 years), an average PSA value of 0.9 (range 0.15–9.1), and normal TRUS and DRE results, plus a group of unaffected men (n=92) enrolled in an ongoing NCI prostate cancer chemoprevention trail, who were free of clinically evident prostate cancer as assessed by DRE and PSA (<3).
P values comparing each group with controls, using Armitage’s test for trend. Control data: 6 (1.4%) with mutation, 417 (98.6%) with no mutation.
P values comparing each group with controls, using Armitage’s test for trend. Control data: 1 (0.2%) with mutation, 422 (99.8%) with no mutation.
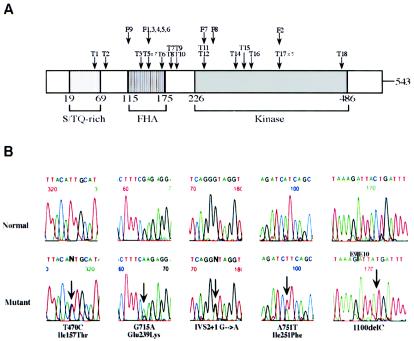
Figure 1.
CHEK2 germline mutations in prostate cancers. A, Mutations found in the CHEK2 gene. “T” indicates the clinic or sporadic prostate tumor samples (numbers as shown in table 1), and “F” indicates families with prostate cancer in which CHEK2 mutations were identified (numbers as indicated in fig. 2). B, Sequence analysis shows the five CHEK2 germline mutations identified in families with familial prostate cancer. DNA sequence analyses were performed on either genomic DNA (first four pairs of panels) or cDNA (right-most panels). Sequences are presented in the 5′→3′ direction, and arrows mark the location of each mutation. The upper panels depict the regions from wild-type alleles and the lower panels show the respective sequences with the mutations. All mutations were detected with genomic DNA and were confirmed with cDNA.
To investigate whether the CHEK2 mutations are also present in familial prostate cancer, we screened two affected members from each of 149 families with familial prostate cancer collected at the Mayo Clinic (Berry et al. 2000). Five different CHK2 mutations in nine families were identified (table 1; fig. 1). Three were missense mutations—one in exon 3 (T470C, Ile157Thr) and two in exon 5 (G715A, Glu239Lys and A751T, Ile251Phe). The other two were frameshift mutations, including the 1100delC mutation and a splice-site mutation in intron 2 (IVS2+1G→A). All five mutations changed amino acids in either the FHA (forkhead homology-associated) or the kinase activation domain of CHK2, which have previously been shown to be important for protein-protein interaction and phosphorylation of p53 in DNA-damage–signaling (Durocher et al. 2000; Shieh et al. 2000; Li et al. 2002). The presence of these mutations in such important functional domains further suggested that these CHK2 mutations could be deleterious.
To evaluate the association between the CHEK2 mutations and prostate cancer risk, we screened a group of unaffected men (n=423). This group was comprised of two individual sets (table 1). One set contained 331 population-based unaffected control men (Wang et al. 2001), and the other was comprised of 92 control men free of evidence of prostate cancer. Within this control group, two different mutations among six individuals were detected: Arg180Cys (n=1) and Ile157Thr (n=5) (table 1). For the six unaffected men with CHEK2 alterations, there was no evidence of disease at the time of blood collection. However, the mean age of these individuals at the time of collection was only 59.6 years (range 45.5–67.0 years), much younger than 71 years, the mean age at diagnosis of prostate cancer for whites in the United States (Bell et al. 1999). Although it is possible that these individuals may develop prostate cancer or other malignancies occurring in LFS or LFS-like syndromes later in life, it is also likely that the Ile157Thr alteration represents a polymorphism rather than a causative mutation.
