Abstract
We performed the first population-based clinical and molecular genetic study of Leber hereditary optic neuropathy (LHON) in a population of 2,173,800 individuals in the North East of England. We identified 16 genealogically unrelated families who harbor one of the three primary mitochondrial DNA (mtDNA) mutations that cause LHON. Two of these families were found to be linked genetically to a common maternal founder. A de novo mtDNA mutation (G3460A) was identified in one family. The minimum point prevalence of visual failure due to LHON within this population was 3.22 per 100,000 (95% CI 2.47–3.97 per 100,000), and the minimum point prevalence for mtDNA LHON mutations was 11.82 per 100,000 (95% CI 10.38–13.27 per 100,000). These results indicate that LHON is not rare but has a population prevalence similar to autosomally inherited neurological disorders. The majority of individuals harbored only mutant mtDNA (homoplasmy), but heteroplasmy was detected in ∼12% of individuals. Overall, however, ∼33% of families with LHON had at least one heteroplasmic individual. The high incidence of heteroplasmy in pedigrees with LHON raises the possibility that a closely related maternal relative of an index case may not harbor the mtDNA mutation, highlighting the importance of molecular genetic testing for each maternal family member seeking advice about their risks of visual failure.
Introduction
Leber hereditary optic neuropathy (LHON [MIM 535000]) characteristically presents with subacute, painless bilateral visual failure in young adults, and it results from the focal degeneration of the retinal ganglion cell layer and optic nerve (Leber 1871; Seedorff 1985). More than 95% of individuals with LHON harbor one of three mtDNA “primary” point mutations, all of which affect genes encoding complex I subunits of the respiratory chain (Mackey et al. 1996). The G11778A mutation results in an arginine-to-histidine substitution at amino acid position 340 (R340H) of the ND4 subunit (Wallace et al. 1988); the G3460A mutation causes an A52T substitution in the ND1 subunit (Howell et al. 1991; Huoponen et al. 1991); and the T14484C mutation causes a M64V substitution in the ND6 subunit (Johns et al. 1992a; Mackey and Howell 1992). Other mtDNA mutations are thought to cause LHON, but these appear to be relatively rare within the population (Man et al. 2002). The most striking feature of LHON is the marked incomplete penetrance and sex-specific bias of this mitochondrial disorder. Only ∼50% of the males and ∼10% of the females who harbor one of these mutations actually develop the optic neuropathy (Harding et al. 1995; Riordan-Eva et al. 1995). This pattern clearly indicates that other genetic and/or environmental factors must be important in modulating the phenotypic expression of LHON (Howell 1999).
The three primary pathogenic mutations were all identified more than a decade ago. Although the clinical features of LHON have been described in several case series, there are no well-established prevalence figures for genetically confirmed cases in the general population (Man et al. 2002). Many of the previous molecular studies of large cohorts of patients with LHON were performed in tertiary referral centers with a special interest in mitochondrial disorders. Patients came from wide and overlapping geographical areas and were referred by a large number of clinicians. The complex referral patterns and the lack of patient follow-up confounded any attempt to derive robust prevalence figures for LHON. It is also possible that the methods of case ascertainment influenced the observed frequency of mtDNA heteroplasmy in these LHON cohorts. To address these problems, we performed a rigorous population-based clinical and molecular study of LHON in a defined geographical region, the North East of England.
Subjects, Material, and Methods
Study Population
The North East Government Office region of England includes Tyne and Wear, Durham, Northumberland, and Teesside (fig. 1). We used the most recently published census data for the North East Government Office region to determine the point prevalence on June 30, 1998 (midyear period); 2,173,800 children and adults <65 years of age were living in the region, 48% of them being male (Office for National Statistics 2000). This region has been relatively stable in terms of migratory flux, and it consists principally of white individuals of Northern European extraction (Vanderpump et al. 1995). The epidemiological component of this study was limited to adults below pensionable age, for two reasons. First, although it is well recognized that LHON may present in the 7th and 8th decades, this is extremely rare (Nikoskelainen et al. 1996). Furthermore, it can be difficult to distinguish between LHON and optic atrophy secondary to other causes, such as anterior ischemic optic neuropathy or glaucoma among the elderly, even if there is a clear family history of LHON. Second, the group that is <65 years of age corresponds to persons of “working age,” a census group that is defined by the Office for National Statistics for the United Kingdom (Office for National Statistics 2000).
