Abstract
Rheumatoid arthritis (RA) is a chronic inflammatory joint disease with a complex etiology in which environmental factors within a genetically susceptible host maneuver the innate and adaptive arms of the immune system toward recognition of autoantigens. This ultimately leads to joint destruction and clinical symptomatology. Despite the identification of a number of disease-susceptibility regions across the genome, RA’s major genetic linkage remains with the major histocompatibility complex (MHC), which contains not only the key immune-response class I and class II genes but also a host of other loci, some with potential immunological relevance. Inside the MHC itself, the sole consistent RA association is that with HLA-DRB1, although this does not encode all MHC-related susceptibility. Indeed, in a set of Japanese patients with RA and a control group, we previously reported the presence of a second RA-susceptibility gene within the telomeric human leukocyte antigen (HLA) class III region. Using microsatellites, we narrowed the susceptibility region to 70 kb telomeric of the TNF cluster, known to harbor four expressed genes (IκBL, ATP6G, BAT1, and MICB). Here, using numerous single-nucleotide polymorphisms (SNPs) and insertion/deletion polymorphisms, we identify the second RA-susceptibility locus within the HLA region, as the T allele of SNP 96452 (T/A), in the promoter region (position −62) of the IκBL gene (P=.0062). This −62T/A SNP disrupts the putative binding motif for the transcriptional repressor, δEF1, and hence may influence the transcription of IκBL, homologous to IκBα, the latter being a known inhibitor of NFκB, which is central to innate immunity. Therefore, the MHC may harbor RA genetic determinants affecting the innate and adaptive arms of the immune system.
Introduction
Rheumatoid arthritis (RA [MIM 180300]) is one of the most common autoimmune diseases with a complex genetic etiology. It is characterized by chronic inflammatory symptoms, such as rheumatoid nodules, vasculitis, and scleritis. From an immunological standpoint, RA has remained a paradigm for the study of autoimmunity. The prevalence of RA has been almost constant in the world population, ranging from 0.3% to 1.0% (Felson 1996). The degree of the genetic component in RA has been estimated on the basis of the relative recurrence risk, λs, for siblings of probands with RA. It is likely that the λs value lies between 5 and 10 (Seldin et al. 1999; Jawaheer et al. 2001). Intense research both in humans and in naturally obtained or genetically engineered rodent models has put forth several interesting pathophysiological models in which it appears that both arms of the immune system, innate and adaptive, intervene in disease pathogenesis within a genetically susceptible host (for reviews, see Feldmann et al. 1996; Jirholt et al. 2001). The association between HLA-DRB1 alleles encoding the “shared epitope” (SE) and susceptibility to RA is widely recognized (Gregersen et al. 1987) despite that the actual mechanism by which these alleles and/or this epitope confers susceptibility to RA remains to be established. In the past few years, several genomewide linkage analyses documented a number of RA-susceptibility loci, scattered across the human genome. Despite some discrepancies (as often occur) between these studies, the chromosome 6p21.3 human leukocyte antigen (HLA) region consistently emerged as the major RA-susceptibility locus in most of them (Cornelis et al. 1998; Jawaheer et al. 2001; MacKay et al. 2002).
The human major histocompatibility complex (MHC), alternatively named “HLA,” encompasses 3.6 Mb of DNA on the short arm of chromosome 6 and is divided into three regions (in order from centromere to telomere): class II, class III, and class I (MHC Sequencing Consortium 1999). The MHC is one of the most (if not the most) densely gene-packed segments of the human genome and contains, besides the antigen-presenting MHC class I and class II molecules, >100 other expressed genes, spread across the entire class II, class III, and class I segments (Shiina et al. 1999). Therefore, it is possible, if not highly probable, that some of these non-HLA genes influence the development of RA, since it is clear that not all patients with RA carry the SE of HLA-DRB1 alleles (Dizier et al. 1993; McDaniel et al. 1995; Teller et al. 1996). In our search for such a putative “second locus,” we recently screened, using a panel of microsatellite markers, the entire HLA region, in a group of Japanese patients with RA and control individuals. This effort led to the identification of a second, HLA-DRB1–independent RA-susceptibility locus at the telomeric end of the HLA class III region, almost 1 Mb away from HLA-DRB1. This was corroborated by independent research that indicated the existence of a second RA-susceptibility locus at the telomeric end of the HLA region (Zanelli et al. 2001; Jawaheer et al. 2002). The 70-kb stretch of DNA that we previously identified is adjacent to the TNF gene cluster (TNFA and LTA) and is bordered by two microsatellites, TNFa and C1-2-A (fig. 1) (Ota et al. 2001b). In this RA critical segment, four expressed genes have thus far been identified (in order from centromere to telomere): IκBL (inhibitor of κB-like protein; also known as “NFKBIL1”), ATP6G (a member of the vacuolar ATPase G subunit family), BAT1 (HLA-B–associated transcript 1; a member of the DEAD-box family of RNA-binding proteins), and MICB (MHC class I chain-related gene B) (Browning and McMichael 1996; MHC Sequencing Consortium 1999; Neville and Campbell 1999; Bahram 2000).
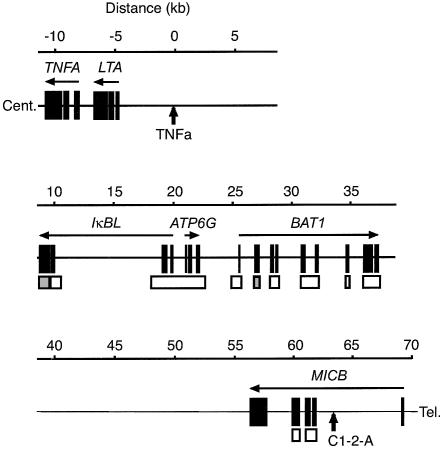
Figure 1.
Physical map of the 70-kb critical segment for the RA-susceptibility gene between TNFa and C1-2-A. Exons are represented by blackened boxes; genomic segments that do or do not contain SNP in the sequenced regions are represented by unblackened boxes or half-blackened boxes, respectively. Arrows indicate transcriptional orientation. The upper bar indicates the distance (in kb) from TNFa. Cent. = centromere; Tel. = telomere.
