Abstract
The human protein CD36 is a major receptor for Plasmodium falciparum–infected erythrocytes and contributes to the pathology of P. falciparum malaria. We performed variation screening of the CD36 gene and examined the possible association between CD36 polymorphisms and the severity of malaria in 475 adult Thai patients with P. falciparum malaria. Accordingly, we identified nine CD36 polymorphisms with a high-frequency (>15%) minor allele. Of these, the frequencies of the −14T→C allele in the upstream promoter region and the −53G→T allele in the downstream promoter region were significantly decreased in patients with cerebral malaria compared to those with mild malaria (P=.016 for −14T→C and P=.050 for −53G→T). The analysis of linkage disequilibrium (LD) between the nine common polymorphisms revealed that there are two blocks with strong LD in the CD36 gene and that the −14T→C and −53G→T polymorphisms are within the upstream block of 35 kb from the upstream promoter to exon 8. Further association testing after the second variation screening in the upstream block indicated that the in3(TG)12 (i.e., 12 TG repeats in intron 3) allele is most strongly associated with the reduction in the risk of cerebral malaria (odds ratio 0.59; 95% confidence interval 0.40–0.87; P=.0069). We found, by reverse-transcriptase PCR amplification, that in3(TG)12 is involved in the nonproduction of the variant CD36 transcript that lacks exons 4 and 5. Since exon 5 of the gene is known to encode the ligand-binding domain for P. falciparum–infected erythrocytes, in3(TG)12 itself or a primary variant on the haplotype with in3(TG)12 may be responsible for protection from cerebral malaria in Thailand. Results of the present study suggest that LD mapping has potential for detecting a disease-associated variant on the basis of haplotype blocks.
Introduction
Plasmodium falciparum malaria remains a major cause of morbidity and mortality in tropical countries, affecting 300 million people and causing more than two million deaths each year. One of the peculiar features of P. falciparum malaria is the adhesion of parasite-infected erythrocytes to capillary endothelia (Miller et al. 1994, 2002). This adhesion contributes to the pathology of P. falciparum malaria, since it causes local microvascular occlusion (Patnaik et al. 1994) and inhibits the immune response to parasites (Urban et al. 1999). A number of receptors have been implicated in cytoadherence of infected erythrocytes—including thrombospondin (Roberts et al. 1985), intercellular adhesion molecule 1 (Berendt et al. 1989), vascular cell adhesion molecule 1 (Ockenhouse et al. 1992), chondroitin sulfate A (Rogerson et al. 1995), platelet/endothelial cell adhesion molecule 1/CD31 (Treutiger et al. 1997), and CD36. Since almost all adherence-positive P. falciparum strains and isolates bind to CD36 (Newbold et al. 1997), CD36 plays the predominant role in the adherence of infected erythrocytes (Ockenhouse et al. 1989, 1991).
CD36 is an 88-kDa glycoprotein expressed on various cells, such as platelets, erythroblasts, adipocytes, monocytes, dendritic cells, macrophages, and vascular endothelial cells. CD36 is involved in a variety of functions in lipid transport, immune regulation, hemostasis, and angiogenesis (Greenwalt et al. 1992; Febbraio et al. 2001). The adhesion of P. falciparum–infected erythrocytes to CD36 may be of benefit to malaria parasites by the sequestration of infected erythrocytes from circulation and by the inhibition of the immune response to parasites (Urban et al. 1999). Pain et al. (2001a) have reported that platelet-mediated clumping of P. falciparum–infected erythrocytes is strongly associated with severe malaria and that CD36 expression is required for such clumping. In contrast, CD36 on monocytes and macrophages has been reported to be necessary for the CD36-dependent phagocytosis of P. falciparum–infected erythrocytes (McGilvray et al. 2000). Thus, CD36 (MIM 173510) is a candidate gene for influencing the outcome of malaria infection.
The human CD36 gene is encoded by 16 exons extending across >46 kb on chromosome 7q11.2 (Armesilla and Vega 1994; Sato et al. 2002). A marked feature of this gene is the presence of two alternative and independent first exons and their promoters, separated by ∼14 kb (Sato et al. 2002). Recently, the 1264T→G stop mutation, causing CD36 deficiency, was found to be associated with protection from severe malaria in Africa (Pain et al. 2001), whereas a contrasting report demonstrates the association between the 1264T→G polymorphism and susceptibility to cerebral malaria (Aitman et al. 2000). Although possible associations between CD36 polymorphisms and severe malaria have been examined in African patients with malaria, CD36 polymorphisms have, to our knowledge, never been studied in Asian patients.
In the present study, we first analyzed 1264T→G and screened for variations in all 16 exons and two promoter regions of CD36 in Thai patients with malaria. A possible association between the identified polymorphisms and the severity of malaria was examined in 272 adult patients with severe P. falciparum malaria and 203 patients with mild malaria. To our knowledge, this is the first association analysis of CD36 polymorphisms detected by systematic variation screening of the CD36 gene.
Patients and Methods
Patients
A total of 272 adult patients with severe P. falciparum malaria (108 patients with cerebral malaria and 164 patients with noncerebral severe malaria) and 203 adult patients with mild P. falciparum malaria (control individuals), all of whom live in northwestern Thailand, were enrolled in the present study. All of them underwent treatment at the Hospital for Tropical Diseases, Faculty of Tropical Medicine, Mahidol University.
Clinical manifestations of malaria were classified as follows. Cerebral malaria was characterized by coma, positive blood smear for the asexual form of P. falciparum, and exclusion of other causes of coma. Severe, but not cerebral, malaria (i.e., noncerebral severe malaria) was characterized by one of the following symptoms: high parasitemia (>100,000 parasites/ml), hypoglycemia (glucose <22 nmol/liter), severe anemia (hematocrit <20% or hemoglobin <7.0 g/dl), and increased serum creatinine levels (>3.0 mg/dl). Mild malaria was characterized by a positive blood smear, fever without other causes of infection, and no manifestations of severe malaria as described above.
All individuals were ⩾13 years of age, and the mean ages of the patients with severe malaria and mild malaria were both 25.5 years. Genomic DNA was extracted from peripheral-blood leukocytes by using a QIAamp Blood Kit (Qiagen). The present study was approved by the institutional review board of the Faculty of Tropical Medicine, Mahidol University, and informed consent was obtained from all patients.