Figure 1.
Map of the British Isles, showing the North East Government Office region of England
Case Ascertainment
For >10 years, patients who present with unexplained visual failure or suspected LHON have been referred, as part of routine clinical practice, to the Northern Genetics Service based in Newcastle upon Tyne by clinicians throughout the region. Diagnostic mitochondrial genetic analysis was then performed within the Department of Neurology, The Medical School, University of Newcastle upon Tyne. We prospectively collected clinical data and DNA samples from these individuals throughout the region during the ∼12-year period from January 1990 to May 2002. Before DNA analysis, all affected individuals were assessed clinically by an ophthalmologist or neurologist who documented subacute visual failure and excluded structural, metabolic, toxic, and inflammatory causes. A clinical diagnosis of LHON was confidently made in some cases, but a large number of patients with unexplained optic neuropathy were also referred for investigation and were included in the present study. We were deliberately overinclusive in our selection criteria, to ensure optimal ascertainment of LHON cases. We restricted our molecular genetic studies to the three primary mtDNA mutations (G11778A, G3460A, and T14484C) because these mutations account for the vast majority (>95%) of cases and are an undisputed cause of LHON (Mackey et al. 1996; Howell et al. 1998). Other mutations are rare, most have been described in only one pedigree, and not all of these mutations are acknowledged to be primarily responsible for the optic neuropathy (Man et al. 2002). After the definite diagnosis of LHON in a proband, further details about other family members were actively sought, and, if the latter consented, they underwent further investigation by a clinician and provided blood samples for DNA analysis. Affected and at-risk individuals were included only if they lived within the North East Government Office region of England.
Mutation Detection
Total genomic DNA was extracted from peripheral blood leukocytes, using established methods. The three primary LHON mutations (G11778A, G3460A, and T14484C) were detected either by direct mtDNA sequencing or RFLP analysis of PCR fragments (Riordan-Eva and Harding 1995; Black et al. 1996).
Noncoding Control Region Sequencing
The noncoding control region of the mtDNA genome (D loop) was sequenced in the index patient by PCR amplification of three overlapping segments, using forward and reverse M13-tagged primer pairs (Andrews et al. 1999). PCR products were purified (Qiagen) and sequenced bidirectionally on an ABI 377 DNA sequencer by the standard dideoxy chain–termination procedure, using Big-Dye terminator kits (Perkin Elmer). The raw sequence data were compared with the revised Cambridge reference sequence (CRS) (Andrews et al. 1999), using Factura (v. 1.2) and Sequence Navigator (v. 1.0) softwares (Perkin Elmer). This allowed identification of the nucleotide changes present in the D loop.
Quantification of Heteroplasmy
Heteroplasmy was determined in each individual by primer extension assay (PEA), as described elsewhere (Fahy et al. 1997). A 700-bp PCR fragment encompassing each LHON mutation was generated with the M13-tagged primers used for sequencing and with AmpliTaq DNA polymerase (Perkin Elmer) under the following conditions: 95°C denaturation for 5 min, followed by 30 cycles of 95°C for 1 min, 58°C for 1 min, and 72°C for 1 min; the last cycle involved a final extension step at 72°C for 10 min. Residual nucleotides in the PCR mixtures were dephosphorylated by adding 1 U of calf intestinal alkaline phosphatase (CIAP) and 5.0 μl of ×10 CIAP buffer and incubating for 30 min at 37°C in a thermal cycler. After this step, 1.1 μl of 0.25 M EDTA (pH 8.0) was added, and the CIAP was denatured at 75°C for 10 min. Double-stranded PCR fragments were purified with QIAquick columns (Qiagen) and then were used as templates for PEA. Fluorescence-labeled oligonucleotide primers have been designed that bind to the mtDNA template adjacent to the mutation site (table 1). The principle of PEA is simply to extend this primer for a variable length, depending on the presence or absence of the LHON mutation, by using a specific mixture of chain-extending deoxynucleotides and chain-terminating dideoxynucleotides. Primer extension reactions were performed in a total volume of 8.0 μl containing 1.5 μl of the purified PCR template, 40 fmol of primer, 200 pmol of each of the relevant dNTPs and ddNTPs, 1.0 U of thermosequenase DNA polymerase, and 1.5 mM MgCl2 in 1× PCR buffer. After an initial denaturation step at 95°C for 2 min, the reaction conditions comprised 20 cycles of 95°C for 20 s and 55°C for 40 s. A 2.0-μl sample of each PEA was mixed with 1.5 μl deionized formamide, 0.5 μl ROX-350 size standard (Perkin Elmer), and 1.5 μml of EDTA loading buffer (Perkin Elmer); the sample was denatured at 95°C for 5 min before immediate quenching on ice. The wild-type and mutant products were accurately sized by electrophoretic separation in a 12% denaturing polyacrylamide gel, using an ABI 373 genetic analyzer. The relative-fluorescence signals of the PEA products were then analyzed by Genescan (v. 3.0) and Genotyper (v. 2.1) software to quantify the relative allele proportions and, hence, the level of heteroplasmy.