In the present study, to pinpoint the RA-susceptibility sequence within these four candidate genes, we conducted association analysis using relevant intragenic SNPs and insertion/deletion polymorphisms (indels). One SNP in the promoter/enhancer region of the IκBL gene was found to be most strongly associated with RA, hence providing genetic evidence that the MHC-based IκBL gene is involved in the development of RA and, from its similarity to the IκBα gene, indirectly suggesting that the NFκB pathway is potentially involved in susceptibility to arthritis.
Subjects and Methods
Subjects
One hundred sixteen patients with RA (19 males and 97 females) (the same patients studied in our microsatellite-based association mapping) (Ota et al. 2001b) and 100 healthy control individuals (43 males and 57 females) were enrolled in this investigation. Patients and control individuals were Japanese. All patients were diagnosed according to the American Rheumatism Association’s criteria (Arnett et al. 1988). All subjects (patients and control individuals) agreed to blood donation and DNA analysis. Genomic DNA from eight of the unaffected control individuals was subjected to a “saturation” sequence-variation identification, of SNPs and indels, as described below (see the “Identification and Genotyping of SNPs and Indels” subsection).
PCR and Sequencing Primers for Identification and Detection of Sequence Variations
Primers for DNA amplification and sequencing were designed manually, according to the genomic sequence of the 70-kb RA target segment between the two microsatellites (TNFa and C1-2-A) in the HLA class III region (Ota et al. 2001b). Repetitive sequences were masked using the RepeatMasker software (RepeatMasker Web Server). Nineteen primer pairs (table 1) were designed, to separately amplify the genomic segments of the IκBL, ATP6G, and BAT1 genes—namely all of their promoter/enhancer regions, all of their exons, certain introns (introns 1 and 2 of IκBL, introns 1 and 2 of ATP6G, and introns 3, 5, and 8–11 of BAT1), and the 3′-flanking intergenic regions of ATP6G and BAT1 (table 1 and fig. 1). The same primers were also used in the sequencing of PCR products.
Table 1.
Primers Used for PCR and Sequencing
|
Sequences(5′→3′) |
||
| Region Amplified | Forward | Reverse |
| IκBL: | ||
| Promoter | GGACAACAACAGGGACAGATC | CTCGTTGCTGCAGTCCTC |
| Promoter | GGAGACACTCCAGGCTGG | TCCTACGATAGTCTTCTTCCGTC |
| Exon 1 | GAAATTGAATATCATGTACCCGG | CACAGTTCACTTCCGTCCTC |
| Intron 1 | CCATGGAACTCTTGGGCT | TCTGCCGGGTACATGATATTC |
| Exon 2 | ACCAGCTTATTTCTCAACTATTGG | CCAAGGCTGAAGTCCTGAC |
| Intron 2 | GGCGAAAACCCATCTCTTC | ACCAATAGTTGAGAAATAAGCTGGT |
| Intron 2 | GAAGAAATCGGTGTAGGCTGTTG | GTCAAAGAATTTGGGCACTGC |
| Exon 3 | GCAGCTGTGGATAGCAGT | AGTCCCAGCTAACTTCTGCTC |
| Exon 4 | GATGAAAACCACAGCAATGG | ACAGGTGATGCCTCCCATG |
| ATP6G: | ||
| Exon 1, intron 1 | GATGAGATTGGGAGAGACACTCG | AGTCACCCTTACACACCTCACTAG |
| Exon 2, intron 2, 3′-flanking region | AGCGAGAGCACGAATTCC | GTGGTGGTAATAGTATCACAGGG |
| BAT1: | ||
| Exon 1, promoter | GGAATGTAGTATAACCCTCAAGCC | TAAGGAAATAGCGAACCAACTAGG |
| Exon 2 | GTGAAGGCTGTGCTCGTG | CTGAGCAACGACAAACACATC |
| Exons 3 and 4, intron 3 | TCTGGAAGTTGGCAAGAACC | TCAACACTCTGTTACACCACAGC |
| Exon 5, intron 5 | AATTGGTTTAGCTCAAACAGAGTG | CACCATATAGAATTGCCAAAGATC |
| Exon 6, intron 5 | TGGACATAGGCCCCATAAGTC | CCTTCTTGGCACTTGAATGAC |
| Exon 7 | AAGCACCTCTGATGGAGTTATTC | CATGACCTATGTGATGGGATTTAG |
| Exons 8–10, introns 8–10 | TCTCCCACGTAATGTCTCTCAC | TGCCCATGACCTATGTGATG |
| Exon 11, introns 10 and 11 | CAGTGAGTACTGATCTCATGAAACC | GCTCTGCAGTCTTAGTCCCATTC |
PCR Amplification of Genomic DNA
Genomic DNA from the eight unrelated healthy individuals described above (see the “Subjects” subsection) was amplified using the 19 primer pairs, to detect genetic variations in this RA critical genomic segment. The 20 μl of reaction mixture contained genomic DNA (2 ng), standard PCR buffer, dNTPs (0.1 mM each), AmpliTaq Gold (0.1 μl; Applied Biosystems), and the primer pair (250 nM each primer). PCRs were performed in a GeneAmp PCR System 9700 (Applied Biosystems) with an initial denaturation at 95°C for 9 min, followed by 40 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min and a final step at 72°C for 7 min. For all amplicons, 6 μl of PCR product was run on a 1.5% agarose gel.
Identification and Genotyping of SNPs and Indels
PCR products were used for direct sequencing. Sixty nanograms of each PCR product was treated with shrimp alkaline phosphatase (2 U; Amersham) and exonuclease I (10 U; Amersham) at 37°C for 15 min, followed by incubation at 80°C for 15 min for enzymatic inactivation. Sequencing reactions were performed using the ABI Prism BigDye Terminator kit (Applied Biosystems) in a GeneAmp PCR System 9700 with an initial denaturation at 96°C for 30 s, followed by 50 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Products were analyzed on an ABI Prism 3700 multicapillary sequencer (Applied Biosystems). Genetic diversity in the MICB gene was detected by direct sequencing of PCR products with the same PCR and sequencing primers as described elsewhere (Ando et al. 1997; Ota et al. 2001a). Three SNPs in the upstream region of the TNFA gene (at nucleotide positions −1031, −863, and −857, in order from the TNFA transcription initiation site) were also genotyped by direct sequencing of PCR products amplified using the primer set described elsewhere (Matsushita et al. 1999).
