Abstract
In 1978, Sohar et al. described a strikingly peculiar syndrome in two Israeli sisters. These young women responded to environmental temperatures of 18°C–7°C with profuse sweating on large segments on their back and chest. Both had additional abnormalities, including a high-arched palate, nasal voice, depressed nasal bridge, inability to fully extend their elbows, and kyphoscoliosis. We have observed this disorder in two Norwegian brothers. Genomewide screening in the two families, followed by saturation marker studies and linkage analysis, identified a 1.4-Mb homozygous candidate region on chromosome 19p12. The maximum multipoint LOD score was 4.22. In both families, DNA sequencing of 25 genes within the candidate region identified potentially deleterious CRLF1 sequence variants that were not found in unaffected control individuals. Our findings confirm that the cold-induced sweating syndrome is an autosomal recessive disorder that is probably caused by impaired function of the CRLF1 gene, and they suggest important developmental functions for human CRLF1.
Introduction
The cold-induced sweating syndrome (CISS [MIM 272430]) was first described by Sohar et al. (1978). Two Israeli sisters experienced profuse sweating, induced by cool surroundings, on large segments of their back and chest. They also had some additional abnormalities, including a high-arched palate, nasal voice, depressed nasal bridge, inability to fully extend their elbows, and kyphoscoliosis. Their parents shared a common grandfather, suggesting that the observed condition represented a novel syndrome inherited as an autosomal recessive trait. Since this initial description, no confirming case of CISS with reference to the original publication has been described. Thus, the reported disorder is probably very rare.
We have observed a clinical phenotype in two Norwegian brothers that is similar to the one described in the Israeli sisters. No parental consanguinity was known, but genealogical studies revealed several shared ancestors, the closest of which was found nine generations back. Thus, also in the affected Norwegian brothers, homozygosity for a mutant gene inherited from a common ancestor constituted a likely mechanism for this disorder. Exploiting this unique situation, we employed a combination of coarse-scale homozygosity mapping, based on the Israeli inbred sibship with a common great-grandfather, and finer-scale localization, based on the Norwegian sibship with distant common ancestors. Thus, on the basis of only four patients, we have identified the candidate chromosomal segment, the candidate gene, and the likely causative mutations.
Material and Methods
Genotyping
Genomic DNA was isolated from whole blood by using an ABI 341 Nucleic Acid Extractor (PE Applied Biosystems). A genomewide scan was performed using a set of 400 microsatellite markers with an average spacing of 10 cM (ABI Prism Linkage Mapping Set MD, version 2). PCR and pipetting were performed using the ABI Catalyst 800 Turbo Lab station. The PCR products were analyzed using an ABI 310 Genetic Analyzer and the Genescan Analysis software (PE Applied Biosystems). High-density mapping was performed by employing markers identified in various databases (see the GenLink, Cooperative Human Linkage Center, Entrez Genome, Genome Database, and Center for Medical Genetics, Marshfield Medical Research Foundation, Web sites). On the basis of chromosome 19 draft sequences from Lawrence Livermore National Laboratories (see the LLNL Human Genome Center Web site), anonymous repeated (CA) sequences were identified, primers were constructed, and amplified fragments were probed for variants in the Norwegian nuclear family. The PCR primer sequences and population heterozygosity frequencies of these markers are given in table Atable A (online only).
Table A.
Novel Chromosome 19 Markers
| Marker | PCR Product Sizea | HeterozygosityFrequencyb | Forward and Reverse PCR Primers(5′→3′) |
| M1 | 185 | ND | AAAGTTAAGGATGAGATCTT |
| CCACAGGAAAGTACTTATCA | |||
| M2 | 143–168 | .67 | ACCCTTAAGTGCACAATTAC |
| TGTGTTTGGGTGGGTTGTAT | |||
| M3 | 163–173 | .63 | AATGGTGCTTTCAACTATT |
| AAGTCGATAAGTGTGTGCAT | |||
| M4 | 167–186 | .42 | CTGAGACATGAGAATTGCAT |
| CACGACACTTGGTTGATTTT | |||
| M5 | 183–209 | .75 | CTGATTCTGCAGTGTGAC |
| GGAGGCAGAGGTTGCAGC | |||
| M6 | 168–193 | .62 | TGCACTCCAGCCTAGGTAAG |
| GGGAGACATTAGCCGGTGTC | |||
| M7 | 188 | ND | GCACTCCAGCCTAGGTA |
| CGTTTTAGCATTTTATGTCA | |||
| M8 | 146–162 | .74 | AAAGCACAGGCTAAAT |
| ACTTTGTTTTTAAACTCCAC | |||
| M11 | 226–228 | .15 | AAAGTGGGCGGTTTGTC |
| ACGGGCAGGTTGAACTGTTA | |||
| M1A | 237–245 | ND | CCAGGACCTCAGACATTT |
| ATAGTCCCAGCTACTTCG | |||
| M3A | 220–230 | ND | TGTGATGGCACCACTATA |
| CCAGCTAATTTTTGTATTAT | |||
| M4A | 170–177 | ND | GGGGGCTGGCCACAC |
| GGGACTCTTGGGGTCCATCT | |||
| M5A | 187–219 | ND | CCCCCACTAATCAGCTAC |
| TGTGGGCAGGTGGAATCTAC | |||
| M6A | 196–212 | ND | CAGGTGATTTCATACACGC |
| CTTCATCACAGGCAACCTAA |
Sizing of the PCR product was performed using an ABI 310 genetic analyzer, a POP 4 polymer, and the TAMRA 500 size standard (PE Applied Biosystems).