Genotyping of the 1264T→G Polymorphism
The 1264T→G polymorphism was analyzed by PCR direct sequencing with forward primer ex10F (5′-AGTTCAGGTTCCTGGAATGC-3′) and reverse primer ex10R (5′-ATGGACTGTGCTACTGAGGT-3′). The PCR products of 90 samples randomly chosen from patients with severe and mild malaria were used for direct sequencing with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Nucleotide Sequencing
Variation screening of all 16 exons with neighboring introns and two promoter regions of CD36 was performed by PCR direct sequencing. For the promoter regions, sequences spanning positions −1 through −773 (nucleotides 1183-1955 in GenBank clone AF434768), for the upstream promoter, and positions −1 through −708 (nucleotides 3608–4316 in GenBank clone AF434767), for the downstream promoter, were analyzed, since these regions have been reported to include putative regulatory elements for transcriptional factors. The first screening panel included 16 patients with malaria. In the second screening, to increase the probability of detecting any rare variants, we increased the numbers of screened patients with malaria for exons 1, 2, 3, 4, 5, 6, and 7 and the upstream and downstream promoter regions to 32, 32, 32, 54, 36, 36, 32, 36, and 36, respectively. Each fragment amplified by PCR from genomic DNA was sequenced on both strands with an ABI Prism 3100 Genetic Analyzer.
Genotyping of the Identified Polymorphisms
The identified polymorphisms were named tentatively for convenience (table 1). Each polymorphism was genotyped by one of five methods: direct sequencing (for the 5′flankingIndel1, 5′flankingSNP1, and 5′UTRSNP2 polymorphisms), PCR-SSCP (for the −88C→G, 5′flankingSNP3, 5′UTRSNP4, 669C→T, in7SNP6, and in11SNP8 polymorphisms), fluorescence-resonance energy transfer (FRET) (for the in3SNP5 polymorphism), PCR-RFLP analysis (for the in8SNP7 polymorphism), or electrophoretic gel–based detection of allele-length difference (for the 3′UTRIndel2 and in14Indel3 polymorphisms). The PCR primers and the annealing temperatures are listed in table 1.
Table 1.
Overview of CD36 Polymorphisms Identified in Thai
| Polymorphism | Nucleotide Substitution (Position)a | GenotypingMethod | Sequenceb | Forward and Reverse Primers | AnnealingTemperature(°C) |
| First screening: | |||||
| 5′flankingIndel1 | −136insCAAA (upstream promoter) | Sequencing | ggagaggttaacaaaCAAAcactggctgcaaga | 5′-TGTTGTTATGTGGTTCCTAG-3′, 5′-CAAAAGTCAGATCAAAGTAG-3′ | 52 |
| 5′flankingSNP1 | −14T→C (upstream promoter) | Sequencing | atgtacagcagtgatT/Ctgacccagcacttg | 5′-TGTTGTTATGTGGTTCCTAG-3′, 5′-CAAAAGTCAGATCAAAGTAG-3′ | 52 |
| 5′UTRSNP2 | 59T→A (exon 1a) | Sequencing | tcccaactagcattcT/Agaaagtgcattaaca | 5′-TGTTGTTATGTGGTTCCTAG-3′, 5′-CAAAAGTCAGATCAAAGTAG-3′ | 52 |
| 5′flankingSNP3 | −53G→T (downstream promoter) | PCR-SSCP | ctcttctctttttttG/Tggggggggagggggt | 5′-CTTTCAATTCCTCTGGCAAC-3′, 5′-AAGTCCTACACTGCAGTCC-3′ | 58 |
| 5′UTRSNP4 | 158C→A (exon 2) | PCR-SSCP | ctgactcatcagttcA/Ctttcctgtaaaattca | 5′-TGATATTAGAGAGTGTCCC-3′, 5′-ATAAGCAAGTATGGTAACAG-3′ | 56 |
| in3SNP5 | IVS3-6T→C (intron 3) | PCR-FRET | aacttattttcttttT/Ccatagcaagttgtcc | 5′-TAAATTATGTTGACTGTTGC-3′, 5′-ACTCACTCACCTGTACG-3′ | 52 |
| in7SNP6 | IVS7+103C→T (intron 7) | PCR-SSCP | catcaacctatagaaC/Tagacctggttataat | 5′-TGTATCTTCTTTGTCACAGC-3′, 5′-TAACACAACCATAACCTGTG-3′ | 56 |
| in8SNP7 | IVS8-156A→G (intron 8) | PCR-RFLP | ccagcatgcctggccA/Ggttatttctttttac | 5′-TATGATCTGGCTACCTAATG-3′, 5′-CATTTACTTCTGTTGCCAG-3′ | 58 |
| in11SNP8 | IVS11-150A→C (intron 11) | PCR-SSCP | tgcatgtgtatataaA/Ctataactatatatgc | 5′-AAACCTTGACATTCGATTGG-3′, 5′-CATAGAAATATTTGGGCAGC-3′ | 56 |
| 3′UTRIndel2 | 1946-61delGCACAAATAAAGCACT (exon 14) | PCR | accattgtaacaataGCACAAATAAAGCACTtgtgccaaagttgtc | 5′-AATATTATCACGCAGATCAC-3′, 5′-AATTGAGAAATGAACCAGTC-3′ | 53 |
| in14Indel3 | IVS14-419delGGGTTGAGA (intron 14) | PCR | cttgtcttgaGGGTTGAGAagcacccttc | 5′-GAACTTGGTCTGAAGGGTG-3′, 5′-AGGCACATAAGAAGGGTGC-3′ | 60 |
| Second screening: | |||||
| −88C→G | −88C→G (downstream promoter) | PCR-SSCP | accacacactgggatC/Gtgacactgtagagtg | 5′-ATGTTAGGGGAAACTCAGC-3′, 5′-AAAGAGAAGAGAAAGCACTC-3′ | 56 |
| 669C→T | 669C→T (Ser127Leu) (exon 5) | PCR-SSCP | tcgaaccttcactatC/Tagttggaacagaggc | 5′-CGTTAGTTTGCTAGAGACC-3′, 5′-TCTTCTAATGCAGTCGATTC-3′ | 54 |
The nucleotide sequence described by Sato et al. (2002) was used as the reference sequence of the recently described exon 1a and upstream promoter region; the nucleotide sequence described by Armesilla and Vega (1994) was used as the reference sequence of other regions.