Microsatellite Genotyping
For the determination of the number of repeats within C1-2-A and TNFa microsatellites, forward primers were 5′-end labeled with the fluorescent reagent 6-FAM (Genset). PCR primers and amplification conditions were identical to those described elsewhere (Jongeneel et al. 1991; Nedospasov et al. 1991; Udalova et al. 1993; Tamiya et al. 1998). On PCR completion, products were denatured at 100°C for 5 min, were mixed with formamide-containing stop buffer, and were electrophoresed on capillary gels with a size-standard marker of GS500 ROX through use of an ABI Prism 3700 capillary sequencer (Applied Biosystems). Fragment sizes were automatically assigned by the GeneScan software (Applied Biosystems).
Statistical Analyses for Allelic Association and Haplotype Analysis
Disease associations with SNP and indel markers were initially assessed by the single-allele χ2 test for a 2×2 contingency table, to compare each allele frequency in patients with RA versus control individuals. Allele frequencies were estimated by direct counting. When any expected number of allele counts in the 2×2 contingency table was <5, the P value was directly calculated by Fisher’s exact test. The corrected P value, Pc, was not calculated for SNP and indel markers, because they have only two as the number of alleles. In microsatellite markers, as described elsewhere (Ota et al. 2001b), each phenotype frequency was compared, in patients with RA versus control individuals, by the single-allele χ2 test for a 2×2 contingency table. The Pc value was calculated for them as the P value multiplied by the number of alleles. The odds ratio and the 95% CI were also calculated for all markers. Pairwise haplotype frequencies were calculated from two-locus–genotype data, by the maximum-likelihood estimates, through use of the EH (estimate haplotype frequencies) program (Terwilliger and Ott 1994) (Web Resources of Genetic Linkage Analysis). Haplotype frequencies were compared in patients with RA versus control individuals and were evaluated, on the basis of the haplotype counts calculated from the estimated haplotype frequencies, by using the 2×4 or 2×2 contingency table. To calculate haplotype frequencies for adjacent-pairwise combinations for SNPs (i.e., pairwise haplotypes between adjacent SNPs), we employed only SNPs with >5% of allele frequencies in each of two alleles both in patients with RA and in control individuals. Any allele combination for two SNPs next to each other created four haplotypes. Frequencies of these haplotypes then were compared, in the global test with 3 df, by using the 2×4 contingency table. For association study of pairwise haplotypes between an adjacent microsatellite and SNP, the 2×2 contingency table was used. To examine whether genotype frequencies in the populations are in Hardy-Weinberg equilibrium, we performed the exact test of Hardy-Weinberg proportion for multiple alleles, as provided by Genepop software package (see Genepop on the Web) (Guo and Thompson 1992). P values >.05 were accepted as nonsignificant deviation from Hardy-Weinberg equilibrium.
Results
Identification of Sequence Variations in the Candidate Segment for RA
Our previous study had reached the limits of the tools available at that time, such that we could not refine the target region further than the 70 kb bordered by the two informative microsatellites (TNFa and C1-2-A) at the telomeric end of the HLA class III region (fig. 1). Hence, our first objective here was to identify sequence variations within this 70-kb candidate segment, to pinpoint the causative gene among the four that are known to be embedded in this segment—namely IκBL, ATP6G, BAT1, and MICB (Browning and McMichael 1996; MHC Sequencing Consortium 1999; Neville and Campbell 1999; Bahram 2000). With regard to MICB, a large number of intragenic SNPs have previously been reported (Ando et al. 1997), with a provisional allele number of 16 (designated as “MICB*001–MICB*016”) (Bahram 2000). For the detection of sequence variations in the three other genes (IκBL, ATP6G, and BAT1), PCR products from their promoter/enhancer regions, the totality of their exons, some of their introns (introns 1 and 2 of IκBL, introns 1 and 2 of ATP6G, and BAT1 introns 3, 5, and 8–11), and the 3′-flanking intergenic regions (ATP6G and BAT1) (table 1 and fig. 1) were amplified using genomic DNA from eight unaffected unrelated individuals and were subjected to nucleotide sequence analysis. In total, 12.2 kb of the 29.0 kb of genomic DNA that contains IκBL and links it to BAT1 was sequenced in each individual. Consequently, a total of 36 SNPs and three indels were identified (table 2). This corresponds to a density of one variation per every 685 bp, on average, in this 29.0-kb region. Among them, eight SNPs (86446, 94619, 94636, 94678, 95429, 96652, a4909, and a13808) and two indels (a13402 and a13736) are novel variations identified in the present study. The other SNPs and one indel have been reported previously (Allcock et al. 1999a, 2001) or were already submitted to dbSNP (see the Single-Nucleotide Polymorphism Web site). Of all the SNPs reported here, three are within exons; these are SNPs a13736 and a5160, in BAT1 3′ UTR and exon 4, respectively, and SNP 86352, in IκBL exon 3 (with the second and third of these SNPs being synonymous).
Table 2.