Allele frequencies were determined in 50 Norwegian blood donors. ND = not determined.
Linkage Analysis
Two-point and multipoint linkage analyses were performed using the Mlink and Linkmap programs of the Fastlink software package (Cottingham et al. 1993). Initially, single-point LOD scores were used to identify all regions consistent with a recessive pattern of inheritance in both families. Those regions were then subjected to fine mapping with additional genetic markers. After identification of the candidate region on chromosome 19p12, multipoint analysis (limited to three markers by running time of the software) was performed, to determine the statistical significance of the finding. We used a disease-allele frequency of 0.001 and penetrances of 0 for carriers and noncarriers and 0.99 for homozygous affected individuals. In the initial genome scan, equal allele frequencies were assumed, but, for the fine mapping in the Norwegian family, allele frequencies were determined in 50 unaffected Norwegian control individuals. We also used a method based on the theoretical work of Durham and Feingold (1997), to estimate the probability that the homozygosity observed in the Norwegian pedigree is a false positive (i.e., a chance occurrence, not caused by linkage to the disease locus). We applied equations 1 and 4 of Durham and Feingold (1997) and considered all inbreeding loops of different lengths to be independent, resulting in a conservative P value.
DNA Sequencing and Mutation Detection
PCR primers for amplification of exons and flanking intron sequences in the 1.4-Mb region were designed using the Oligo 6.3 software (Molecular Biology Insights). PCR amplification was performed under standard conditions, using AmpliTaq Gold (PE Applied Biosystems) or Taq polymerase (Qiagen). After amplification, the PCR products were treated with SAP/exonuclease I (Amersham), were sequenced using the ABI Prism BigDye terminator and sequencing kit version 2, and were analyzed on an ABI 3100 Genetic Analyzer (PE Applied Biosystems). A list of our SNP findings is given in table Btable B (online only). Sequencing primers are available on request. DNA sequences were analyzed using the Staden software package (Bonfield et al. 1998).
Table B.
SNPs Identified
| Gene | SNP | NCBI GIa |
| FKBP8 | g.159C→T | 15307595 |
| g.2349C→T | ||
| BSMAP | g.3145 T→C | 11427558 |
| g.7728C→G | ||
| FLJ11078 | g.10C→T | 13633557 |
| g.3378C→G | ||
| g.5702T→C | ||
| g.5850G→A | ||
| KIAA0616 | g.91943A→C | 13633557 |
| g.92007T→C | ||
| COMP | g.6837C→T | 17456204 |
| RENT1 | g.13912G→A | 13633557 |
| g.18700C→T | ||
| g.28535T→C | ||
| LASS1 | g.26782C→T | 17456204 |
| COPE | g.6334A→G | 13633557 |
| g.8180C→G | ||
| g.8217A→G | ||
| g.18945A→G | ||
| FLJ10432 | g.2144T→C | 13633557 |
| g.4668delT | ||
| g.5477dup32 | ||
| g.8806C→T | ||
| HOMER-3 | g.669C→T | 12742013 |
| g.7258G→A | ||
| g.7825A→G | ||
| KIAA0365 | g.30955C→T | 13633557 |
| FLJ20422 | g.30C→T | 13633557 |
| g.5458_5459insC | ||
| g.5425A→G | ||
| g.16602G→A | ||
| MEF2B | g.44306C→T | 17456204 |
| g.45579C→T | ||
| RFXANK | g.1870A→G | 13633557 |
| CSPG3 | g.22827G→A | 2627293 |
| g.27986A→G | ||
| g.27762A→G | ||
| PBX4 | g.49199C→T | 17456204 |
| KOX6 | g.448A→C | 17456204 |
| CNTF (11q) | g.31734G→C | 16184682 |
| g.31776A→G | ||
| g.33339A→G | ||
| g.35039A→G |
For details, see GenBank.
Control Samples
DNA samples were obtained from 200 ostensibly healthy local Norwegian blood donors and 50 Israeli matched control individuals of an ethnic background similar to the patients.
Screening Tests for the CRLF1 Sequence Variants
The c.844_845delGT mutation was verified by PCR amplification by forward (5′-GCAGAGGGAAGAGGAGGAAAACAGA-3′) and reverse (5′-CACACCACTATGCGACAGAATGAG-3′) primers of exon 5 of the CRLF1 gene, followed by analysis of the fluorescence-labeled PCR product on an ABI 310 Genetic Analyzer. The R81H mutation destroys a natural HhaI restriction site in exon 2 of the CRLF1 gene, and this formed the basis for a rapid screening for this mutation. Exon 2 of the CRLF1 gene was PCR amplified using the exon 2 forward (5′-ATTTAACCCAACTGATCTCTACCTT-3′) and reverse (5′-TGAAAGACCTGCATAGCCAT-3′) primers, followed by digestion by HhaI and separation on an 3% Nusieve agarose gel (FMC). HhaI digestion of the PCR-amplified products from normal chromosomes gives two bands (286 and 290 bp), whereas no cutting of the PCR product amplified from mutant chromosomes was observed. We found no simple way of detecting L374R, and a search for this variant was performed by sequencing the proper exon.