Among the mutations detected, the frequency of the Ile157Thr mutation did not appear to differ between case (1.6%) and control (1.18%) individuals. We therefore tested the significance of our mutation data with and without this alteration. A global test using Fisher’s exact test showed a significant difference among all of the groups (P=.002). When the Ile157Thr mutation was omitted, the P value was <.0001. Each of the four case groups was then compared individually with the pooled control groups. With all of the data included, only the first unselected group and the sporadic case group showed a statistically significant increase in the frequency of CHEK2 mutations, compared with the control group (table 1). When the Ile157Thr mutation was excluded, each of the three nonfamilial groups demonstrated statistically significant increases (P<.0001, .0003, and .008, respectively). In both analyses, the frequency of CHEK2 mutations in the familial group was not statistically different than the control group. When the mutations are broken down into four different categories (1100delC, all truncating mutations, all missense mutations, and all missense mutations except Ile157Thr), the associations between the mutations and prostate cancer risk are still significant, with the exception of 1100delC. However, the numbers within each category are too small to allow conclusions to be drawn. The 1100delC mutation has been proposed to confer a low penetrant risk associated with breast cancer risk. Whether it is also a risk factor for prostate cancer or other cancers remains to be elucidated.
CHEK2 Mutations Present in Families with Familial Prostate Cancer
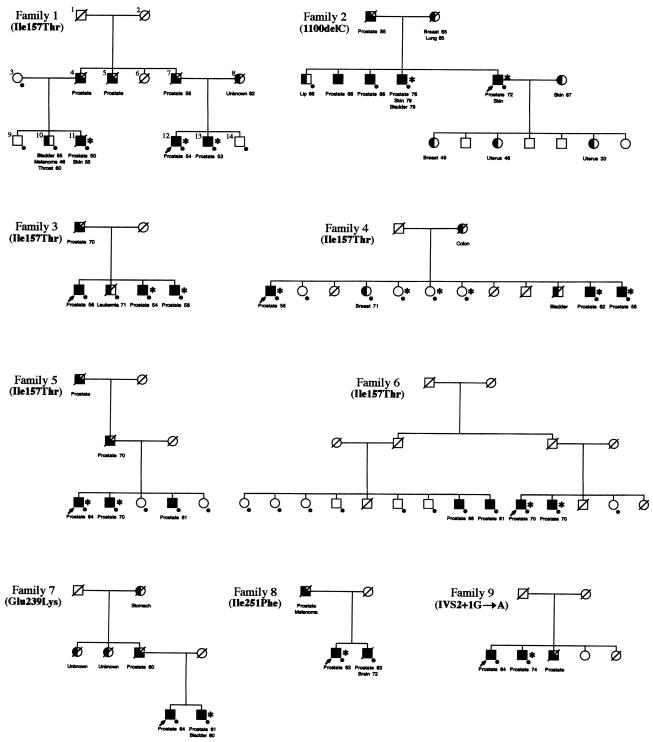
To determine whether CHEK2 mutations cosegregated with prostate cancer in the nine families (families 1–9 in fig. 2), we analyzed the DNA from all available family members for CHEK2 mutations, including both affected and unaffected individuals. Families 1, 3, 4, 5, and 6 had the Ile157Thr mutation. This alteration was present in all affected individuals in two of the five families (families 1 and 4). Family 1 had six prostate cancer cases in two generations. The Ile157Thr mutation was present in all three affected men, including two brothers (individuals 12 and 13) and their cousin (individual 11), and was absent from three unaffected male siblings (individuals 9, 10, and 14). Although not tested directly, the proband’s father (individual 7) and paternal uncle (individual 4) are also expected to be carriers of this mutation, since both are affected and have affected sons with a mutation. In family 4, all the individuals affected with prostate cancer carried the Ile157Thr mutation. However, three sisters also carried the mutation but had no evidence of cancer.
Figure 2.
Segregation of CHEK2 mutations in nine families with prostate cancer (families 1–9). Where known, the individual’s age is indicated to the right side of each cancer. A dot (●) is present at the lower right corner of the symbol if a blood sample was available and was analyzed. An asterisk (*) to the right of the symbol indicates the presence of the CHEK2 mutation carriers in each family. The individual indicated with a dot but without an asterisk has tested negative for CHEK2 mutation. Arrows (↗) indicate probands. Squares denote males; circles denote females; completely blackened symbols denote patients with prostate cancer for whom pathology records were available; 3/4 blackened symbols denote patients with prostate cancer for whom records were unavailable; 1/2 blackened symbols denote patients with other types of cancer; all symbols with a diagonal denote deceased individuals. The cancer type for each individual is shown underneath each symbol.