Nucleotide Variations Identified in and around the Second Susceptibility Region for RA and Results of Association Study Using These Variations[Note]
|
Allele Frequencyd |
||||||||
| SNP Name (dbSNP Accession Number)a | Location | Relative Distanceb(kb) | SNP Allelec | Patients (2n=232) | Control Individuals (2n=200) | Odds Ratio (95% CI) | χ2e | Pf |
| 69321 (rs1799724) | TNFA promoter | −7.0 | G/A | .302 | .185 | 1.90 (1.21–3.00) | 7.85 | .0051 |
| 69327 (rs1800630) | TNFA promoter | −7.0 | G/T | .828 | .800 | 1.20 (.74–1.95) | .54 | .4617 |
| 69495 (rs1799964) | TNFA promoter | −6.9 | A/G | .815 | .785 | 1.20 (.75–1.93) | .59 | .4415 |
| 86352 (rs2230365) | IκBL exon 3 | 10.0 | C/T | .703 | .665 | 1.19 (.79–1.79) | .70 | .4016 |
| 86446 (ss4480593) | IκBL intron 2 | 10.1 | T/C | .996 | .995 | 1.16 (.07–18.76) | … | .9999 |
| 86481 (rs2239707) | IκBL intron 2 | 10.1 | C/T | .625 | .595 | 1.13 (.77–1.67) | .41 | .5236 |
| 86616 (rs3093949) | IκBL intron 2 | 10.3 | C/T | .720 | .640 | 1.45 (.96–2.17) | 3.16 | .0754 |
| 86695 (rs2857604) | IκBL intron 2 | 10.3 | G/A | .302 | .200 | 1.73 (1.11–2.70) | 5.86 | .0155 |
| 86948 (rs2857605) | IκBL intron 2 | 10.6 | T/C | .224 | .220 | 1.02 (.65–1.61) | .01 | .9178 |
| 94619 (ss4480594) | IκBL intron 2 | 18.3 | C/T | .155 | .155 | 1.00 (.59–1.69) | .00 | .9961 |
| 94636 (ss4480595) | IκBL intron 2 | 18.3 | G/A | 1.000 | .995 | … | … | .4630 |
| 94678 (ss4480596) | IκBL intron 2 | 18.3 | T/C | .858 | .840 | 1.15 (.68–1.95) | .26 | .6067 |
| 95429 (ss4480597) | IκBL intron 2 | 19.1 | C/G | .853 | .845 | 1.07 (.63–1.81) | .06 | .8065 |
| 95993 (rs2071591) | IκBL intron 1 | 19.6 | G/A | .720 | .630 | 1.51 (1.01–2.26) | 3.97 | .0463 |
| 96029 (rs2239708) | IκBL intron 1 | 19.7 | A/T | .853 | .845 | 1.07 (.63–1.81) | .06 | .8065 |
| 96452 (rs2071592) | IκBL promoter | 20.1 | T/A | .720 | .595 | 1.75 (1.17–2.61) | 7.48 | .0062 |
| 96652 (ss4480598) | IκBL promoter | 20.3 | A/G | .155 | .115 | 1.41 (.81–2.48) | 1.47 | .2254 |
| 96714 (ss4480599) | IκBL promoter | 20.4 | C/G | .970 | .930 | 2.42 (.96–6.12) | 3.68 | .0549 |
| 96818 (ss4480600) | IκBL promoter | 20.5 | A insertion | .082 | .055 | 1.53 (.71–3.30) | 1.20 | .2728 |
| 97923 (rs2523502) | ATP6G intron 2 | 21.6 | A/T | .082 | .070 | 1.19 (.58–2.43) | .22 | .6425 |
| 98228 (rs2523503) | ATP6G intron 2 | 21.9 | G/T | .828 | .800 | 1.20 (.74–1.95) | .54 | .4617 |
| 98385 (rs2239705) | ATP6G intron 2 | 22.0 | C/T | .293 | .185 | 1.83 (1.16–2.88) | 6.82 | .0090 |
| 98988 (rs2071593) | 3′-flanking region of ATP6G | 22.6 | C/T | .845 | .835 | 1.08 (.64–1.80) | .08 | .7810 |
| 99067 (rs2071594) | 3′-flanking region of ATP6G | 22.7 | C/G | .720 | .625 | 1.54 (1.03–2.31) | 4.41 | .0357 |
| a1682 (rs2239528) | BAT1 promoter | 25.3 | C/T | .772 | .770 | 1.01 (.64–1.58) | .00 | .9695 |
| a1785 (rs2523505) | BAT1 promoter | 25.4 | G/C | 1.000 | .990 | … | … | .2138 |
| a1820 (rs2523506) | BAT1 promoter | 25.5 | C/A | .823 | .795 | 1.20 (.74–1.94) | .56 | .4549 |
| a2008 (rs2239527) | BAT1 promoter | 25.7 | G/C | .703 | .615 | 1.48 (.99–2.21) | 3.68 | .0550 |
| a4722 (rs2071595) | BAT1 intron 2 | 28.4 | C/G | .159 | .160 | 1.07 (.64–1.81) | .00 | .9883 |
| a4909 (ss4480601) | BAT1 intron 3 | 28.6 | G/C | 1.000 | .990 | … | … | .2138 |
| a4930 (rs2523511) | BAT1 intron 3 | 28.6 | A/G | .078 | .055 | 1.45 (.67–3.14) | .87 | .3496 |
| a4983 (rs2523512) | BAT1 intron 3 | 28.6 | C/T | .828 | .805 | 1.16 (.71–1.89) | .37 | .5448 |
| a5040 (rs2516393) | BAT1 intron 3 | 28.7 | G/T | .082 | .075 | 1.10 (.54–2.23) | .07 | .7907 |
| a5093 (rs2071596) | BAT1 intron 3 | 28.7 | C/T | .711 | .660 | 1.27 (.84–1.91) | 1.31 | .2522 |
| a5136 (rs933208) | BAT1 intron 3 | 28.8 | A/C | .806 | .785 | 1.14 (.71–1.82) | .29 | .5883 |
| a5160 (rs1129640) | BAT1 exon 4 | 28.8 | G/A | .073 | .055 | 1.36 (.62–2.97) | .59 | .4417 |
| a7809g (rs2075580) | BAT1 intron 5 | 31.5 | G/C | ND | ND | ND | ND | ND |
| a8086 (rs929138) | BAT1 intron 5 | 31.7 | G/A | .319 | .210 | 1.76 (1.14–2.97) | 6.49 | .0108 |
| a13047 (rs2516478) | BAT1 intron 9 | 36.7 | T/C | .828 | .800 | 1.20 (.74–1.95) | 1.38 | .2402 |
| a13402 (ss4480602) | BAT1 intron 10 | 37.1 | CT insertion | ND | ND | ND | ND | ND |
| a13736 (ss4480603) | BAT1 exon 11 (3′ UTR) | 37.4 | T deletion | .875 | .855 | 1.19 (.68–2.06) | .37 | .5432 |
| a13808 (ss4480604) | 3′-flanking region of BAT1 | 37.5 | G/A | .996 | .995 | 1.16 (.07–18.68) | … | .9999 |
Note.— ND = not determined.
The name of each SNP is derived from nucleotide position of the SNP as detected starting from the first base on GenBank accession number AP000505 (no prefix) or AP000506 (prefixed by “a”). For SNPs in dbSNP, see the Single Nucleotide Polymorphism Web site.
Distance from the TNFa microsatellite.
A nucleotide on the left-hand side of a slash mark (/) is a more frequent allele in the control individuals. Each SNP allele is represented by the nucleotide sequence of the sense strand of each gene.