The nucleotides denoted by capital letters indicate the polymorphisms; the nucleotide to the left of a slash mark (/) is the more frequently observed allele at the SNP site in Thai patients with malaria.
The 5′flankingIndel1, 5′flankingSNP1, and 5′UTRSNP2 polymorphisms were genotyped by direct sequencing of a single 560-bp PCR product encompassing these sites. Genotyping of −88C→G, 5′flankingSNP3, 5′UTRSNP4, 669C→T, in7SNP6, and in11SNP8 was performed by PCR-SSCP; the samples of those genotypes that have been confirmed by direct sequencing were used as references in the PCR-SSCP analysis. Genotyping of in8SNP7 was performed using the PCR-RFLP method; after PCR, the amplified products containing the polymorphic site were digested with MspI for 3 h and were analyzed on a 10% polyacrylamide gel. Genotyping for in3SNP5 was performed by the FRET method, using a LightCycler (Roche Diagnostics), according to the manufacturer's instructions; the sequences of the fluorescein and LCRed probes used were 5′-ACTTGCTATGGAAAAGAAAA-3′ and 5′-AAGTTTGGGTTATGTCTTTGTAGTAAGTACAGGCACTTCTGGAAGATTGT-3′, respectively; and, after amplification, genotypes were determined by melting-curve analysis. The 3′UTRIndel2 and in14Indel3 polymorphisms were genotyped by the amplification of a fragment containing the deleted region, followed by the measurement of the allele size by gel electrophoresis with a 10% polyacrylamide gel.
Genotyping of the TG Repeat Polymorphism
Genotyping of the dinucleotide repeat, in3(TG)n (i.e., the TG repeat polymorphism in intron 3), of the CD36 gene was performed by PCR, followed by use of an ABI 377 Automated Sequencer (PE Applied Biosystems). The sequence of primers comprised a forward primer (5′-ATTTATAAAAGAAGTTGCAC-3′, with FAM-labeling at the 5′ end) and a reverse primer (5′-GTTTACCTACAATTTAATAA-3′). The PCR fragments were sized using the GeneScan Analysis Software (version 2.1; PE Applied Biosystems), as described in the manufacturer’s manual. A control sample with a known repeat number was tested as a quality control on each gel.
Identification of Variant CD36 Transcript by RT-PCR
Total RNA was purified from peripheral-blood mononuclear cells (PBMC) from unaffected Japanese volunteers by using the RNeasy Mini Kit (Qiagen) and was then reverse transcribed into cDNA. The cDNA amplifications for detecting the variant CD36 transcript were performed in a total volume of 50 μl containing 1.5 μl cDNA, 0.2 μM forward primer ex3-6F (5′-CAATTAAAAAGGCTGCATCC-3′), 0.2 μM reverse primer ex7R (5′-TATGTGTCGATTATGGCAAC-3′), 0.4 mM dNTPs, 2.5 mM MgCl2, and 2 U Taq polymerase. Primer ex3-6F was designed to bind to the junction sequence of exons 3 and 6, which exclusively amplify the variant CD36 cDNAs that lack exons 4 and 5, even though they are present at low levels. The condition for the amplification of the variant CD36 cDNAs consisted of an initial denaturation at 96°C for 10 min, followed by 40 cycles of denaturation at 96°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s. It has been reported that various alternative splicing transcripts of CD36 were observed in human PBMC, whereas exons 2 and 3 were observed in all identified transcripts (Kern et al. 1999). Therefore, to confirm the presence of the CD36 transcript, we amplified CD36 cDNAs through exons 2 and 3 by using forward primer ex2F (5′-GCTGTTGATTTGTGAATAAG-3′) and reverse primer ex3R (5′-TGTCTTCTGGATAAGCAGG-3′). The condition for the amplification consisted of an initial denaturation at 96°C for 10 min, followed by 33 cycles of denaturation at 96°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. Glyceraldehyde phosphate dehydrogenase (GAPDH) cDNAs were amplified as internal controls (Møller et al. 2000).
Statistical Analyses
The χ2 test, Fisher’s exact test, and χ2 test for trend (Armitage's trend test) were used to compare the frequencies of the CD36 polymorphisms among the three groups of patients with malaria. Fisher’s exact test was used when one or more cells in a 2×2 contingency table contained a value of less than five. Conformity of the genotype proportion to Hardy-Weinberg equilibrium was examined for each polymorphism in each group of patients with malaria.
To assess the extent of pairwise linkage disequilibrium (LD) between polymorphisms, we calculated Lewontin’s D′ (Lewontin 1964) by using the program LDfinder (developed by J.O.), in which haplotype frequencies between two biallelic polymorphisms are estimated on the basis of an expectation-maximization algorithm (Excoffier and Slatkin 1995). Throughout the present article, D′ and haplotype frequencies were calculated only for polymorphisms with a minor-allele frequency >15%. D′ was further assessed for 40 and 33 publicly available polymorphisms in the region between intron 3 and intron 14 of CD36 in 24 African-descent (AD) Americans and in 23 European-descent (ED) Americans, respectively, on the basis of genotype data from the UW-FHCRC Variation Discovery Resource (National Heart, Lung, and Blood Institute Program for Genomic Applications). Pairwise D′ was plotted using the GOLD program (see the GOLD Home Page) (Abecasis and Cookson 2000). For each pair of polymorphisms, D′ was plotted at the Cartesian coordinates corresponding to the polymorphism location on the physical map.
Frequencies of haplotypes defined by 10 polymorphisms were estimated on the basis of a maximum-likelihood method with an expectation-maximization algorithm (Excoffier and Slatkin 1995). The calculation was performed using the program Arlequin (Schneider et al. 2000).