Four families (families 2, 7, 8, 9) had mutations other than Ile157Thr. Family 2 is a family with multiple cancers, including five prostate cancers, two breast cancers, two uterine cancers, three skin cancers, one lung cancer, one bladder cancer, and one lip cancer, in six males and five females in three generations (fig. 2). The proband harbors the 1100delC mutation. Analysis of the available DNA from three affected men and one unaffected man of this family revealed the mutation in two affected men but not in the unaffected brother. The proband’s daughters, one of whom was diagnosed with breast cancer and two of whom were diagnosed with uterine cancer, were not available for study. In the other three families, the CHEK2 mutation was detected in only one of the two affected brothers. Overall, analysis of CHEK2 mutations in available family members from all nine families revealed that 17 of 25 (68%) affected men harbored CHEK2 mutations, whereas none of the unaffected men (n=8) carried the mutation (fig. 2).
To test for cosegregation of CHEK2 mutations with prostate cancer, we performed linkage analyses under the assumption of an autosomal dominant model (Smith et al. 1996) and no recombination between the underlying susceptibility locus with CHEK2. Although seven of nine families showed evidence against cosegregation, we could rule out only cosegregation with a highly penetrant effect; we cannot rule out a weakly penetrant effect with our data.
Mutant CHEK2 Altered Protein Expression
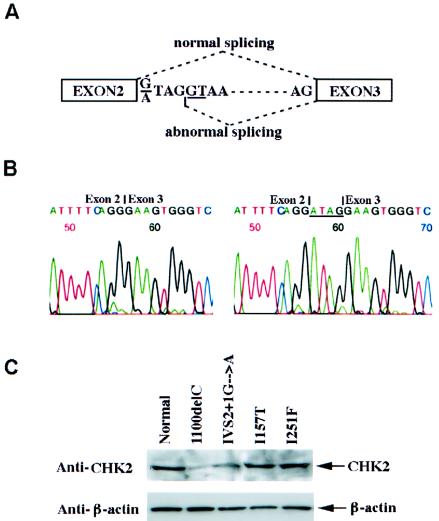
The functional importance of the CHEK2 mutations in prostate cancer development was explored by examination of the mutant gene products. RT-PCR analysis of the EBV-transformed cell lines that were established from the leukocytes of each proband confirmed that all CHEK2 mutations were present in their transcripts, including the 1100delC mutation (fig. 1). The splice-site mutation (IVS2+1G→A) results in a 4-bp insertion due to an abnormal splicing using an alternative splice donor site in intron 2 (fig. 3A and 3B). This mutation creates a premature termination codon in exon 3 and eliminates part of FHA domain and the entire kinase activation domain of CHEK2. Western blot analysis of the two frameshift mutations in the patients’ cell lines showed dramatic reduction of CHEK2 protein levels in both cases (fig. 3C). Reduction of CHEK2 protein has been shown to reduce the kinase activity of CHEK2 in response to DNA damage (Matsuoka et al. 2001). Altogether, our data provide evidence that some of the CHEK2 mutations identified in the patients with prostate cancer whom we studied lead to disruption of CHEK2 expression.
Figure 3.
Abnormal splicing and abnormal protein syntheses of the two CHEK2 frameshift mutations. A, Schematic representation of the abnormal splice for the IVS2+1G→A mutant. A 4-bp insertion is created in the mutant transcript because of the usage of the new splice donor site (underlined). B, Sequences of the wild-type (left) and mutant (right) CHEK2 transcripts (between exons 2 and 3) from the cell line established from the affected men carrying the IVS2+1G→A germline mutation. C, Western blot analyses showing the reduction of CHEK2 in the cell lines carrying the frameshift mutations, compared with the normal lymphocyte cells and the cells carrying CHEK2 missense mutations.
Discussion
In the present study, we identified 18 unique germline CHEK2 mutations among 28 (4.8%) individuals with prostate cancer and in nine families with familial prostate cancer. With the exception of two mutations (1100delC and Ile157Thr) that were previously reported in LFS (Bell et al. 1999), all CHEK2 mutations identified in the present study are unique to prostate cancer. Moreover, some of the mutations presumed to be deleterious are represented by two new truncation mutations (IVS2+1G→A and Glu239Stop), which are predicted to lose their kinase activities.