Clinical Findings
A brief clinical description of the Israeli sisters has been given elsewhere (Sohar et al. 1978). Both sisters noted the cold-induced sweating at age 16–17 years, shortly after menarche. Their problem has now persisted unchanged for 25 years. The sweating reaction to cold exposure always starts at the same point, in the presternal region in one and in the left hand in the other. It quickly spreads to the rest of the affected areas, distributed as patches, above the waist, on the chest and back. In the affected areas, no sweating occurs at warm temperatures or during febrile episodes. No medical treatment or remedy has so far been helpful in relieving this socially embarrassing disorder. Renewed x-ray examinations show that both sisters have thoracolumbar scoliosis, moderate (30°–35°) in the older sister and less pronounced in the younger sister. Neither of these patients had feeding difficulties in the newborn period. Both sisters have four children, none of whom are affected with this disorder.
The Norwegian brothers were born at term, after uneventful pregnancies. The older would not suckle in the neonatal period and was admitted, dehydrated, to the hospital at 5 d old. He was fed first by a nasogastric tube and subsequently by a special sucking device intended for newborn lambs. Because of continued severe feeding problems, complicated by bronchopulmonary and urinary tract infections, he was treated in the neonatal ward for 3 mo. His younger brother was admitted at 1 d old, primarily because of respiratory problems. Also, this newborn baby did not suckle spontaneously and had to be fed in ways similar to those used for his older brother. Both have problems with fully opening their mouths, rendering ordinary dental work difficult. While playing in the snow, the older brother has repeatedly experienced frostbite in his hands, which was, on one occasion, severe, requiring professional treatment. Likewise, he can hold his palms in a flame or put his hands in boiling water without any sensory pain.
Both of these Norwegian boys have severe progressive kyphoscoliosis. In the younger brother, an S-shaped scoliosis rapidly progressed over a period of 6 mo, at age 13 years. At that time, the major curve measured 47°, and the kyphosis measured 70° (Cobb angle [Cobb 1948]). Posterior-spine surgery was performed, the deformity was corrected, and thoracic vertebrae 3–11 were fused. However, with time, severe kyphosis developed above the fused part. The older brother was first seen by an orthopedic surgeon at age 18 years. He then had both a severe kyphosis (Cobb angle 90°) and scoliosis that was somewhat less pronounced. Also, this patient underwent spine surgery, but a combined anterior and posterior approach was chosen because of severe stiffness. Posterior instrumentation was done, and fusion between thoracic vertebrae 2 and 12 was achieved with satisfactory correction.
The procedures performed on these brothers are considered to be very painful in the postoperative period. However, it was noted that the boys had unusually low demand for pain-relieving medication and seemingly were not bothered by the postoperative pain. During surgery on the older brother, the surgeon noted unusually lightly colored muscle (“like chicken meat”). A biopsy was performed, and the finding was described as “muscular atrophy.”
Both brothers have short hands with pronounced clinodactyly and tapering of fingers. They cannot fully extend their elbows (30° deficit). Also, their toes are somewhat short, and both have flat feet. They have insufficient activity of facial muscles, leading to expressionless faces; instead of a smile, a grin results. Their sweating problem was noted at ∼7 years of age. The patchwise distribution of affected areas much resembles those described in the Israeli sisters. These areas do not sweat at warm temperatures, during fever episodes, or during exercise. The mother sometimes had to cool her overheated children by putting their feet in cold water. Subtropical environment does not bother these patients. They can stay in bright sunlight without feeling the heat and have no desire to take their clothes off for cooling.
Genealogical Studies
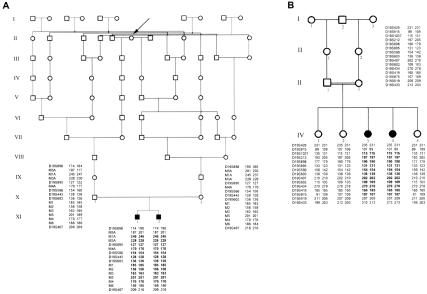
Until the 20th century, the majority of the Norwegian population was attached to a single locality throughout life. A cumulative population-inbreeding coefficient has been estimated at 0.0027 (Gedde-Dahl 1973). Many rural communities in Norway have produced printed local histories, often including volumes of painstakingly gathered genealogical data for each farm, back to the first church records and censuses (17th century). For communities without such printed sources, online data from national censuses and church records were used (see the Digitalarkivet Web site). These tools made it possible to identify, by name, all of the Norwegian brothers' 32 ancestors, five generations back, all of whom were born in the first half of the 19th century. All ancestral lines were pursued as far back as possible. Of the 16 ancestors in each line, 3 in the paternal line and 8 in the maternal line originated from the same rural community. It was possible to trace the majority of these ancestral branches back to the 17th century, and it was possible to trace some branches even further back. The first common ancestor was identified nine generations back (fig. 1A, indicated by the arrow). Another five, six, three, two, and one new common ancestral couples were identified 10, 11, 12, 13, and 14 generations back, respectively. Nearly 1,000 ancestors were identified by name, and a very complex pedigree emerged.
Figure 1.
Pedigree of Norwegian (A) and Israeli (B) families. Blackened symbols indicate patients with CISS. In the Norwegian family, the first common ancestor was found nine generations back (arrow). Observed marker homozygosity is shown in boldface.
Results
Linkage Analysis
When genomewide screening was performed, using markers with an average spacing of 10 cM, three possible candidate regions could not readily be excluded. Ambiguous results owing to limited heterozygosity were obtained in regions on chromosomes 3q and 21. The results of a total of 12 additional markers, distributed between the screening markers, excluded these regions as true candidates. However, on chromosome 19, the Israeli sisters and the Norwegian brothers had inherited, from their parents, a pair of common segments spanning <60 Mb and <48 Mb, respectively (table 1). Within the 42-Mb overlapping candidate segment, the Israeli sisters were homozygous for only two nonadjacent markers, D19S221 and D19S414. Homozygosity was not observed for any of the initial screening markers in this region (D19S221-D19S414) in the Norwegian sibship. Detailed mapping of this region showed that the two Israeli sisters were homozygous for a segment, encompassing 28.8 Mb, that was not revealed by the screening markers (fig. 1 and table 1). Markers D19S221 and D19S414 were both positioned outside this segment. Subsequently, saturation marker studies, as well as SNPs detected by DNA sequencing, demonstrated a 1.4-Mb region of homozygosity within the candidate segment in the Norwegian brothers (fig. 1 and table 1).