Listed frequencies are higher in the patients than in the control individuals.
P values <.05 that have been accepted as statistically significant are underlined. The P values of the SNPs for which there are no data given for the odds ratio and/or χ2 were directly calculated by Fisher's exact test because the expected numbers in the 2×2 table were <5.
Association studies were not performed.
Association Study Using SNPs and Indels
Allele frequencies of a total of 35 SNPs and two indels among the variations identified above (see the “Identification of Sequence Variations in the Candidate Segment for RA” subsection) in the IκBL, ATP6G, and BAT1 genes (table 2) were compared between the patient and unaffected control groups. As a result, statistically significant (P<.05) positive associations with the disease were observed for six SNPs, as shown in table 2. Among these, SNP 96452, a T/A substitution in the IκBL promoter region (at nucleotide position −62 from the IκBL transcription start site), was most significantly associated with RA with a P value of .0062. The allele frequencies of SNP 96452 were almost the same between females and males among both the patients and the control individuals (data not shown). The MICB gene was also genotyped for association study. Although the MICB*002 allele revealed the most remarkable difference in allele frequency between the patient and control groups, it did not reach statistical significance (χ2=2.9; 1 df; P=.088). Three SNP markers (SNPs 69321, 69327, and 69495) in the upstream region of the TNFA gene (nucleotide positions −1031, −863, and −857, respectively, in order from the TNFA transcription initiation site), 5 kb centromerically from our 70-kb candidate segment (fig. 1) (Matsushita et al. 1999; Ota et al. 2001b), were also included in genotyping. The previous finding of a significant association of SNP 69321, in the promoter/enhancer region of the TNFA gene, was confirmed (Seki et al. 1999). However, this does not represent a primary association but is due to linkage disequilibrium with DRB1 alleles or SEs, as described elsewhere (Higuchi et al. 1998; Matsushita et al. 1999; Seki et al. 1999; Ota et al. 2001b). Furthermore, the two microsatellites, C1-2-A and TNFa, which define the boundaries of our critical segment, were also confirmed to be significantly associated with the disease (alleles 113 and 242, respectively) (table 3) despite the fact that we dealt here with an unaffected population that was different from that in our previous study (Ota et al. 2001b). Finally, all polymorphic markers investigated here followed the Hardy-Weinberg equilibrium in both patients and control individuals.
Table 3.
Association of TNFa and C1-2-A Alleles with RA
|
Phenotype Frequency |
|||||||
| Microsatellite Name (No. of Alleles) | Most Strongly Associated Allele | Patients (n=116) | Control Individuals (n=100) | Odds Ratio (95% CI) | χ2 | P | Pc |
| TNFa (13) | 113 | .353 | .180 | 2.98 (1.91–4.64) | 12.02 | .00053 | .0068 |
| C1-2-A (13) | 242 | .629 | .450 | 2.68 (1.82–3.94) | 13.52 | .00024 | .0031 |
Association Study of Pairwise Haplotype
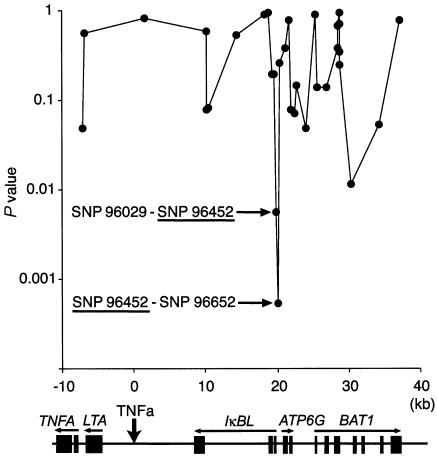
Frequencies of pairwise haplotypes consisting of C1-2-A or TNFa and disease-associated SNPs alleles were then investigated for comparison between the patient and control groups, as listed in table 4. Both allele 113 of TNFa and allele 242 of C1-2-A revealed the strongest association when combined with SNP 96452 (TNFa P=.0089; C1-2-A P=.000085), as expected. To pinpoint the susceptible area more precisely, we calculated the frequencies of all possible haplotypes with adjacent-pairwise combinations for 34 SNP markers within the critical segment, as well as for three SNP markers (SNPs 69321, 69327, and 69495) in the upstream region of the TNFA gene, and these frequencies were then compared between the patients and control individuals (fig. 2). The haplotype frequencies of all adjacent-pairwise combinations were compared in the global test with 3 df. Remarkably strong associations were observed for two haplotypes containing SNP 96452, namely SNP 96029–SNP 96452 (P=.0056) and SNP 96452–SNP 96652 (P=.00054). Taken together, these data clearly define SNP 96452 in the IκBL promoter/enhancer region as the second intra-MHC–located RA-susceptibility locus.
Table 4.
Haplotype Association between TNFa or C1-2-A Microsatellites and SNPs
|
Microsatellite |
SNP |
Estimated Haplotype Frequency |
||||||
| Name | Allele | Name | Allele | Patients (2n=232) | Control Individuals (2n=200) | Odds Ratio (95% CI) | χ2 | P |
| TNFa | 113 | 69321 | A | .156 | .084 | 2.02 (1.11–3.69) | 5.20 | .023 |
| TNFa | 113 | 86695 | A | .156 | .088 | 1.91 (1.06–3.46 ) | 4.52 | .034 |
| TNFa | 113 | 95993 | C | .177 | .090 | 2.17 (1.22–3.87) | 6.85 | .0089 |
| TNFa | 113 | 96452 | T | .177 | .090 | 2.17 (1.22–3.87) | 6.85 | .0089 |
| TNFa | 113 | 98385 | T | .146 | .083 | 1.89 (1.03–3.48) | 4.15 | .042 |
| TNFa | 113 | 99067 | C | .175 | .093 | 2.08 (1.17–3.70) | 6.21 | .013 |
| TNFa | 113 | a8086 | T | .150 | .087 | 1.85 (1.02–3.36) | 4.00 | .046 |
| C1-2-A | 242 | 69321 | A | .292 | .168 | 2.04 (1.29–3.24) | 9.21 | .0024 |
| C1-2-A | 242 | 86695 | A | .292 | .171 | 2.00 (1.27–3.17) | 8.74 | .0031 |
| C1-2-A | 242 | 95993 | C | .388 | .221 | 2.24 (1.47–3.41) | 14.01 | .00018 |
| C1-2-A | 242 | 96452 | T | .388 | .213 | 2.34 (1.53–3.58) | 15.45 | .000085 |
| C1-2-A | 242 | 98385 | T | .288 | .160 | 2.12 (1.33–3.38) | 9.93 | .0016 |
| C1-2-A | 242 | 99067 | C | .388 | .214 | 2.32 (1.52–3.54) | 15.12 | .00010 |
| C1-2-A | 242 | a8086 | T | .297 | .167 | 2.10 (1.33–3.33) | 10.00 | .0016 |
Figure 2.