Results
Lack of the 1264T→G Polymorphism in Thai Patients with Malaria
In recent years, the role that CD36 polymorphisms play in patients with P. falciparum malaria has been a major subject of research. There are two contradictory reports on the association between the CD36 polymorphism (1264T→G) and the severity of malaria in African populations (Aitman et al. 2000; Pain et al. 2001). However, 1264T→G was not detected in 180 chromosomes in the present study. We therefore concluded that this polymorphism does not significantly contribute to the severity of malaria in Thai.
First Screening and Identified Polymorphisms
In the first screening, 11 polymorphisms were identified: 8 SNPs, 1 insertion, and 2 deletions (SNP1–SNP8 and Indel1–Indel3 in fig. 1 and table 1). Of these 11 polymorphisms, 9 were common and had a minor-allele frequency >15%.
Figure 1.
Graphical representation of identified polymorphisms and the TG repeat in intron 3, in relation to the exon-intron structure of the human CD36 gene.
Table 2 summarizes the allele frequencies of CD36 polymorphisms in 475 Thai patients with malaria. Among these polymorphisms, the frequency of the C allele of 5′flankingSNP1 was found to be decreased in patients with cerebral malaria compared to that in patients with mild malaria (odds ratio [OR] 0.62; 95% CI 0.42–0.92; P=.016). The frequency of the T allele of 5′flankingSNP3 was moderately decreased in patients with cerebral malaria compared to that in patients with mild malaria (OR=0.68; 95% CI 0.47–1.0; P=.050). No statistically significant difference was observed for the other polymorphisms either between patients with cerebral malaria and mild malaria or between patients with noncerebral severe malaria and mild malaria. None of the polymorphisms in each group deviated from expectations based on Hardy-Weinberg equilibrium at a significance level of .05.
Table 2.
Allele Frequencies of CD36 Polymorphisms in Thai Patients with Malaria
|
No. (%) with |
|||
| Polymorphism and Allele | Cerebral Malaria (2n=216) | Noncerebral Severe Malaria (2n=328) | Mild Malaria (2n=406) |
| 5′flankingIndel1: | |||
| Insertion | 5 (2.3) | 12 (3.7) | 13 (3.2) |
| Common | 211 (97.7) | 316 (96.3) | 393 (96.8) |
| 5′flankingSNP1:a | |||
| C allele | 46 (21.3) | 90 (27.4) | 123 (30.3) |
| T allele | 170 (78.7) | 238 (72.6) | 283 (69.7) |
| 5′UTRSNP2: | |||
| A allele | 71 (32.9) | 108 (32.9) | 116 (28.6) |
| T allele | 145 (67.1) | 220 (67.1) | 290 (71.4) |
| 5′flankingSNP3:b | |||
| T allele | 49 (22.7) | 92 (28.0) | 122 (30.0) |
| G allele | 167 (77.3) | 236 (72.0) | 284 (70.0) |
| 5′UTRSNP4: | |||
| C allele | 74 (34.3) | 116 (35.4) | 132 (32.5) |
| A allele | 142 (65.7) | 212 (64.6) | 274 (67.5) |
| in3SNP5: | |||
| C allele | 91 (42.1) | 119 (36.3) | 145 (35.7) |
| T allele | 125 (57.9) | 209 (63.7) | 261 (64.3) |
| in7SNP6: | |||
| T allele | 48 (22.2) | 63 (19.2) | 75 (18.5) |
| C allele | 168 (77.8) | 265 (80.8) | 331 (81.5) |
| in8SNP7: | |||
| G allele | 102 (47.2) | 161 (49.1) | 190 (46.8) |
| A allele | 114 (52.8) | 167 (50.9) | 216 (53.2) |
| in11SNP8: | |||
| C allele | 101 (46.8) | 160 (48.8) | 188 (46.3) |
| A allele | 115 (53.2) | 168 (51.2) | 218 (53.7) |
| 3′UTRIndel2: | |||
| Deletion | 11 (5.0) | 17 (5.2) | 27 (6.7) |
| Common | 205 (95.0) | 311 (94.8) | 379 (93.3) |
| in14Indel3: | |||
| Deletion | 102 (47.2) | 159 (48.5) | 200 (49.3) |
| Common | 114 (52.8) | 169 (51.5) | 206 (50.7) |
| −88C→G: | |||
| G allele | 2 (.9) | 3 (.9) | 5 (1.2) |
| C allele | 214 (99.1) | 325 (99.1) | 401 (98.8) |
| 669C→T: | |||
| T allele | 1 (.5) | 2 (.6) | 5 (1.2) |
| C allele | 215 (99.5) | 326 (99.4) | 401 (98.8) |
OR=0.62; 95% CI 0.42–0.92; χ2=5.77, and P=.016 (cerebral malaria vs. mild malaria, by χ2 test based on a 2×2 contingency table).
OR=0.68; 95% CI 0.47–1.0; χ2=3.83, and P=.050 (cerebral malaria vs. mild malaria, by χ2 test based on a 2×2 contingency table).
The 5′flankingSNP1 and 5′flankingSNP3 polymorphisms showed a significant association with protection from cerebral malaria. However, such an association can be caused by LD between the polymorphisms and a primary associated polymorphism in CD36. To determine the extent of LD in the CD36 gene, we analyzed pairwise D′ between the nine polymorphisms common in Thai patients with malaria.
Structure of LD in CD36
The pairwise D′ between the nine polymorphisms with a high-frequency (>15%) minor allele in Thai patients with malaria is shown in figure 2a. There was no marked difference in D′ between patients with cerebral and mild malaria. Therefore, the LD profile in patients with malaria does not seem to be affected by the differences in allele frequency. Figure 2a suggests the presence of two blocks of strong LD in the CD36 gene. The boundary between the two blocks exists around in7SNP6, and 5′flankingSNP1 and 5′flankingSNP3 are located in the upstream block. Thus, the upstream block is considered to contain the primary polymorphism associated with protection from cerebral malaria. The prediction of the precise boundary between the two blocks is important, because we can then determine the end of the region where the primary polymorphism exists. However, the boundary cannot be inferred precisely, because only a small number of polymorphisms were available for the calculation of D′ in this region.
Figure 2.