Table 1.
Genotypes of Chromosome 19 Markers
|
Genotypeb in Pedigree |
||||||||||||||||||||
| Markera | X-1 | X-2 | XI-1 | XI-2 | III-2 | IV-1 | IV-2 | IV-3 | IV-4 | IV-5 | ||||||||||
| D19S209 |
251 | 251 | 247 | 243 | 251 | 243 | 251 | 247 | 246 | 250 | 248 | 250 | 248 | 250 | 244 | 246 | 248 | 250 | 244 | 250 |
| D19S216 |
265 | 269 | 271 | 267 | 265 | 271 | 265 | 271 | 259 | 265 | 265 | 265 | 267 | 265 | 265 | 259 | 267 | 265 | 265 | 265 |
| D19S884 |
107 | 97 | 107 | 107 | 107 | 107 | 107 | 107 | 103 | 107 | 91 | 107 | 97 | 107 | 97 | 103 | 97 | 107 | 91 | 107 |
| D19S221 |
88 | 104 | 104 | 106 | 88 | 104 | 88 | 104 | 98 | 98 | 93 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 93 | 98 |
| D19S226 |
254 | 254 | 246 | 250 | 254 | 246 | 254 | 246 | 240 | 254 | 254 | 254 | 238 | 254 | 238 | 240 | 238 | 240 | 254 | 254 |
| D19S929 | 251 | 253 | 251 | 253 | 251 | 251 | 251 | 251 | 253 | 249 | 251 | 249 | 249 | 249 | 249 | 253 | 249 | 253 | 251 | 249 |
| D19S841 | 245 | 245 | 235 | 235 | 245 | 235 | 245 | 235 | 247 | 243 | 235 | 243 | 243 | 243 | 243 | 247 | 243 | 247 | 235 | 243 |
| D19S588 | 162 | 148 | 170 | 170 | 162 | 170 | 162 | 170 | 149 | 149 | 149 | 149 | 166 | 149 | 166 | 149 | 166 | 149 | 149 | 149 |
| D19S244 | 138 | 109 | 126 | 99 | 138 | 126 | 138 | 126 | 101 | 105 | 105 | 105 | 90 | 149 | 90 | 101 | 90 | 101 | 105 | 105 |
| D19S930 | 181 | 196 | 183 | 181 | 181 | 183 | 181 | 183 | 187 | 183 | 198 | 183 | 189 | 149 | 189 | 187 | 189 | 187 | 198 | 183 |
| D19S899 | 102 | 109 | 104 | 112 | 102 | 104 | 102 | 104 | 103 | 105 | 101 | 105 | 107 | 149 | 107 | 103 | 107 | 103 | 101 | 105 |
| D19S410 | 155 | 168 | 173 | 155 | 155 | 173 | 155 | 173 | 162 | 154 | 154 | 154 | 171 | 149 | 171 | 162 | 171 | 162 | 154 | 154 |
| D19S579 | 163 | 179 | 179 | 171 | 163 | 179 | 163 | 179 | 175 | 175 | 175 | 175 | 171 | 149 | 171 | 175 | 171 | 175 | 175 | 175 |
| D19S429 | 231 | 235 | 239 | 231 | 235 | 239 | 235 | 239 | 231 | 231 | 231 | 231 | 235 | 149 | 235 | 231 | 235 | 231 | 231 | 231 |
| D19S915 | 109 | 101 | 107 | 111 | 109 | 107 | 109 | 107 | 89 | 109 | 89 | 109 | 107 | 149 | 107 |
89 |
107 |
89 |
89 | 109 |
| D19S1037 | 119 | 119 | 123 | 123 | 119 | 123 | 119 | 123 | 115 | 131 | 135 | 131 | 115 | 149 | 115 | 115 | 115 | 115 | 135 | 131 |
| D19S212 | 193 | 197 | 193 | 205 | 193 | 193 | 193 | 193 | 197 | 205 | 193 | 205 | 197 | 149 | 197 | 197 | 197 | 197 | 193 | 205 |
| D19S460 | 128 | 130 | 128 | 122 | 130 | 128 | 130 | 128 | 128 | 122 | 130 | 122 | 128 | 149 | 128 | 128 | 128 | 128 | 130 | 122 |
| D19S898 | 174 | 184 | 190 | 180 | 174 | 190 | 174 | 190 | 190 | 179 | 177 | 179 | 190 | 149 | 190 | 190 | 190 | 190 | 177 | 179 |
| M6A | 196 | 212 | 202 | 198 | 196 | 202 | 196 | 202 | ||||||||||||
| M5A | 187 | 117 | 201 | 203 |
187 |
201 |
187 |
201 |
||||||||||||
| M1A | 245 | 247 | 245 | 237 | 245 | 245 | 245 | 245 | ||||||||||||
| M3A | 228 | 230 | 228 | 220 | 228 | 228 | 228 | 228 | ||||||||||||
| D19S895 | 127 | 122 | 127 | 127 | 127 | 127 | 127 | 127 | 131 | 123 | 133 | 123 | 131 | 123 | 131 | 131 | 131 | 131 | 133 | 123 |
| M4A | 170 | 177 | 170 | 170 | 170 | 170 | 170 | 170 | ||||||||||||
| D19S566 | 154 | 160 | 154 | 156 | 154 | 154 | 154 | 154 | 154 | 142 | 156 | 142 | 154 | 142 | 154 | 154 | 154 | 154 | 156 | 142 |
| D19S443 | 128 | 128 | 128 | 128 | 128 | 128 | 128 | 128 | 126 | 126 | 126 | 126 | 126 | 126 | 126 | 126 | 126 | 126 | 126 | 126 |
| D19S603 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 | 136 |
| M1 | 185 | 185 | 185 | 185 | 185 | 185 | 185 | 185 | ||||||||||||