Table 1.
Primers and Nucleotide Combinations Used for Primer Extension Reaction[Note]
| Gene | Base | Substitution | Sequence(5′→3′) | Strand | 5′Label | Length(bp) | Product Size(bp) | dNTP | ddNTP |
| ND1 | 3460 | G→A | GCTCTTTGGTGAAGAGTTTTATGG | Heavy | FAM | 24 | W = 26 M = 25 | C | G and T |
| ND4 | 11778 | G→A | AGTCCTTGAGAGAGGATTATGATG | Heavy | ROX | 24 | W = 26 M = 25 | C | G and T |
| ND6 | 14484 | T→C | CTGTAGTATATCCAAAGACAACCA | Light | HEX | 24 | W = 25 M = 27 | C | A and T |
Note.— Primers were synthesized by MWG Biotech, with ROX (red), HEX (green) and FAM (blue) fluorescent tags at the 5′ end. W = wild-type; M = mutant; dNTPs = deoxynucleotides; ddNTPs = dideoxynucleotides.
Statistical Analysis
Group comparisons were performed using the χ2 test, Student’s t test, and one-way analysis of variance, with 95% CI being determined as appropriate (Altman 1991).
Results
Minimum Prevalence of LHON
We identified 16 genealogically unrelated pedigrees with LHON with at least one affected family member (table 2). In total, we identified 82 individuals with visual failure attributable to LHON in these families. Seventy of these individuals were clinically affected and alive during the midyear period of 1998, giving a minimum point prevalence of 3.22 per 100,000 for the population (95% CI 2.47–3.97 per 100,000). When adult males were considered separately, the minimum point prevalence was 7.11 per 100,000 (95% CI 5.49–8.73 per 100,000). Two of the 16 pedigrees—11778-(A) and 11778-(C)—shared identical sequences in the noncoding D-loop region (table 2), suggesting that they both descended from a common maternal ancestor who harbored the G11778A mutation. The remaining families had distinctly different D-loop sequences, indicating that the primary LHON mutations arose on a number of different occasions within the North East of England. The relative frequency of each primary LHON mutation was as follows: G11778A, 60% (9 of 15 genetically independent maternal pedigrees); G3460A, 33% (5 pedigrees); and T14484C, 7% (1 pedigree). We were able to establish a de novo mutation in only one family with an affected individual harboring the G3460A mutation. The mutation could not be identified in a blood sample from the patient's mother, who shared an identical noncoding region sequence with the proband.
Table 2.