Association studies between patients with sporadic disease and unaffected control individuals indicated an increased risk of developing prostate cancer in men harboring CHEK2 mutations. The risk appears to be higher when the Ile157Thr mutation is excluded. In contrast, the frequency of CHEK2 mutations in the familial group was not significantly different from that in the control group (table 1). Although the small sample size may account for this, the finding may reflect the presence of more-highly-penetrant genes in the familial group, compared with the other groups. In addition, the patients having CHEK2 mutations in the familial cases may themselves represent phenocopies—that is, the prostate cancer in these patients may be due to CHEK2 mutations and not due to other highly penetrant susceptibility genes segregating within the family. We recognize that well-designed epidemiological studies of large sample sets will be necessary to determine the relevance of these mutations in families with familial prostate cancer.
Germline CHEK2 mutations were first reported by Bell et al. (1999) in patients with classic LFS and wild-type p53. LFS is a highly penetrant familial cancer syndrome, classically associated with germline mutations of TP53. The spectrum of cancers in this syndrome includes breast cancer, soft tissue sarcoma, brain tumors, osteosarcoma, leukemia, and adrenocortical carcinoma (Birch et al. 1984, 1990). Recently, the germline 1100delC mutation was identified in noncarriers of BRCA1 or BRCA2 mutations from families with breast cancer, the primary cancer in LFS (Meijers-Heijboer et al. 2002). This mutation is thought to confer a low penetrance for breast cancer. Allinen et al. (2001) screened the CHEK2 gene in 79 Finnish families with hereditary breast cancer that did not have mutations in BRCA1, BRCA2, or TP53. However, they found only the Ile157Thr alteration, which was also present in 6.5% of control DNA samples. To date, other than somatic CHEK2 mutations, there is no germline CHEK2 mutation reported in other primary tumors of LFS (Miller et al. 2002). It is important to point out that there was no evidence, on the basis of published criteria, that the nine families with familial prostate cancer in which we detected germline CHEK2 mutations had LFS or LFL syndrome (Li et al. 1988; Birch et al. 1994). Ascertainment of these families included collection of family history through telephone interviews and construction of formal pedigrees.
The most common CHEK2 mutation identified in our study was Ile157Thr. The role of this mutation, however, is controversial, even though both genetic and biochemical data from previous studies suggest that this mutation is deleterious (Bell et al. 1999; Falck et al. 2001; Li et al. 2002). On the other hand, this mutation was found in 2.1% (2/95) of healthy population control individuals in Finland and was proposed as a polymorphism (Vahteristo et al. 2001). Other reports also indicate that this mutation is relatively common in normal healthy control individuals (Allinen et al. 2001; Meijers-Heijboer et al. 2002). In our current study, the frequency of this variant was not significantly different among the several groups of samples tested (1.21% for the sporadic prostate cancer groups, 2.34% for familial prostate cancer, 1.18% for unaffected control groups) (table 1). Whether this functionally related CHEK2 variant confers susceptibility to prostate cancer, or even to other cancers, remains to be clarified.
The presence of CHEK2 mutations in prostate cancer highlights the importance of the integrity of the DNA-damage–signaling pathway in prostate cancer development. The fact that mutations in BRCA1 and BRCA2, two other proteins in this pathway, confer an increased risk of prostate cancer further supports this notion (Gayther et al. 2000). Moreover, the recently developed genomic instability-based transgenic mouse model for prostate cancer demonstrated the presence of a similar phenotype of early stages of human prostate cancer and that the genomic instability could be an early event in this disease (Voelkel-Johnson et al. 2000). Overall, our data provide new genetic evidence for the involvement of the DNA-damage–signaling pathway in prostate cancer development. Although the mechanism by which CHEK2 mutations contribute to the development of prostate cancer remains unclear, future studies will add to the observations in the present report. The finding of germline mutations in CHEK2 in both sporadic and familial prostate cancer may facilitate early diagnosis of this cancer and may provide additional insights into the biology of this malignancy, for future therapeutic applications.