Association study of pairwise haplotypes. Frequencies of all possible haplotypes with adjacent-pairwise combinations were calculated for 31 SNP markers (the frequencies of which were >5% both in the patients and in the control individuals) within the 70-kb RA critical segment between C1-2-A and TNFa, as well as for 3 SNP markers (SNPs 69321, 69327, and 69495) in the upstream region of the TNFA gene, were compared between the patients and control individuals. P values are dotted at the midpoint between two markers. The horizontal bar indicates the distance (in kb) from TNFa. A physical map at bottom shows the location of genes in this region; symbols are the same as in figure 1.
Genotype Frequency of SNP 96452 Alleles
Genotype frequencies of SNP 96452 alleles were investigated in the patient and unaffected control groups (table 5). The frequency of the major genotype with a T/T homozygosity was significantly higher in patients with RA (P=.0088) with an odds ratio of 2.08 (95% CI 1.41–3.07) as compared to control individuals. Accordingly, this T allele is likely to increase the risk of the prevalence of RA in the case of homozygosity, with a recessive trait.
Table 5.
Genotype Association between SNP 96452 and RA
|
Genotype Frequency |
|||||
| Genotype | Patients (n=116) | Control Individuals (n=100) | Odds Ratio (95% CI) | χ2 | P |
| T/T | .520 | .340 | 2.08 (1.41–3.07) | 6.86 | .0088 |
| T/A | .412 | .510 | .65 (.45–.96) | 2.38 | .12 |
| A/A | .080 | .150 | .48 (.25–.90) | 2.85 | .091 |
Discussion
Several recent genomewide scans in various populations have confirmed the eminence of MHC genetics with respect to RA susceptibility (Cornelis et al. 1998; Jawaheer et al. 2001; MacKay et al. 2002). We have recently documented the existence of a second, HLA-DRB1–independent, intra-MHC RA-susceptibility locus (Ota et al. 2001b). In the present study, to identify this second locus (previously mapped to a 70-kb segment bordered by two microsatellites at the telomeric end of the HLA class III region), we systematically performed allele, pairwise haplotype, and genotype association analyses of RA with respect to a total of 35 SNPs and two indels identified in the IκBL, ATP6G, and BAT1 genes, as well as with respect to MICB alleles. This investigation collectively indicates that the T allele of SNP 96452 (T/A substitution) located in the IκBL promoter region, at nucleotide position −62 from the IκBL transcription start site, is implicated in the development of RA. This conclusion was also supported by the fact that the disease-associated alleles of the two microsatellites, C1-2-A and TNFa, which defined both ends of the RA-susceptibility locus (Ota et al. 2001b), were in linkage disequilibrium with the T allele of SNP 96452.
Although the biological function of the putative IκBL protein is, at this point, mostly speculative, its similarity to IκBα in the sequence (Browning and McMichael 1996) and the cellular localization (Semple et al. 2002), which regulates the nuclear localization of NFκB (a nuclear factor that stimulates the transcription of many immunologically and inflammatory relevant genes, including cytokines such as the closely linked TNF-α; for review, see Ghosh et al. 1998), presumes analogous function. To investigate the functional consequence of our SNP, which lies within the putative regulatory region of IκBL, we scanned the DNA segment containing and extending this site through use of the TFSearch program (see the TFSearch Web site). This search revealed that the nucleotide sequence surrounding the RA-susceptible allele—that is, the T allele of SNP 96452 (−62T) (CTCCACCTGCG)—is indeed a putative binding site for the transcription factor δEF1, as previously proposed by Allcock et al. (2001). In contrast, the nonsusceptible allele, the A allele of SNP 96452 (−62A), is likely to disrupt this motif for δEF1. In fact, examination of 41 known δEF-binding sites showed that all contained a T nucleotide at this position (Sekido et al. 1994). Because δEF1 is a transcription repressor involved in skeletal and T-cell development (Takagi et al. 1998), it is reasonable to speculate that −62T/A may somehow affect the transcriptional promoter activity, altering the immune response in the development of RA. Allcock et al. (2001) did investigate the effect that the −62T/A SNP has on the transcription level of IκBL mRNA by using Epstein-Barr virus–transformed B-lymphoblastoid cells, but they did not observe any difference of IκBL mRNA level between the A and T alleles at position −62. However, it is likely that B-cell lines do not represent the best in vitro model for the recapitulation of the affected inflammatory joints in RA, and further analysis in different cell lines and, more importantly, in situ may reveal the biological consequence of this promoter dimorphism. de la Concha et al. (2000) reported an association between a structural SNP at amino acid position 224 of the IκBL molecule, which is predicted to be the protein kinase C phosphorylation site (Albertella et al. 1994), and susceptibility to ulcerative colitis. These findings raise the possibility that the IκBL protein interacts with NFκB, so transcription-level changes depending on the promoter-based diallelic polymorphism may affect diverse immunological processes and may predispose one to a number of major autoimmune diseases linked to the MHC. It is perhaps relevant that a second susceptibility locus for multiple sclerosis, as well as for type I diabetes and celiac disease, has also been mapped to this telomeric part of the MHC (Allcock et al. 1999b; Lie et al. 1999a, 1999b).