mtDNA D-Loop Sequences for Pedigrees with LHON in the North East of England[Note]
| Pedigree | D-Loop Nucleotide Changes |
| 3460-(A) | A16183C, C16184C, T16189C, T16519C, A263G, CCC303ins (hetero), C311ins |
| 3460-(B) | A16343G, G16390A, T16519C, A73G, C150T, A263G, CC303ins, C311ins |
| 3460-(C) | C16167T, G16274A, T16304C, A73G, A263G, C303ins, C311ins, C456T |
| 3460-(D) | T16519C, A263G, C303ins, C311ins |
| 3460-(E) | T16224C, T16311C, T16519C, A73G, T146C, T152C, A263G, C311ins, C324T |
| 11778-(A) | C16069T, T16126C, C16193T, C16278T, A73G, C150T, T152C, A263G, C295T, CC303ins, C311ins, C353ins, T489C |
| 11778-(B) | C16167T, C16179T, A73G, A263G, T283C, CC303ins, C311ins |
| 11778-(C) | C16069T, T16126C, C16193T, C16278T, A73G, C150T, T152C, A263G, C295T, CC303ins (hetero), C311ins, C353ins, T489C |
| 11778-(D) | T152C, T193C, A263G, C303ins, C311ins, AC515-516del |
| 11778-(E) | T16224C, T16311C, T16519C, A73G, C150T, A263G, C311ins, C497T |
| 11778-(F) | T16224C, T16311C, T16519C, A73G, T146C, T152C, A263G, C311ins, C494del |
| 11778-(G) | T16519C, A263G, C 311 ins, AC515-516del |
| 11778-(H) | T16362C, T152C, T239C, A263G, C303ins, C311ins, AAA335-337del |
| 11778-(K) | C16069T, T16126C, T16519C, A73G, G185A, A188G, T195C, G228A, A263G, C295T, CC303ins (hetero), C311ins, C642T, T489C, AC515-516del |
| 11778-(L) | C16069T, T16126C, C16278T, C16287T, A16293G, C16380T, A73G, C150T, T152C, A153G, C295T, C311ins, T489C, C568ins |
| 14484-(A) | C16069T, T16126C, C16261T, T16519C, A73G, T146C, G185A, A188G, C222T, G228A, A263G, C295T, C311ins, C462T, T489C |
Note.— All nucleotide positions refer to that of the CRS mtDNA light strand. Positions of insertions and deletions in homopolymeric tracts correlate to the position of the first residue of the respective repeat sequence.
Minimum Prevalence of LHON Mutations
Pedigree analysis identified a total of 257 living maternal relatives who either harbored a primary pathogenic mutation or were direct maternal descendants of a woman who was homoplasmic for the LHON mutation. The combined minimum point prevalence of LHON mutations in the population was estimated at 11.82 per 100,000 (95% CI 10.38–13.27 per 100,000).
Sex-Specific Distribution, Penetrance, and Age at Onset
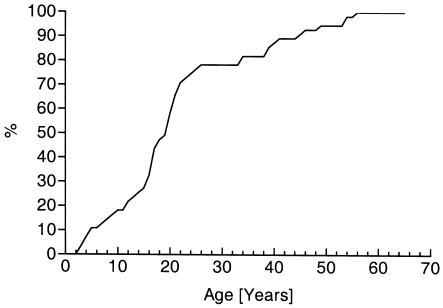
We observed the characteristic predominance of affected males and sex-specific penetrance of LHON (table 3). The mean age at onset was 22.0 years (SD 13.6), and >95% of patients were affected at the age of ⩽50 years. There was no statistically significant difference in age at onset between males and females (T=-0.45, P>.05) or between the three primary LHON mutations (F=0.51, P>.05). Therefore, we pooled the clinical data to generate a cumulative distribution curve (fig. 2).
Table 3.
Sex-Specific Bias and Penetrance of the Primary LHON Mutations
|
No. of Affected |
Penetrance (95% CI) among |
||||
| Mutation | Men | Women | Sex Ratio (95% CI) | Men | Women |
| G3460A | 19 | 11 | 1.73:1 (.78–4.02) | 48.7 (32.4–65.2) | 28.2 (15.0–44.9) |
| G11778A | 41 | 8 | 5.13:1 (2.37–12.66) | 50.6 (39.3–61.9) | 8.5 (3.71–6.1) |
| T14484C | 3 |
0 |
NAa | 37.5 (8.5–75.5) | 0 (0–45.9) |
| Overall | 63 | 19 | 3.32:1 (1.96–5.87) | 49.2 (40.3–58.2) | 13.7 (8.4–20.5) |
NA = Not applicable.
Figure 2.
Cumulative age at onset of LHON in the North East of England.
Incidence of Heteroplasmy
At least one heteroplasmic individual was identified in 40% of the families with G3460A and in 33% of the families with G11778A. No heteroplasmic individuals were identified in the single T14484C family. Overall, 33% of families with LHON in the North East of England included at least one individual who was heteroplasmic for the primary LHON mutation. When individuals were considered separately, 9% of those with the G3460A mutation and 16% of those with the G11778A mutation were heteroplasmic, giving an overall frequency of 12% for individuals who harbor primary LHON mutations in the North East of England. None of the affected individuals harbored <40% mutant mtDNA in their blood leukocytes.