Acknowledgments
We thank the members of the families for their cooperation. We thank Dr. Junjie Chen of oncology department at Mayo Clinic, for his gift of CHEK2 antibodies and helpful advice. We thank Dr. Robert B. Jenkins of the Division of Experimental Pathology at Mayo Clinic, for the EBV-transformed cell lines. This work was supported by the Mayo Clinic and the Mayo Clinic Cancer Center, by U. S. Public Health Service Specialized Programs of Research Excellence grant CA 91956, National Institutes of Health grants CA 72818 and DK 58859, Department of Defense grant DAMD17-02-1-0093, and by the Merck Research Laboratory.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CHEK2 [accession number XM_009898])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for prostate cancer [MIM 300200], HPC1 [MIM 601518], HPC2/ELAC2 [MIM 605367], BRCA1 [MIM 113705], TP53 [MIM 191170], LFS [MIM 151623], ataxia telangiectasia and ATM [MIM 208900], androgen receptor [MIM 313700], BRCA2 [MIM 600185], CHEK2 [MIM 604373], and Cdc25A [MIM 116974])
References
- Allinen M, Huusko P, Mantyniemi S, Launonen V, Winqvist R (2001) Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br J Cancer 85:209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA (1999) Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528–2531 [DOI] [PubMed] [Google Scholar]
- Berry R, Schroeder JJ, French AJ, McDonnell SK, Peterson BJ, Cunningham JM, Thibodeau SN, Schaid DJ (2000) Evidence for a prostate cancer-susceptibility locus on chromosome 20. Am J Hum Genet 67:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JM, Hartley AL, Blair V, Kelsey AM, Harris M, Teare MD, Jones PH (1990) Identification of factors associated with high breast cancer risk in the mothers of children with soft tissue sarcoma. J Clin Oncol 8:583–590 [DOI] [PubMed] [Google Scholar]
- Birch JM, Hartley AL, Marsden HB, Harris M, Swindell R (1984) Excess risk of breast cancer in the mothers of children with soft tissue sarcomas. Br J Cancer 49:325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JM, Hartley AL, Tricker KJ, Prosser J, Condie A, Kelsey AM, Harris M, Jones PH, Binchy A, Crowther D (1994) Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res 54:1298–1304 [PubMed] [Google Scholar]
- Boddy MN, Furnari B, Mondesert O, Russell P (1998) Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280:909–912 [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Higashimoto Y, Popoff IJ, Gaarde WA, Basrur V, Potapova O, Appella E, Fornace AJ Jr (2001) Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411:102–107 [DOI] [PubMed] [Google Scholar]
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, et al (2002) Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet 30:181–184 [DOI] [PubMed] [Google Scholar]
- Caspari T (2000) How to activate p53. Curr Biol 10:R315–R317 [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB (2000) The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell 6:1169–1182 [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842–847 [DOI] [PubMed] [Google Scholar]
- Fan Z, Chakravarty P, Alfieri A, Pandita TK, Vikram B, Guha C (2000) Adenovirus-mediated antisense ATM gene transfer sensitizes prostate cancer cells to radiation. Cancer Gene Ther 7:1307–1314 [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343:692–695 [DOI] [PubMed] [Google Scholar]
- Gayther SA, de Foy KA, Harrington P, Pharoah P, Dunsmuir WD, Edwards SM, Gillett C, Ardern-Jones A, Dearnaley DP, Easton DF, Ford D, Shearer RJ, Kirby RS, Dowe AL, Kelly J, Stratton MR, Ponder BA, Barnes D, Eeles RA (2000) The frequency of germ-line mutations in the breast cancer predisposition genes BRCA1 and BRCA2 in familial prostate cancer. The Cancer Research Campaign/British Prostate Group United Kingdom Familial Prostate Cancer Study Collaborators. Cancer Res 60:4513–4518 [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW (2000) DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824–1827 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411:366–374 [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254 [DOI] [PubMed] [Google Scholar]
- Koivisto PA, Rantala I (1999) Amplification of the androgen receptor gene is associated with P53 mutation in hormone-refractory recurrent prostate cancer. J Pathol 187:237–241 [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF Jr, Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, Miller RW (1988) A cancer family syndrome in twenty-four kindreds. Cancer Res 48:5358–5362 [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ (2002) Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9:1045–1054 [DOI] [PubMed] [Google Scholar]