Pattern of LD in CD36. Pairwise LD between polymorphisms, as measured on the basis of D′, is represented. The graphs are adjusted for physical distance. Regions of high and low degrees of LD are shown in red and blue, respectively. a, Pairwise D′ in Thai patients with cerebral malaria (upward triangle) and mild malaria (downward triangle). b, Pairwise D′ in ED (upward triangle) and AD (downward triangle) Americans.
To determine whether the two-LD-block structure observed in Thai is common, we next analyzed the higher-resolution LD profiles of the CD36 gene in AD and ED Americans, on the basis of genotype data from the UW-FHCRC Variation Discovery Resource. In the genomic region of CD36, extending 29,894 bp (from exon 3 to intron 14), we selected 40 and 33 polymorphisms with a high-frequency (>15%) minor allele in AD and ED Americans, respectively. The average distances between adjacent polymorphisms were 705.3 bp in AD Americans and 855.4 bp in ED Americans. The profiles of D′ between these polymorphisms are shown in figure 2b. Two blocks of strong LD were again observed in both populations. The regions with the abrupt LD breakdown were almost identical in the two populations and extended 3 kb around exon 8 of the CD36 gene. From figures 2a and 2b, we concluded that there are two blocks of strong LD in the CD36 gene; these are the upstream block, spanning ⩾35 kb from the upstream promoter to intron 7, and the downstream block, spanning ⩾8 kb from intron 8 to intron 14. Since 5′flankingSNP1 and 5′flankingSNP3 were in the upstream block, we then performed further screening, for rare variations in this block, and we performed genotyping of in3(TG)n.
Second Screening and Identified Polymorphisms
Two polymorphisms were newly identified in the second screening of exons 1–7 with neighboring introns and upstream and downstream promoter regions. The −88C→G polymorphism was detected in the downstream promoter region. The 669C→T polymorphism, causing a nonconservative amino acid change (Ser127Leu), was identified in exon 5. However, no significant association was observed between these polymorphisms and the severity of malaria (table 2). Throughout the present study, five novel polymorphisms have been identified: 5′flankingIndel1 (GenBank accession number AB089812), 5′flankingSNP1 (AB088854), 5′UTRSNP2 (AB088853), −88C→G (AB088855), and 669C→T (AB088852).
The TG Repeat Polymorphism
We analyzed in3(TG)n in Thai patients with malaria. The in3(TG)n polymorphism is a known polymorphic marker in the CD36 gene (Lipsky et al. 1994) and has been reported to be linked with CD36 deficiency (Kashiwagi et al. 1995). As shown in table 3, the number of TG repeats ranged from 11 to 19 in Thai patients with malaria. Among the eight identified alleles, in3(TG)12, in3(TG)13, and in3(TG)16 were dominant in the studied population (at frequencies of 32.0%, 31.5%, and 24.6%, respectively, in patients with mild malaria). The frequency of in3(TG)12 was significantly higher in patients with mild malaria than in those with cerebral malaria (OR=0.59; 95% CI 0.40–0.87; P=.0069) (table 3). There was a significant reduction in the risk of cerebral malaria in genotypes with in3(TG)12 (P=.0097, by χ2 test for trend) (table 4). Note that in3(TG)12 showed a smaller P value than did 5′flankingSNP1 and 5′flankingSNP3. Neither allele frequency nor genotype frequency showed a significant difference between patients with noncerebral severe malaria and patients with cerebral malaria or mild malaria.
Table 3.
Allele Frequencies of in3(TG)n of CD36 in Thai Patients with Malaria
|
No. (%) with |
|||
| Allele | Cerebral Malaria (2n=216) | Noncerebral Severe Malaria (2n=328) | Mild Malaria (2n=406) |
| in3(TG)11 | 10 (4.6) | 18 (5.5) | 16 (3.9) |
| in3(TG)12a | 47 (21.8) | 97 (29.6) | 130 (32.0) |
| in3(TG)13 | 73 (33.8) | 105 (32.0) | 128 (31.5) |
| in3(TG)14 | 5 (2.3) | 4 (1.2) | 3 (.7) |
| in3(TG)15 | 11 (5.1) | 16 (4.9) | 13 (3.2) |
| in3(TG)16 | 63 (29.2) | 81 (24.7) | 100 (24.6) |
| in3(TG)17 | 6 (2.8) | 7 (2.1) | 13 (3.2) |
| in3(TG)19 | 1 (.4) | 0 | 3 (.7) |
OR=0.59; 95% CI 0.40–0.87; χ2=7.29, and P=.0069 (cerebral malaria vs. mild malaria, by χ2 test based on a 2×2 contingency table).
Table 4.
Genotype Frequencies of in3(TG)12 in Thai Patients with Malaria
|
No. (%) with |
|||
| Genotype | Cerebral Malariaa (n=108) | Noncerebral Severe Malaria (n=164) | Mild Malaria (n=203) |
| in3(TG)12/in3(TG)12 | 7 (6.5) | 16 (9.8) | 24 (11.8) |
| in3(TG)12/others | 33 (30.5) | 65 (39.6) | 82 (40.4) |
| Others/others | 68 (63.0) | 83 (50.6) | 97 (47.8) |
χ2=6.68, and P=.0097 (cerebral malaria vs. mild malaria, by χ2 test for trend).
The TG Repeat Polymorphism and Variant CD36 Transcript Produced by the Skipping of Exons 4 and 5
Among the identified polymorphisms, in3(TG)12 showed the most prominent association with the reduction in the risk of cerebral malaria. If in3(TG)12 directly influences the pathogenesis of cerebral malaria, then the consequence would be an abnormal splicing of the CD36 transcript. A previous study has revealed that alternative splicing transcripts of the CD36 gene were observed in human PBMC (Kern et al. 1999). Of various splicing transcripts, only the variant CD36 transcript produced by the skipping of exons 4 and 5 (fig. 3a) was reported to be expressed on the surface of HEL cells (an erythroleukemia cell line) (Tang et al. 1994). To assess the possible association between the splicing variant and the genotypes of in3(TG)n, we examined their allelic expression pattern by RT-PCR (fig. 3b). Sequencing of the products confirmed three transcripts: the variant transcript produced by the skipping of exons 4 and 5, the transcript containing exons 2 and 3, and the internal control GAPDH. The variant transcript was detected in individuals without in3(TG)12. In contrast, no amplification of the variant transcript was detected in homozygotes for in3(TG)12 or in a heterozygote for in3(TG)11 and in3(TG)12. An intermediate level of the transcript was observed in heterozygotes for in3(TG)11 or in3(TG)12. For the CD36 transcript with exons 2 and 3, there was no difference in expression levels between the genotypes of in3(TG)n. These results suggest that shorter repeat alleles, such as in3(TG)11 and in3(TG)12, are involved in the nonproduction of the variant CD36 transcript lacking exons 4 and 5 but that other alleles are committed to the production of the variant transcript.