Discussion
We have performed the first population-based clinical and molecular genetic epidemiological study of LHON. In contrast to our previous report of mtDNA LHON mutations, which involved patients referred with the most stringent selection criteria (Chinnery et al. 2000), in the present study we optimized ascertainment by also investigating patients with unexplained optic neuropathy. The most striking finding is the high prevalence of LHON within the North East of England. LHON affects at least 1 in 31,054 individuals, and ∼1 in 8,500 harbor a primary LHON mutation. When male adults are considered separately, at least 1 in 14,067 men within this region have visual failure due to LHON. These figures are higher than those reported in the premolecular era (Mackey and Buttery 1992), and they establish LHON as one of the most common mitochondrial disorders, at 3.22 per 100,000 (Chinnery et al. 2000). LHON is approximately as common as blindness due to autosomal dominant optic atrophy (MIM 165500), which has a prevalence of 3.5 per 100,000 (Kjer et al. 1996); however, it is less common than visual impairment due to retinitis pigmentosa (MIM 268000), which affects 21 per 100,000 (Bunker et al. 1984). The prevalence of LHON is comparable to that of inherited neurological disorders such as Huntington disease (MIM 143100) (6.4 per 100,000, according to Morrison et al. [1995]) and to Duchenne muscular dystrophy (MIM 310200) (3.2 per 100,000) and myotonic dystrophy (MIM 160900) (5.0 per 100,000), according to Emery [1991]). This finding has implications for the distribution of health care resources and the management of inherited eye diseases. Individuals with LHON have special requirements because of the characteristic pattern of visual failure, the strict maternal inheritance pattern, and the difficult nature of genetic counseling for unaffected carriers (Man et al. 2002). Some studies have also raised the possibility that specific treatments may enhance the rate of visual recovery in patients with LHON (Mashima et al. 2000).
Before extrapolating these data to other populations, it is important to consider whether the spectrum of disease due to the primary LHON mutations is unusual in the North East of England. The sex-specific bias, penetrance, and age at onset for the primary LHON mutations within our region (table 3) were similar to those reported in other populations (Newman et al. 1991; Johns et al. 1992b, 1993; Harding et al. 1995; Macmillan et al. 1998). There were minor differences in the mean values; however, these did not reach statistical significance, and they are more likely due to the small sample sizes involved (type 1 error). It is well recognized that the frequency of the different primary LHON mutations varies throughout the world. For example, the T14484C mutation is the most common cause of LHON in French-Canadian families (Macmillan et al. 2000), but it is rare in the Japanese population (Mashima et al. 1998). Although we identified only one pedigree with T14484C in the North East of England, this would be predicted on the basis of the proportion of T14484C pedigrees reported in a large multinational cohort of pedigrees with LHON from Northern Europe and Australia (Mackey et al. 1996). It is therefore reasonable to conclude that the observations that we have made in the North East of England do have broader relevance for LHON in other populations. Our analyses identified only one de novo primary LHON mutation. This is in accordance with the established view that de novo LHON mutations are rare (Biousse et al. 1997). To our knowledge, this is the first reported case of the de novo G3460A mutation.
Although there are well-established risks of visual failure in the maternal relatives of a proband with LHON (Harding et al. 1995; Macmillan et al. 1998), our observations highlight the importance of performing molecular genetic analysis on unaffected carriers who seek guidance on their future risk of visual failure or of having affected offspring. The potential pitfalls are illustrated by one of our pedigrees (11778-[A]). Despite the fact that the majority of individuals in this extensive pedigree are homoplasmic for the G11778A mutation in blood, one branch of the family completely lost the mutation, and another branch of the family is heteroplasmic (11778-[C]). In the present study as a whole, at least one in three families who harbor a primary LHON mutation had one individual who was heteroplasmic for the mtDNA mutation. Given that heteroplasmy may influence the risk of visual failure and the risk of maternal transmission of the mtDNA defect (Smith et al. 1993; Black et al. 1996; Chinnery et al. 2001), confident presymptomatic counseling is possible only after molecular genetic testing in the individual concerned. It is then possible to use well-established age- and sex-specific penetrance data for accurate counseling of an individual known to harbor a primary LHON mutation (Harding et al. 1995; Macmillan et al. 1998).