| M2 | 156 | 158 | 156 | 158 | 156 | 156 | 156 | 156 | ||||||||||||
| M3 | 163 | 165 | 163 | 165 | 163 | 163 | 163 | 163 | ||||||||||||
| M5 | 201 | 189 | 201 | 201 | 201 | 201 | 201 | 201 | ||||||||||||
| M4 | 170 | 177 | 170 | 170 |
170 |
170 |
170 |
170 |
||||||||||||
| M6 | 188 | 186 | 186 | 184 | 188 | 186 | 188 | 186 | ||||||||||||
| D19S546 | 314 | 314 | 314 | 314 | 314 | 314 | 314 | 314 | 314 | 284 | 314 | 284 | 314 | 284 | 314 | 314 | 314 | 314 | 314 | 284 |
| D19S407 | 209 | 209 | 216 | 216 | 209 | 216 | 209 | 216 | 202 | 210 | 210 | 210 | 202 | 210 | 202 | 202 | 202 | 202 | 210 | 210 |
| M8 | 155 | 148 | 153 | 148 | 155 | 153 | 155 | 153 | ||||||||||||
| D19S911 | 231 | 231 | 225 | 231 | 231 | 225 | 231 | 231 | 231 | 227 | 229 | 227 | 231 | 227 | 231 | 231 | 231 | 231 | 229 | 227 |
| D19S602 | 103 | 105 | 109 | 103 | 103 | 109 | 103 | 103 | 109 | 103 | 115 | 103 | 109 | 103 | 109 | 109 | 109 | 109 | 115 | 103 |
| D19S925 | 262 | 266 | 266 | 268 | 262 | 266 | 262 | 268 | 266 | 262 | 266 | 262 | 266 | 262 | 266 | 266 | 266 | 266 | 266 | 262 |
| D19S215 | 259 | 243 | 249 | 257 | 259 | 249 | 259 | 257 | 243 | 257 | 249 | 257 | 243 | 257 | 243 | 243 | 243 | 243 | 249 | 257 |
| D19S910 | 239 | 251 | 239 | 239 | 239 | 239 | 239 | 239 | 239 | 239 | 237 | 239 | 239 | 239 | 239 | 239 | 239 | 239 | 237 | 239 |
| D19S401 | 353 | 353 | 345 | 341 | 353 | 345 | 353 | 341 | 345 | 349 | 353 | 349 | 345 | 349 | 345 | 345 | 341 | 345 | 353 | 349 |
| D19S568 | 256 | 246 | 256 | 268 | 256 | 256 | 256 | 268 | 246 | 246 | 256 | 246 | 246 | 246 | 246 | 246 | 246 | 246 | 256 | 246 |
| D19S434 | 269 | 273 | 273 | 269 | 269 | 273 | 269 | 269 | 270 | 278 | 270 | 278 | 270 | 278 | 270 | 270 | 270 | 270 | 270 | 278 |
| D19S1036 | 208 | 208 | 208 | 204 | 208 | 208 | 208 | 204 | 208 | 204 | 204 | 204 | 208 | 204 | 208 | 208 | 208 | 208 | 204 | 204 |
| Centromere | ||||||||||||||||||||
| D19S419 | 165 | 167 | 167 | 163 | 165 | 167 | 165 | 163 | 165 | 165 | 165 | 165 | 165 | 165 | 165 | 165 | 165 | 165 | 165 | 165 |
| D19S931 | 159 | 161 | 157 | 157 | 159 | 157 | 159 | 157 | 156 | 160 | 164 | 160 | 156 | 160 | 156 | 156 | 156 | 156 | 164 | 160 |
| D19S870 | 256 | 251 | 256 | 251 | 256 | 256 | 256 | 251 | 251 | 256 | 251 | 256 | 251 | 256 | 251 | 251 | 251 | 251 | 251 | 256 |
| D19S222 | 231 | 231 | 231 | 231 | 231 | 231 | 231 | 231 | 239 | 237 | 237 | 237 | 239 | 237 | 239 | 239 | 239 | 239 | 237 | 237 |
| D19S920 | 213 | 213 | 209 | 209 | 213 | 209 | 213 | 209 | 209 | 213 | 209 | 213 | 209 | 213 | 209 | 209 | 209 | 209 | 209 | 213 |
| D19S932 | 141 | 129 | 137 | 135 | 141 | 137 | 141 | 135 | 129 | 135 | 129 | 135 | 129 | 135 | 129 | 129 | 129 | 129 | 129 | 135 |
| D19S875 | 91 | 111 | 105 | 103 | 91 | 105 | 91 | 103 | 107 | 109 | 91 | 109 | 107 | 109 | 107 |
107 |
107 |
107 |
91 | 109 |
| D19S919 | 213 | 209 | 209 | 209 | 213 | 209 | 213 | 209 | 209 | 209 | 211 | 209 | 207 | 209 | 207 | 209 | 207 | 209 | 211 | 209 |
| D19S433 | 196 | 196 | 200 | 215 | 196 | 200 | 196 | 215 | 213 | 203 | 199 | 203 | 213 | 203 | 213 | 213 | 213 | 213 | 199 | 203 |
| (continued) | ||||||||||||||||||||
| D19S405 | 113 | 115 | 107 | 115 | 113 | 107 | 113 | 115 | 113 | 113 | 111 | 113 | 113 | 113 | 113 | 113 | 113 | 113 | 111 | 113 |
| D19S882 | 279 | 277 | 279 | 267 | 279 | 279 | 279 | 267 | 279 | 279 | 267 | 279 | 279 | 279 | 279 | 279 | 279 | 279 | 267 | 279 |
| D19S414 |
181 | 167 | 167 | 167 | 181 | 167 | 181 | 167 | 186 | 184 | 182 | 184 | 186 | 184 | 186 | 186 | 186 | 186 | 182 | 184 |
| D19S225 | 173 | 173 | 167 | 171 | 173 | 167 | 173 | 171 | 169 | 175 | 171 | 175 | 169 | 175 | 169 | 169 | 169 | 169 | 171 | 175 |