In summary, we pinpoint the MHC class III–located IκBL protein as the second, class II–independent, intra-MHC RA-susceptibility locus. Although little is known about the function of IκBL per se, its similarity to IκBα infers similar function (i.e., involvement in the silencing of NFκB functionality). NFκB, a pivotal nuclear factor, activates the transcription of many important genes involved in immune and inflammatory responses (for review, see Ghosh et al. 1998) and is piloted by a variety of extracellular signals, most importantly those that trigger the innate immune system. Hence, the MHC may encode RA predisposition through the innate and adaptive components of the immune system. Hence, the recent report of the involvement of both innate and adaptive effectors of the immune system in a genetically engineered animal model of RA may be relevant to this observation (Ji et al. 2002). Finally, further analysis may implicate structural or regulational IκBL diversity in other MHC-linked autoimmune diseases. The emergence of IκBL from the densely packed MHC class III loci may now help to focus our attention on its biology, a goal we shall pursue through the creation of IκBL-transgenic and -knockout mice, which may open novel pathophysiological insights, as well as therapeutic areas of intervention for RA and other autoimmune conditions.
Acknowledgments
We thank Rie Sato for technical assistance and Dr. Akira Oka for helpful suggestions. H.I. and S.B. acknowledge funding from INSERM/Japan Society for the Promotion of Science. S.B. wishes to thank INSERM/Centre National de la Recherche Scientifique/Ministère de la Recherche for support received through the Séquençage à Grande Échelle program.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (accession numbers AP000505 and AP000506)
- Genepop on the Web, http://wbiomed.curtin.edu.au/genepop/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RA [MIM 180300])
- RepeatMasker Web Server, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker
- Single Nucleotide Polymorphism, http://www.ncbi.nlm.nih.gov/SNP/ (for newly identified SNPs and indels [accession numbers ss4480593, ss4480594, ss4480595, ss4480596, ss4480597, ss4480598, ss4480601, ss4480602, ss4480603, and ss4480604], the SNP and indel previously reported without dbSNP accession numbers [ss4480599 and ss4480600, respectively], and other SNPs already in dbSNP)
- TFSearch: Searching Transcription Factor Binding Sites, http://www.cbrc.jp/research/db/TFSEARCH.html
- Web Resources of Genetic Linkage Analysis, http://linkage.rockefeller.edu/
References
- Albertella MR, Campbell RD (1994) Characterization of a novel gene in the human major histocompatibility complex that encodes a potential new member of the I kappa B family of proteins. Hum Mol Genet 3:793–799 [DOI] [PubMed] [Google Scholar]
- Allcock RJ, Baluchova K, Cheong KY, Price P (2001) Haplotypic single nucleotide polymorphisms in the central MHC gene IKBL, a potential regulator of NF-kappaB function. Immunogenetics 52:289–293 [DOI] [PubMed] [Google Scholar]
- Allcock RJ, Christiansen FT, Price P (1999a) The central MHC gene IKBL carries a structural polymorphism that is associated with HLA-A3,B7,DR15. Immunogenetics 49:660–665 [DOI] [PubMed] [Google Scholar]
- Allcock RJ, de la Concha EG, Fernandez-Arquero M, Vigil P, Conejero L, Arroyo R, Price P (1999b) Susceptibility to multiple sclerosis mediated by HLA-DRB1 is influenced by a second gene telomeric of the TNF cluster. Hum Immunol 60:1266–1273 [DOI] [PubMed] [Google Scholar]
- Ando H, Mizuku N, Ota M, Yamazaki M, Ohno S, Goto K, Miyata Y, Wakisaka K, Bahram S, Inoko H (1997) Allelic variations of the human MHC class I chain-related gene B (MICB). Immunogenetics 46:499–508 [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, et al (1998) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31:315–324 [DOI] [PubMed] [Google Scholar]
- Bahram S (2000) MIC genes: from genetics to biology. Adv Immunol 76:1–60 [DOI] [PubMed] [Google Scholar]
- Browning M, McMichael A (1996) HLA and MHC: genes, molecules and function. BIOS Scientific, Oxford [Google Scholar]
- Cornelis F, Faure S, Martinez M, Prud'Homme JF, Fritz P, Dib C, Alves H, et al (1998) New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci USA 95:10746–10750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Concha EG, Fernandez-Arquero M, Lopez-Nava G, Martin E, Allcock RJ, Conejero L, Paredes JG, Diaz-Rubio M (2000) Susceptibility to severe ulcerative colitis is associated with polymorphism in the central MHC gene IKBL. Gastroenterology 119:1491–1495 [DOI] [PubMed] [Google Scholar]
- Dizier MH, Eliaou JF, Babron MC, Combe B, Sany J, Clot J, Clerget-Darpoux F (1993) Investigation of the HLA component involved in rheumatoid arthritis (RA) by using the marker association-segregation χ2 (MASC) method: rejection of the unifying-shared-epitope hypothesis. Am J Hum Genet 53:715–721 [PMC free article] [PubMed] [Google Scholar]
- Feldmann M, Brennan FM, Maini RN (1996) Rheumatoid Arthritis. Cell 85: 307–310 [DOI] [PubMed] [Google Scholar]
- Felson DT (1996) Epidemiology of the rheumatic diseases. In: Koopman WJ (ed) Arthritis and allied conditions: a textbook of rheumatology, 13th ed. Williams & Wilkins, Philadelphia, PA, pp 3–34 [Google Scholar]
- Ghosh S, May MJ, Kopp EB (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16:225–260 [DOI] [PubMed] [Google Scholar]
- Gregersen PK, Silver J, Winchester RJ (1987) The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30:1205–1213 [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372 [PubMed] [Google Scholar]
- Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, Itoh K (1998) Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens 51:605–612 [DOI] [PubMed] [Google Scholar]