Figure 3.
The variant CD36 transcript produced by exon skipping. a, Schematic representation of alternative splicing of CD36, causing the variant CD36 transcript. b, Expression of the variant CD36 transcript that lacks exons 4 and 5 was examined by RT-PCR with cDNAs from human PBMC. Numbers above each lane refer to the genotypes of in3(TG)n; for example, “16/16” is homozygous for in3(TG)16. In each case, PCR products of the expected sizes were observed for the variant CD36 transcript that lacks exons 4 and 5 (top), the CD36 transcript from exons 2 and 3 (middle), and GAPDH (bottom). The PCR products were separated on 10% polyacrylamide gel and were stained by Syber Gold (Molecular Probes).
Haplotype Estimation
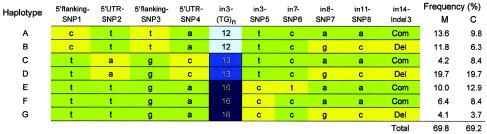
In the CD36 gene, 14 polymorphisms were identified. Of these polymorphisms, 10 (shown in boldface in fig. 1) were used for haplotype estimation in Thai patients with malaria. Figure 4 shows seven haplotypes, each of which consist of 10 polymorphisms that have been inferred to occur at a frequency >3%. These seven haplotypes accounted for ∼70% of all haplotypes estimated in the studied population. The C allele of 5′flankingSNP1, the T allele of 5′flankingSNP3, and in3(TG)12 were found to be together on haplotypes A and B.
Figure 4.
Overview of the seven most common haplotypes estimated in 203 patients with mild malaria and 108 patients with cerebral malaria. The green boxes represent major alleles, and the yellow boxes represent minor alleles at biallelic sites. M = patients with mild malaria; C = patients with cerebral malaria.
Seven common haplotypes were inferred in Thai patients with malaria, whereas only three major haplotypes (A + B, C + D, and E + F + G) were observed in the region from 5′flankingSNP1 to in3SNP5. Interestingly, these three haplotypes could also be distinguished by means of the alleles of the TG repeat. Two haplotypes (A + C + E + F and B + D + G) were inferred in the region from in8SNP1 to in14Indel2. These results indicate that there are two blocks with low haplotype diversity in the CD36 gene.
Although we cannot statistically compare the estimated frequency of haplotype A or haplotype B between patients with cerebral and mild malaria, the estimated frequencies of both haplotypes were decreased in patients with cerebral malaria compared to those in patients with mild malaria. This observation supports the hypothesis that in3(TG)12 itself or one or more primary variants on the haplotype with in3(TG)12 may be responsible for protection from cerebral malaria in Thailand.
Discussion
Our study showed a significant association between polymorphisms of the human CD36 gene and the severity of malaria. Analysis of LD profiles indicated that, in the CD36 gene, there are two blocks with strong LD and the polymorphism (or polymorphisms) that provides protection from cerebral malaria is in the upstream block, spanning 35 kb. The in3(TG)n polymorphism in this block showed a promising association with protection from cerebral malaria, and it has been suggested that in3(TG)n is involved in the production of the variant CD36 transcript without exons 4 and 5.
In the present study, in3(TG)12 was shown to be most strongly associated with the significant reduction in the risk of cerebral malaria. Although the functional significance of in3(TG)n remains unclear, one of the TG repeat alleles is known to be linked with the platelet-specific regulation of CD36 (Kashiwagi et al. 1995). Our results suggest that in3(TG)12 is involved in the nonproduction of the variant CD36 transcript but that other alleles may produce the transcript, causing the expression of the CD36 isoform. The splicing out of exons 4 and 5 maintains the reading frame, and the translated product is expected to yield the CD36 isoform that lacks 103 amino acid residues (residues 41–143). Such an alternative splicing is likely to alter the binding affinity of P. falciparum–infected erythrocytes for CD36, because exons 3, 5, and 6 of CD36 encode the ligand-binding domains at amino acid positions 8–21, 97–110, and 145–171, (Asch et al. 1993; Baruch et al. 1999), respectively. The CD36 isoform that lacks 103 amino acid residues is suggested to be expressed on the surface of HEL cells (Tang et al. 1994). Furthermore, the transient expression of the variant CD36 cDNA in COS-1 cells (a monkey kidney cell line) showed that COS-1 cells expressing the CD36 isoform do not bind to the monoclonal antibody OKM5 (Tang et al. 1994), which specifically blocks CD36-mediated adherence of P. falciparum–infected erythrocytes (Barnwell et al. 1985). Thus, the epitope recognized by OKM5 is altered by the lack of amino acids, suggesting that the binding region for P. falciparum–infected erythrocytes should also be affected in the CD36 isoform. A recent study has revealed that CD36 on monocytes and macrophages plays a crucial role in the CD36-dependent phagocytosis of P. falciparum–infected erythrocytes and in protective immunity against malaria (McGilvray et al. 2000). Considering these earlier results together with our results, we can hypothesize that in3(TG)12, which does not produce the CD36 isoform, is associated with protection from cerebral malaria because of the efficient phagocytosis mediated by intact CD36.
The other, shorter allele, in3(TG)11, was also suggested to be involved in the nonproduction of the variant CD36 transcript, although in3(TG)11 did not show a significant association with protection from cerebral malaria. Because of low frequency of this allele, the sample size in the association test might have been too small for the detection of statistically significant difference, even if in3(TG)11 is truly associated with protection from cerebral malaria. Thus, further investigation will be required in order to address this issue.