| D19S868 | 194 | 198 | 198 | 198 | 194 | 198 | 194 | 198 | 179 | 190 | 190 | 190 | 179 | 190 | 179 | 179 | 179 | 179 | 190 | 190 |
| D19S416 | 168 | 170 | 166 | 172 | 168 | 166 | 168 | 172 | 168 | 168 | 168 | 168 | 168 | 168 | 168 | 168 | 168 | 168 | 168 | 168 |
| D19S425 | 251 | 259 | 271 | 251 | 251 | 271 | 251 | 251 | 251 | 261 | 251 | 261 | 265 | 261 | 265 | 251 | 265 | 251 | 251 | 261 |
| D19S224 | 249 | 233 | 233 | 251 | 249 | 233 | 249 | 251 | 251 | 253 | 251 | 253 | 251 | 253 | 251 | 251 | 251 | 251 | 251 | 253 |
| D19S220 |
283 | 289 | 279 | 285 | 283 | 279 | 283 | 285 | 289 | 279 | 281 | 279 | 279 | 279 | 279 | 289 | 279 | 289 | 281 | 279 |
| D19S420 |
110 | 90 | 100 | 106 | 110 | 100 | 110 | 106 | 106 | 98 | 110 | 98 | 94 | 98 | 94 | 106 | 94 | 106 | 110 | 98 |
| D19S902 |
259 | 244 | 250 | 240 | 259 | 250 | 259 | 240 | 249 | 249 | 239 | 249 | 239 | 249 | 239 | 249 | 239 | 249 | 247 | 249 |
| D19S571 |
287 | 287 | 287 | 313 | 287 | 287 | 287 | 313 | 310 | 310 | 310 | 310 | 310 | 310 | 310 | 310 | 310 | 310 | 310 | 310 |
| D19S418 |
96 | 92 | 92 | 86 | 96 | 92 | 96 | 92 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 | 90 |
| D19S877 | 250 | 244 | 240 | 242 | 250 | 240 | 250 | 240 | 261 | 261 | 240 | 261 | 261 | 261 | 240 | 261 | 240 | 261 | 240 | 261 |
| D19S254 | 140 | 132 | 112 | 112 | 132 | 112 | 140 | 112 | 112 | 132 | 112 | 132 | 128 | 132 | 128 | 132 | 112 | 112 | 112 | 112 |
| D19S210 |
177 | 185 | 177 | 179 | 177 | 177 | 177 | 177 | 177 | 175 | 175 | 175 | 177 | 175 | 175 | 177 | 177 | 177 | 177 | 175 |
| D19S890 | 278 | 280 | 282 | 274 | 280 | 282 | 278 | 274 | 280 | 280 | 276 | 280 | 288 | 280 | 288 | 280 | 276 | 280 | 276 | 280 |
Markers are ordered according to NCBI Map Viewer, build 30 (see the Entrez Genome Web site). ABI screening kit markers are underlined; the markers beginning with “M” were established in our laboratory (for details, see table Atable A).
Regions of homozygosity are boxed. Boldface italic numerals refer to the shared chromosomal segments in the Norwegian brothers; roman numerals refer to the pedigrees in figure 1.
We performed multipoint linkage analysis, to determine the statistical significance of the observed homozygosity. For the Israeli family, the maximum multipoint LOD score within the shared segment was 2.47. Unfortunately, the complexity of the Norwegian pedigree imposed computational constraints, limiting the analysis to three markers at a time. Within the Norwegian family, we obtained a maximum four-point LOD score of 1.75, on the basis of markers D19S895, D19S566, and D19S603. In the absence of ancestral genotypes, this is an underestimate of the true LOD score (see the “Discussion” section). Hence, we used the method of Durham and Feingold (1997) to directly estimate the probability that, in the Norwegian pedigree, we would find such an identical-by-descent (IBD) segment by chance (i.e., without linkage to the disease locus). We estimate this genomewide probability as p<0.02. This value is statistically significant by itself, and the LOD score of 4.22 for the two families combined further supports the significance of the finding.
Identification and DNA Sequencing of Candidate Genes
Chromosome 19 is unusually gene rich. More than 50 confirmed and hypothetical genes reside within this 1.4-Mb candidate region (NCBI Map Viewer, build 30 [see the Entrez Genome Web site]). No detrimental mutations were detected in the coding sequences of the first 24 sequenced genes (table Btable B).