In conclusion, the present study shows that LHON is not a rare disorder and that it is essential to put into place the necessary resources for the optimal management of these patients and their families. Heteroplasmy is common in families with LHON, and molecular genetic analysis is mandatory for all maternal relatives seeking guidance about the risks of visual failure for themselves and their offspring.
Acknowledgments
This work was supported by the Wellcome Trust (P.F.C. and D.M.T.), the Medical Research Council (D.M.T.), and the PPP Healthcare Trust (P.Y.W.M.). We wish to thank all the patients and their family members for their participation in this study. We gratefully acknowledge our colleagues from the Departments of Neurology, Ophthalmology, and Human Genetics in the North East of England for referring suspected cases of LHON, and we are particularly grateful for the work performed by Sally Anne Lynch, Sharon McDonnell, and Grace Aherne.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
References
- Altman D (1991) Practical statistics for medical research. Chapman Hall, London [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147–147 [DOI] [PubMed] [Google Scholar]
- Biousse V, Brown MD, Newman NJ, Allen JC, Rosenfeld J, Meola G, Wallace DC (1997) De novo 14484 mitochondrial DNA mutation in monozygotic twins discordant for Leber's hereditary optic neuropathy. Neurology 49:1136–1138 [DOI] [PubMed] [Google Scholar]
- Black GC, Morten K, Laborde A, Poulton J (1996) Leber's hereditary optic neuropathy: heteroplasmy is likely to be significant in the expression of LHON in families with the 3460 ND1 mutation. Br J Ophthalmol 80:915–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH (1984) Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol 97:357–365 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Andrews RM, Turnbull DM, Howell N (2001) Leber hereditary optic neuropathy: does heteroplasmy influence the inheritance and expression of the G11778A mitochondrial DNA mutation? Am J Med Genet 98:235–243 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Johnson MA, Wardell TM, Singh-Kler R, Hayes C, Brown DT, Taylor RW, Bindoff LA, Turnbull DM (2000) The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol 48:188–193 [PubMed] [Google Scholar]
- Emery AE (1991) Population frequencies of inherited neuromuscular diseases: a world survey. Neuromuscul Disord 1:19–29 [DOI] [PubMed] [Google Scholar]
- Fahy E, Nazarbaghi R, Zomorrodi M, Herrnstadt C, Parker WD, Davis RE, Ghosh SS (1997) Multiplex fluorescence-based primer extension method for quantitative mutation analysis of mitochondrial DNA and its diagnostic application for Alzheimer's disease. Nucleic Acids Res 25:3102–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE, Sweeney MG, Govan GG, Riordan-Eva P (1995) Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Hum Genet 57:77–86 [PMC free article] [PubMed] [Google Scholar]
- Howell N (1999) Human mitochondrial diseases: answering questions and questioning answers. Int Rev Cytol 186:49–116 [DOI] [PubMed] [Google Scholar]
- Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull DM (1991) Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet 49:939–950 [PMC free article] [PubMed] [Google Scholar]
- Howell N, Bogolin C, Jamieson R, Marenda DR, Mackey DA (1998) mtDNA mutations that cause optic neuropathy: how do we know? Am J Hum Genet 62:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huoponen K, Vilkki J, Aula P, Nikoskelainen EK, Savontaus ML (1991) A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy. Am J Hum Genet 48:1147–1153 [PMC free article] [PubMed] [Google Scholar]
- Johns DR, Heher KL, Miller NR, Smith KH (1993) Leber's hereditary optic neuropathy: clinical manifestations of the 14484 mutation. Arch Ophthalmol 111:495–498 [DOI] [PubMed] [Google Scholar]
- Johns DR, Neufeld MJ, Park RD (1992a) An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy. Biochem Biophys Res Commun 187:1551–1557 [DOI] [PubMed] [Google Scholar]
- Johns DR, Smith KH, Miller NR (1992b) Leber's hereditary optic neuropathy: clinical manifestations of the 3460 mutation. Arch Ophthalmol 110:1577–1581 [DOI] [PubMed] [Google Scholar]