- Jawaheer D, Li W, Graham RR, Chen W, Damle A, Xiao X, Monteiro J, Khalili H, Lee A, Lundsten R, Begovich A, Bugawan T, Erlich H, Elder JT, Criswell LA, Seldin MF, Amos CI, Behrens TW, Gregersen PK (2002) Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am J Hum Genet 71:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawaheer D, Seldin MF, Amos CI, Chen WV, Shigeta R, Monteiro J, Kern M, Criswell LA, Albani S, Nelson JL, Clegg DO, Pope R, Schroeder HW Jr, Bridges SL Jr, Pisetsky DS, Ward R, Kastner DL, Wilder RL, Pincus T, Callahan LF, Flemming D, Wener MH, Gregersen PK (2001) A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet 68:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FM, Boackle SA, Takahashi K, Holers VM, Walport M, Gerard C, Ezekowitz A, Carroll MC, Brenner M, Weissleder R, Verbeek JS, Duchatelle V, Degott C, Benoist C, Mathis D (2002) Arthritis critically dependent on innate immune system players. Immunity 16:157–168 [DOI] [PubMed] [Google Scholar]
- Jirholt J, Lindqvist AB, Holmdahl R (2001) The genetics of rheumatoid arthritis and the need for animal models to find and understand the underlying genes. Arthritis Res 3:87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongeneel CV, Briant L, Udalova IA, Sevin A, Nedospasov SA, Cambon-Thomsen A (1991) Genetic polymorphism in the human tumor necrosis factor region and relation to extended HLA haplotypes. Proc Natl Acad Sci USA 88:9717–9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie BA, Sollid LM, Ascher H, Ek J, Akselsen HE, Ronningen KS, Thorsby E, Undlien DE (1999a) A gene telomeric of the HLA class I region is involved in predisposition to both type 1 diabetes and coeliac disease. Tissue Antigens 54:162–168 [DOI] [PubMed] [Google Scholar]
- Lie BA, Todd JA, Pociot F, Nerup J, Akselsen HE, Joner G, Dahl-Jorgensen K, Ronningen KS, Thorsby E, Undlien DE (1999b) The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am J Hum Genet 64:793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S, Brown MA, Bell J, Silman A, Ollier W, Wordsworth P, Worthington J (2002) Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum 46:632–639 [DOI] [PubMed] [Google Scholar]
- Matsushita M, Tsuchiya N, Nakayama T, Ohashi J, Shibue T, Shiota M, Oka T, Yamane A, Tokunaga K (1999) Allele typing of human TNFA 5′-flanking region using polymerase chain reaction-preferential homoduplex formation assay (PCR-PHFA): linkage disequilibrium with HLA class I and class II genes in Japanese. Tissue Antigens 54:478–484 [DOI] [PubMed] [Google Scholar]
- McDaniel DO, Alarcon GS, Pratt PW, Reveille JD (1995) Most African-American patients with rheumatoid arthritis do not have the rheumatoid antigenic determinant (epitope). Ann Intern Med 123:181–187 [DOI] [PubMed] [Google Scholar]
- MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. Nature 401:921–923 [DOI] [PubMed] [Google Scholar]
- Nedospasov SA, Udalova IA, Kuprash DV, Turetskaya RL (1991) DNA sequence polymorphism at the human tumor necrosis factor (TNF) locus: numerous TNF/lymphotoxin alleles tagged by two closely linked microsatellites in the upstream region of the lymphotoxin (TNF-beta) gene. J Immunol 147:1053–1059 [PubMed] [Google Scholar]
- Neville MJ, Campbell RD (1999) A new member of the Ig superfamily and a V-ATPase G subunit are among the predicted products of novel genes close to the TNF locus in the human MHC. J Immunol 162:4745–4754 [PubMed] [Google Scholar]
- Ota M, Katsuyama Y, Bahram S, Naruse T, Inoko H (2001a) MICA and MICB allele determination by sequence-based typing (SBT). In: Tilanus MG (ed) Proceedings of the Thirteenth International Histocompatibility Workshop and Conference. International Histocompatibility Working Group, Seattle, WA, pp TM27-1–TM27-5 [Google Scholar]
- Ota M, Katsuyama Y, Kimura A, Tsuchiya K, Kondo M, Naruse T, Mizuki N, Itoh K, Sasazuki T, Inoko H (2001b) A second susceptibility gene for developing rheumatoid arthritis in the human MHC is localized within a 70-kb interval telomeric of the TNF genes in the HLA class III region. Genomics 71:263–270 [DOI] [PubMed] [Google Scholar]
- Seki N, Kamizono S, Yamada A, Higuchi T, Matsumoto H, Niiya F, Kimura A, Tsuchiya K, Suzuki R, Date Y, Tomita T, Itoh K, Ochi T (1999) Polymorphisms in the 5′-flanking region of tumor necrosis factor-α gene in patients with rheumatoid arthritis. Tissue Antigens 54:194–197 [DOI] [PubMed] [Google Scholar]
- Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H (1994) The delta-crystallin enhancer-binding protein δEF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol 14:5692–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Amos CI, Ward R, Gregersen PK (1999) The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum 42:1071–1079 [DOI] [PubMed] [Google Scholar]
- Semple JI, Brown SE, Sanderson CM, Campbell RD (2002) A distinct bipartite motif is required for the localization of inhibitory κB-like (IκBL) protein to nuclear speckles. Biochem J 361:489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Tamiya G, Oka A, Takishima N, Yamagata T, Kikkawa E, Iwata K, Tomizawa M, Okuaki N, Kuwano Y, Watanabe K, Fukuzumi Y, Itakura S, Sugawara C, Ono A, Yamazaki M, Tashiro H, Ando A, Ikemura T, Soeda E, Kimura M, Bahram S, Inoko H (1999) Molecular dynamics of MHC genesis unraveled by sequencing analysis of the 1,796,938-bp HLA class I region. Proc Natl Acad Sci USA 96:13282–13287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Moribe H, Kondoh H, Higashi Y (1998) δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 125:21–31 [DOI] [PubMed] [Google Scholar]
- Tamiya G, Ota M, Katsuyama Y, Shiina T, Oka A, Makino S, Kimura M, Inoko H (1998) Twenty-six new polymorphic microsatellite markers around the HLA-B, -C and -E loci in the human MHC class I region. Tissue Antigens 51:337–346 [DOI] [PubMed] [Google Scholar]
- Teller K, Budhai L, Zhang M, Haramati N, Keiser HD, Davidson A (1996) HLA-DRB1 and DQB typing of Hispanic American patients with rheumatoid arthritis: the “shared epitope” hypothesis may not apply. J Rheumatol 23:1363–1368 [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. John Hopkins University Press, Baltimore, pp 188–198 [Google Scholar]
- Udalova IA, Nedospasov SA, Webb GC, Chaplin DD, Turetskaya RL (1993) Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics 16:180–186 [DOI] [PubMed] [Google Scholar]
- Zanelli E, Jones G, Pascual M, Eerligh P, van der Slik AR, Zwinderman AH, Verduyn W, Schreuder GM, Roovers E, Breedveld FC, de Vries RR, Martin J, Giphart MJ (2001) The telomeric part of the HLA region predisposes to rheumatoid arthritis independently of the class II loci. Hum Immunol 62:75–84 [DOI] [PubMed] [Google Scholar]