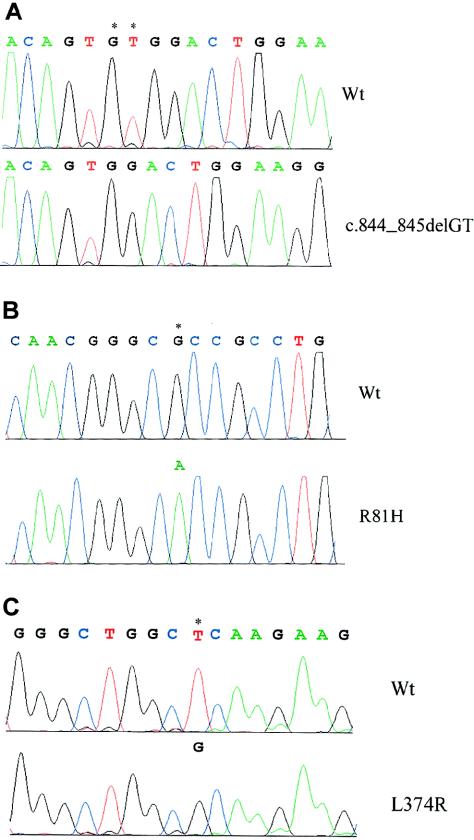
Eventually, DNA sequencing of the cytokine receptor–like factor 1 gene (CRLF1) identified homozygosity for a 2-bp deletion (c.844_845delGT) in exon 5 of the CRLF1 gene in the Norwegian brothers (fig. 2A). Such a frameshift mutation will result in a nonfunctional gene product. In the Israeli sisters, homozygosity for two sequence variants was demonstrated, in codons 81 (CGC→CAC) and 374 (CTC→CGC) (figs. 2B and 2C). Each substitution is predicted to produce amino acid an change, R81H and L374R, respectively. Neither the deletion nor the substitutions were identified among 200 Norwegian and 50 Israeli control individuals, supporting the assumption that these mutations are causally related to the disorder.
Figure 2.
Mutation analysis of CRLF1 in Norwegian and Israeli families. Wt = wild type. A, DNA sequence of CRLF1 exon 5, showing the 2-bp deletion (c.844_845delGT). B, DNA sequence of CRLF1 exon 2, showing the A→G substitution in the second position of codon 81, predicting a change from arginine to histidine (R81H). C, DNA sequence of CRLF1 exon 7, showing the T→G substitution in the second position of codon 374, substituting arginine for leucine (L374R). The sites of mutational changes are indicated by asterisks (*). (Numbering of CRLF1 cDNA here is based on GenBank [accession number NM_004750.2].)
To investigate whether sequence variants in the CRLF1 gene could be commonly encountered in the population, we sequenced the nine exons of the CRLF1 gene in 10 unaffected Norwegian blood donors. No variants were found.
We also sequenced the coding sequence of the functionally related CNTF gene in all four patients, to investigate whether a variant in this gene could influence the CISS phenotype. A predicted serine→glycine variant in CNTF codon 208 (i.e., heterozygous S208G) was identified in both Norwegian brothers. This sequence variant represents a common polymorphism in the Norwegian population (allele frequency 0.27). No sequence variants were identified in the Israeli sisters.
Discussion
Our study demonstrated a 1.4-Mb candidate region of homozygosity (IBD) in the Norwegian patients. Because of computational constraints, we were not able to calculate an exact multipoint LOD score for the Norwegian pedigree. In the absence of ancestral genotypes, the probability that a shared segment is inherited IBD from a common ancestor increases with the number of informative markers contained in the segment; that is, a shared segment containing only three markers has a significant probability of being simply identical by state, whereas a segment containing a large number of shared markers is much more likely to be IBD from a common ancestor. Thus, the maximum four-point LOD score of 4.22 for the two families combined, on the basis of only three markers from within an interval containing 13 shared markers, is an underestimate of the true LOD score. As an alternative approach, we adapted the method of Durham and Feingold (1997) to estimate the probability that the homozygosity observed in the Norwegian pedigree is a false positive. We estimate this probability as p<0.02. Both of the above approaches provide strong support for the hypothesis that the shared segment on chromosome 19 contains the disease locus.
DNA sequencing of the CRLF1 gene identified mutations in both the Norwegian brothers and the Israeli sisters. The 2-bp deletion observed in the Norwegian brothers will result in a frameshift encoding a nonfunctional gene product. The substitutions observed in the Israeli sisters are predicted to produce the amino acid changes R81H and L374R. Although the overall phenotype in the Norwegian brothers and the Israeli sisters was similar, the phenotype in the brothers was more severe (e.g., including feeding difficulties, serious kyphoscoliosis, earlier age at onset of the sweating problem, and reduced pain and temperature sensitivity). These differences may be related to different degrees of functional severity of the observed CRLF1 mutations—namely, a knockout mutation in the Norwegian brothers and the presence of a CRLF1 protein that may have some residual activity in the Israeli sisters. Possibly, other genetic factors (e.g., different sex of the patients) may have also contributed.