Analysis of LD profiles in three populations revealed that there are two blocks with strong LD in the CD36 gene (fig. 2), and the region responsible for protection from cerebral malaria was indicated to be in the 35-kb upstream block, from the upstream promoter region to exon 8. The upstream and downstream promoter regions have been suggested to contain various common regulatory elements of eukaryotic genes (Armesilla et al. 1996; Sato et al. 2002). The 5′ UTR of the CD36 transcript was reported to play a crucial role in the regulation of CD36 expression (Griffin et al. 2001) at the translation level, and its secondary structure was suggested to be involved in the translational efficiency of CD36. In addition, the ligand-binding domain for P. falciparum–infected erythrocytes was encoded by exons in this block. On the basis of our findings and the biological significance of the upstream block of CD36, this block is the most plausible candidate for the region responsible for protection from cerebral malaria. In the present study, no polymorphism that shows a P value smaller than that of in3(TG)12 was identified by variation screening of two promoter regions and all exons with neighboring introns in this block. Therefore, we conclude that in3(TG)12 itself may be responsible for protection from cerebral malaria in Thailand—although we cannot exclude the possibility of the presence of other primary polymorphisms on the haplotype with in3(TG)12, because not all the introns and promoter regions were investigated in the present study.
The LD pattern of the CD36 gene indicated that there are two blocks with strong LD in Thai patients with malaria. To determine whether this structure of LD is observed in other human populations, we analyzed the higher-resolution LD profiles, in AD and ED Americans, that were based on genotype data from the UW-FHCRC Variation Discovery Resource. Both populations showed similar LD profiles with similar breakpoints around exon 8 of CD36. There is considerable evidence that recombination sites in humans are not randomly distributed but are often localized in specific hotspots (Jeffreys et al. 2001; Petes 2001). Jeffreys et al. (2001) observed three blocks with significantly increased D′ in the class II human leukocyte antigen region and determined the crossover rates in the three hotspots; they indicated that the LD profile could be used to predict the locations of putative recombination hotspots. Because of the different population histories of AD and ED Americans and Thai, it seems difficult to explain the present observations in terms of population admixture, subdivision, and bottlenecks. There must be an essential mechanism, probably a recombination hotspot, that underlies the generation of a unique feature of the LD profile of the CD36 gene. In the predicted region of the recombination hotspot, the fraction of GC content (36.0%) was lower than the average (41.0%) for the human genome, and two Alu sequences facing each other in intron 8 of CD36 (nucleotides 21038–21343 and 21631–21938 in GenBank clone AY095373) were found. These observations are consistent with previous reports on the recombination hotspot (Jeffreys et al. 2001; Kong et al. 2002). Thus, we propose the existence of a recombination hotspot within 3 kb of exon 8 of the CD36 gene.
Our results have implications for disease-gene mapping, particularly for genomewide LD testing through use of SNP markers. The potential for success when using such testing depends on haplotype structure in the region where a disease variant is located. We found that in3(TG)12 is located on one of three major haplotypes in the upstream block of LD. These haplotypes are characterized by two SNPs (e.g., 5′flankingSNP1 and 5′UTRSNP2) with a high-frequency minor allele, and each haplotype corresponds to each allele of the TG repeat polymorphism (fig. 4). This observation suggests that similar haplotypes in a block of strong LD were derived from a common ancestral haplotype and have been inherited without recombination. If a disease-associated variant has, by mutation, arisen in such a haplotype block, then the association would be detected easily by the surrounding SNP markers in genomewide LD testing.
Acknowledgments
We sincerely thank the patients who participated in this study. We wish to thank the anonymous reviewers for thoughtful comments on the manuscript. This study was supported by the Core University System Exchange Programme, under the Japan Society for the Promotion of Science and coordinated by the University of Tokyo and Mahidol University; the National Research Council of Thailand; a Grant-in-Aid for Scientific Research on Priority Areas (C) “Medical Science” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to J.O.); and the Genetic Diversity Project, supported by the New Energy and Industrial Technology Development Organization (support to J.O.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for clones [accession numbers AF434767, AF434768 and AY095373] and novel polymorphisms [accession numbers AB089812, AB088852, AB088853, AB088854, and AB088855])
- GOLD Home Page, http://www.sph.umich.edu/csg/abecasis/GOLD/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CD36 [MIM 173510])
- UW-FHCRC Variation Discovery Resource, http://pga.mbt.washington.edu/
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Aitman TJ, Cooper LD, Norsworthy PJ, Wahid FN, Gray JK, Curtis BR, McKeigue PM, Kwiatkowski D, Greenwood BM, Snow RW, Hill AV, Scott J (2000) Malaria susceptibility and CD36 mutation. Nature 405:1015–1016 [DOI] [PubMed] [Google Scholar]
- Armesilla AL, Calvo D, Vega MA (1996) Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J Biol Chem 271:7781–7787 [DOI] [PubMed] [Google Scholar]
- Armesilla AL, Vega MA (1994) Structural organization of the gene for human CD36 glycoprotein. J Biol Chem 269:18985–18991 [PubMed] [Google Scholar]
- Asch AS, Liu I, Briccetti FM, Barnwell JW, Kwakye-Berko F, Dokun A, Goldberger J, Pernambuco M (1993) Analysis of CD36 binding domains: ligand specificity controlled by dephosphorylation of an ectodomain. Science 262:1436–1440 [DOI] [PubMed] [Google Scholar]