- Liu W, Oefner PJ, Qian C, Odom RS, Francke U (1997) Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test 1:237–242 [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF, Jr., Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA (1990) Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250:1233–1238 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ (1998) Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893–1897 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Nakagawa T, Masuda A, Haruki N, Elledge SJ, Takahashi T (2001) Reduced expression and impaired kinase activity of a Chk2 mutant identified in human lung cancer. Cancer Res 61:5362–5365 [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, et al (2002) Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 31:55–59 [DOI] [PubMed] [Google Scholar]
- Melton LJ 3rd (1996) History of the Rochester Epidemiology Project. Mayo Clin Proc 71:266–274 [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71 [DOI] [PubMed] [Google Scholar]
- Miller CW, Ikezoe T, Krug U, Hofmann WK, Tavor S, Vegesna V, Tsukasaki K, Takeuchi S, Koeffler HP (2002) Mutations of the CHK2 gene are found in some osteosarcomas, but are rare in breast, lung, and ovarian tumors. Genes Chromosomes Cancer 33:17–21 [DOI] [PubMed] [Google Scholar]
- Oesterling JE, Jacobsen SJ, Chute CG, Guess HA, Girman CJ, Panser LA, Lieber MM (1993) Serum prostate-specific antigen in a community-based population of healthy men: establishment of age-specific reference ranges. JAMA 270:860–864 [PubMed] [Google Scholar]
- Ostrander EA, Stanford JL (2000) Genetics of prostate cancer: too many loci, too few genes. Am J Hum Genet 67:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH (1995) A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841–847 [DOI] [PubMed] [Google Scholar]
- Rebbeck TR, Walker AH, Zeigler-Johnson C, Weisburg S, Martin AM, Nathanson KL, Wein AJ, Malkowicz SB (2000) Association of HPC2/ELAC2 genotypes and prostate cancer. Am J Hum Genet 67:1014–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RO, Bergstralh EJ, Lieber MM, Jacobsen SJ (2000) Digital rectal examination and prostate-specific antigen abnormalities at the time of prostate biopsy and biopsy outcomes, 1980 to 1997. Urology 56:817–822 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357–360 [DOI] [PubMed] [Google Scholar]
- Sasieni PD (1997) From genotypes to genes: doubling the sample size. Biometrics 53:1253–1261 [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268:1749–1753 [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C (2000) The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev 14:289–300 [PMC free article] [PubMed] [Google Scholar]
- Slager SL, Schaid DJ (2001) Evaluation of candidate genes in case-control studies: a statistical method to account for related subjects. Am J Hum Genet 68:1457–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Rova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Issacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, et al (2001) A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27:172–180 [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen LA, Blomqvist C, Aittomaki K, Nevanlinna H (2001) p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res 61:5718–5722 [PubMed] [Google Scholar]
- Voelkel-Johnson C, Voeks DJ, Greenberg NM, Barrios R, Maggouta F, Kurtz DT, Schwartz DA, Keller GM, Papenbrock T, Clawson GA, Norris JS (2000) Genomic instability-based transgenic models of prostate cancer. Carcinogenesis 21:1623–627 [PubMed] [Google Scholar]
- Wang L, McDonnell SK, Elkins DA, Slager SL, Christensen E, Marks AF, Cunningham JM, Peterson BJ, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, Thibodeau SN (2001) Role of HPC2/ELAC2 in hereditary prostate cancer. Cancer Res 61:6494–6499 [PubMed] [Google Scholar]
- Xu J, Zheng SL, Carpten JD, Nupponen NN, Robbins CM, Mestre J, Moses TY, Faith DA, Kelly BD, Isaacs SD, Wiley KE, Ewing CM, Bujnovszky P, Chang B, Bailey-Wilson J, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB (2001) Evaluation of linkage and association of HPC2/ELAC2 in patients with familial or sporadic prostate cancer. Am J Hum Genet 68:901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]