- Kjer B, Eiberg H, Kjer P, Rosenberg T (1996) Dominant optic atrophy mapped to chromosome 3q region II Clinical and epidemiological aspects. Acta Ophthalmol Scand 74:3–7 [DOI] [PubMed] [Google Scholar]
- Leber T (1871) Ueber hereditaere und congenital angelegte sehnervenleiden. Graefes Arch Ophthalmol 17:249–291 [Google Scholar]
- Mackey DA, Buttery RG (1992) Leber hereditary optic neuropathy in Australia. Aust NZ J Ophthalmol 20:177–184 [DOI] [PubMed] [Google Scholar]
- Mackey D, Howell N (1992) A variant of Leber hereditary optic neuropathy characterized by recovery of vision and by an unusual mitochondrial genetic etiology. Am J Hum Genet 51:1218–1228 [PMC free article] [PubMed] [Google Scholar]
- Mackey DA, Oostra RJ, Rosenberg T, Nikoskelainen E, Bronte-Stewart J, Poulton J, Harding AE, Govan G, Bolhuis PA, Norby S (1996) Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet 59:481–485 [PMC free article] [PubMed] [Google Scholar]
- Macmillan C, Johns TA, Fu K, Shoubridge EA (2000) Predominance of the T14484C mutation in French-Canadian families with Leber hereditary optic neuropathy is due to a founder effect. Am J Hum Genet 66:332–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan C, Kirkham T, Fu K, Allison V, Andermann E, Chitayat D, Fortier D, Gans M, Hare H, Quercia N, Zackon D, Shoubridge EA (1998) Pedigree analysis of French Canadian families with T14484C Leber's hereditary optic neuropathy. Neurology 50:417–422 [DOI] [PubMed] [Google Scholar]
- Man PYW, Turnbull DM, Chinnery PF (2002) Leber hereditary optic neuropathy. J Med Genet 39:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashima Y, Kigasawa K, Wakakura M, Oguchi Y (2000) Do idebenone and vitamin therapy shorten the time to achieve visual recovery in Leber hereditary optic neuropathy? J Neuroophthalmol 20:166–170 [DOI] [PubMed] [Google Scholar]
- Mashima Y, Yamada K, Wakakura M, Kigasawa K, Kudoh J, Shimizu N, Oguchi Y (1998) Spectrum of pathogenic mitochondrial DNA mutations and clinical features in Japanese families with Leber's hereditary optic neuropathy. Curr Eye Res 17:403–408 [DOI] [PubMed] [Google Scholar]
- Morrison PJ, Johnston WP, Nevin NC (1995) The epidemiology of Huntington's disease in Northern Ireland. J Med Genet 32:524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman NJ, Lott MT, Wallace DC (1991) The clinical characteristics of pedigrees of Leber's hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol 111:750–762 [DOI] [PubMed] [Google Scholar]
- Nikoskelainen EK, Huoponen K, Juvonen V, Lamminen T, Nummelin K, Savontaus ML (1996) Ophthalmologic findings in Leber hereditary optic neuropathy, with special reference to mtDNA mutations. Ophthalmology 103:504–514 [DOI] [PubMed] [Google Scholar]
- Office for National Statistics (2000) Populations and Households. In: Matheson J, Edwards G (eds). Vol 35: Regional Trends. Government Statistical Service, London, pp 49–62 [Google Scholar]
- Riordan-Eva P, Harding AE (1995) Leber's hereditary optic neuropathy: the clinical relevance of different mitochondrial DNA mutations. J Med Genet 32:81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan-Eva P, Sanders MD, Govan GG, Sweeney MG, Da Costa J, Harding AE (1995) The clinical features of Leber's hereditary optic neuropathy defined by the presence of a pathogenic mitochondrial DNA mutation. Brain 118:319–337 [DOI] [PubMed] [Google Scholar]
- Seedorff T (1985) The inheritance of Leber's disease: a genealogical follow-up study. Acta Ophthalmologica 63:135–145 [DOI] [PubMed] [Google Scholar]
- Smith KH, Johns DR, Heher KL, Miller NR (1993) Heteroplasmy in Leber's hereditary optic neuropathy. Arch Ophthalmol 111:1486–1490 [DOI] [PubMed] [Google Scholar]
- Vanderpump MPJ, Tunbridge WMG, French JM, Appleton D, Bates D, Clark F, Grimley Evans J, et al (1995) The incidence of thyroid-disorders in the community: a twenty-year follow-up of the Whickham survey. Clin Endocrinol (Oxf) 43:55–68 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJd, Nikoskelainen EK (1988) Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science 242:1427–1430 [DOI] [PubMed] [Google Scholar]