- Barnwell JW, Ockenhouse CF, Knowles DM 2nd (1985) Monoclonal antibody OKM5 inhibits the in vitro binding of Plasmodium falciparum–infected erythrocytes to monocytes, endothelial, and C32 melanoma cells. J Immunol 135:3494–3497 [PubMed] [Google Scholar]
- Baruch DI, Ma XC, Pasloske B, Howard RJ, Miller LH (1999) CD36 peptides that block cytoadherence define the CD36 binding region for Plasmodium falciparum-infected erythrocytes. Blood 94:2121–2127 [PubMed] [Google Scholar]
- Berendt AR, Simmons DL, Tansey J, Newbold CI, Marsh K (1989) Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 341:57–59 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Febbraio M, Hajjar DP, Silverstein RL (2001) CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest 108:785–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwalt DE, Lipsky RH, Ockenhouse CF, Ikeda H, Tandon NN, Jamieson GA (1992) Membrane glycoprotein CD36: a review of its roles in adherence, signal transduction, and transfusion medicine. Blood 80:1105–1115 [PubMed] [Google Scholar]
- Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch AS (2001) A link between diabetes and atherosclerosis: glucose regulates expression of CD36 at the level of translation. Nat Med 7:840–846 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Kashiwagi H, Tomiyama Y, Kosugi S, Shiraga M, Lipsky RH, Nagao N, Kanakura Y, Kurata Y, Matsuzawa Y (1995) Family studies of type II CD36 deficient subjects: linkage of a CD36 allele to a platelet-specific mRNA expression defect(s) causing type II CD36 deficiency. Thromb Haemost 74:758–763 [PubMed] [Google Scholar]
- Kern P, Kolowos W, Hagenhofer M, Frank C, Kalden JR, Herrmann M (1999) Alternatively spliced mRNA molecules of the thrombospondin receptor (CD36) in human PBMC. Eur J Immunogenet 26:337–342 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 120:849–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Ikeda H, Medved ES (1994) A dinucleotide repeat in the third intron of CD36. Hum Mol Genet 3:217 [DOI] [PubMed] [Google Scholar]
- McGilvray ID, Serghides L, Kapus A, Rotstein OD, Kain KC (2000) Nonopsonic monocyte/macrophage phagocytosis of Plasmodium falciparum–parasitized erythrocytes: a role for CD36 in malarial clearance. Blood 96:3231–3240 [PubMed] [Google Scholar]
- Miller LH, Baruch DI, Marsh K, Doumbo OK (2002) The pathogenic basis of malaria. Nature 415:673–679 [DOI] [PubMed] [Google Scholar]
- Miller LH, Good MF, Milon G (1994) Malaria pathogenesis. Science 264:1878–1883 [DOI] [PubMed] [Google Scholar]
- Møller LB, Tumer Z, Lund C, Petersen C, Cole T, Hanusch R, Seidel J, Jensen LR, Horn N (2000) Similar splice-site mutations of the ATP7A gene lead to different phenotypes: classical Menkes disease or occipital horn syndrome. Am J Hum Genet 66:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold C, Warn P, Black G, Berendt A, Craig A, Snow B, Msobo M, Peshu N, Marsh K (1997) Receptor-specific adhesion and clinical disease in Plasmodium falciparum. Am J Trop Med Hyg 57:389–398 [DOI] [PubMed] [Google Scholar]
- Ockenhouse CF, Ho M, Tandon NN, Van Seventer GA, Shaw S, White NJ, Jamieson GA, Chulay JD, Webster HK (1991) Molecular basis of sequestration in severe and uncomplicated Plasmodium falciparum malaria: differential adhesion of infected erythrocytes to CD36 and ICAM-1. J Infect Dis 164:163–169 [DOI] [PubMed] [Google Scholar]
- Ockenhouse CF, Tandon NN, Magowan C, Jamieson GA, Chulay JD (1989) Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science 243:1469–1471 [DOI] [PubMed] [Google Scholar]
- Ockenhouse CF, Tegoshi T, Maeno Y, Benjamin C, Ho M, Kan KE, Thway Y, Win K, Aikawa M, Lobb RR (1992) Human vascular endothelial cell adhesion receptors for Plasmodium falciparum–infected erythrocytes: roles for endothelial leukocyte adhesion molecule 1 and vascular cell adhesion molecule 1. J Exp Med 176:1183–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Ferguson DJP, Kai O, Urban BC, Lowe B, Marsh K, Roberts DJ (2001a) Platelet-mediated clumping in Plasmodium falciparum–infected erythrocytes is a common adhesive phenotype and is assoicated with severe malaria. Proc Natl Acad Sci USA 98:1805–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Urban BC, Kai O, Casals-Pascual C, Shafi J, Marsh K, Roberts DJ (2001b) A non-sense mutation in Cd36 gene is associated with protection from severe malaria. Lancet 357:1502–1503 [DOI] [PubMed] [Google Scholar]
- Patnaik JK, Das BS, Mishra SK, Mohanty S, Satpathy SK, Mohanty D (1994) Vascular clogging, mononuclear cell margination, and enhanced vascular permeability in the pathogenesis of human cerebral malaria. Am J Trop Med Hyg 51:642–647 [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369 [DOI] [PubMed] [Google Scholar]
- Roberts DD, Sherwood JA, Spitalnik SL, Panton LJ, Howard RJ, Dixit VM, Frazier WA, Miller LH, Ginsburg V (1985) Thrombospondin binds falciparum malaria parasitized erythrocytes and may mediate cytoadherence. Nature 318:64–66 [DOI] [PubMed] [Google Scholar]
- Rogerson SJ, Chaiyaroj SC, Ng K, Reeder JC, Brown GV (1995) Chondroitin sulfate A is a cell surface receptor for Plasmodium falciparum–infected erythrocytes. J Exp Med 182:15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato O, Kuriki C, Fukui Y, Motojima K (2002) Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor α and γ ligands. J Biol Chem 277:15703–15711 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin, version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva [Google Scholar]
- Tang Y, Taylor KT, Sobieski DA, Medved ES, Lipsky RH (1994) Identification of a human CD36 isoform produced by exon skipping: conservation of exon organization and pre-mRNA splicing patterns with a CD36 gene family member, CLA-1. J Biol Chem 269:6011–6015 [PubMed] [Google Scholar]
- Treutiger CJ, Heddini A, Fernandez V, Muller WA, Wahlgren M (1997) PECAM-1/CD31, an endothelial receptor for binding Plasmodium falciparum–infected erythrocytes. Nat Med 3:1405–1408 [DOI] [PubMed] [Google Scholar]
- Urban BC, Ferguson DJ, Pain A, Willcox N, Plebanski M, Austyn JM, Roberts DJ (1999) Plasmodium falciparum-infected erythrocytes modulate the maturation of dendritic cells. Nature 400:73–77 [DOI] [PubMed] [Google Scholar]