CRLF1 is a soluble cytokine receptor with homology to type 1 cytokine receptors (Elson et al. 1998). CRLF1 associates with the cardiotrophin-like cytokine, to form a soluble functional heteromeric ligand, and competes with ciliary neurotrophic factor (CNTF) for the binding to the ciliary neurotrophic factor receptor (CNTFR) complex (Elson et al. 2000). The binding of CRLF1 and CNTF to a common receptor—and their apparent functional similarity—led to the dubbing of CRLF1 as “CNTF II” (Lesser and Lo 2000). CNTF exerts a survival-promoting effect on a variety of neuronal cells. However, the use of CNTF as an experimental treatment of patients with motor-neuron disease did not influence the clinical course of this degenerative disorder (Lambert et al. 2001). Furthermore, a null mutation in the CNTF gene occurs as a common variant in the Japanese population and is not associated with any neurological disorder (Takahashi et al. 1994).
To our knowledge, no impaired function of either CRLF1 or any of the other factors constituting the CNTFR complex has so far been implicated in any human disorder. However, some clinical observations in the Norwegian brothers show similarities to observations made in experimental animals and in cell cultures. In the developing mouse embryo, CRLF1 is expressed at multiple sites, including skeletal muscle (Elson et al. 1998; Alexander et al. 1999). CNTFR, the receptor for CRLF1, is primarily expressed in the nervous system (Stockli et al. 1991; DeChiara et al. 1995), but expression is also detected in skeletal muscle (Davis et al. 1991). A muscle biopsy performed during back surgery in one of the Norwegian patients showed atrophic skeletal muscle, possibly contributing to the development of his severe kyphoscoliosis. This may indicate that normal CRLF1 exerts an effect not only on neuronal but also on skeletal-muscle development and survival.
In vitro experiments show that CRLF1 can promote the survival of developing embryonic motor neurons (Elson et al. 2000). Mouse models lacking either the CRLF1, CNTFR, or CNTF function have been constructed. A significant reduction in motor-neuron numbers in brain motor nuclei and in the spinal cord has been observed in mice that lack CNTFR (DeChiara et al. 1995), but no structural anomalies have been observed in mice that lack CNTF (Alexander et al. 1999). The metal used for vertebral fixation in the Norwegian boys precludes renewed magnetic-resonance–imaging studies. However, the preoperative images of the cervicothoracic spine show apparently normal dimensions of the spinal cord in both brothers.
Mice lacking the CRLF1 gene (i.e., NR6−/− mice) were unable to suckle and died of starvation shortly after birth, with their stomachs devoid of milk (Alexander et al. 1999). The newborn mice could open and close their mouths, and no anatomical anomalies were detected on dissection. Alexander et al. (1999) concluded that CRLF1 was indispensible for suckling, but they were unable to identify the mechanism by which its role was mediated. They have put forth a hypothesis that involves either (a) defective recognition or processing of pheromonal signals or (b) defective mechanics of suckling itself. Also, newborn mice that lack CNTFR are unable to feed; in these mice, impaired jaw movements have been observed.
Both Norwegian patients (but not the Israeli patients) had severe feeding problems as newborns, requiring hospitalization and nasogastric feeding. As children, the brothers continued to show no interest in food. They made many excuses to avoid eating and lagged behind in their growth and development. They both have restricted jaw movements, making dental work difficult, but physical restraint is not a major reason for them not to eat. Interestingly, injections of the related CNTF can cause weight loss in animals and humans, likely to work via a leptinlike pathway on appetite centers in the hypothalamus (Lambert et al. 2001). Thus, it is tempting to speculate that the potentially lethal lack of appetite exerted by a knockout mutation in the CRLF1 gene may also, in some way, be mediated through malfunction of appetite-regulation centers.
One possibility is that there is normally a physiological competitive binding of CNTF and CRLF1 to their common receptor, CNTFR, at various stages in development (Elson et al. 2000). In the Norwegian patients with a CRLF1-knockout mutation, the postulated normal balance between the two ligands competing for the same receptor could be impaired. Since no CRLF1 is produced, their common receptor may be stimulated solely by CNTF, exerting a potentially lethal appetite-depressive effect. Interestingly, leptin has recently been shown to act as a skeletal growth factor, with a direct peripheral effect on the mouse mandibular growth center through a mechanism that is as yet unknown (Maor et al. 2002).
Response to cold is a complex interplay of ion channels in both cold-sensitive and cold-insensitive neurons (Viana et al. 2002). Information on gentle cooling is transmitted by a small subpopulation of sensory nerves, whereas others transmit information on noxious cold and pain. Small changes in the balance of channel expression or in the properties of cold-insensitive neurons may transform cold-insensitive neurons into cold-sensitive fibers (McMemy et al. 2002; Peier et al. 2002; Viana et al. 2002). In all four patients with CISS, the parts of the body surface that sweat profusely at cold temperatures were completely dry under circumstances that normally induce sweating (e.g., hot weather, strenuous exercise, and fever). Thus, the sweat glands in the implicated parts of the body remain under neural control but react inversely to environmental temperatures. The Norwegian patients have impaired peripheral sensitivity to pain and temperature, including the direct exposure to subfreezing cold and steaming heat. Thus, further studies of patients with the CRLF1-deficient phenotype may yield information on complex neuronal processing and the interrelationship between various sensory stimuli.
Acknowledgments
We thank V. Marton, for performing clinical examinations; J. Kråkenes and K. Rosendal, for reviewing x-ray films; A. Berg, for reviewing pediatric records; and I. Tjelflaat and G. Matre, for technical assistance. Y. Shinar and E. Rabinovitz (Sheba Medical Center, Tel Hashomer) kindly donated DNA samples from ethnically matched control individuals. L. K. Ashworth (Lawrence Livermore National Laboratories) provided helpful assistance in early phases, when this region on chromosome 19 was insufficiently mapped. This study was supported in part by National Human Genome Research Institute grant HG00008 (to J.M. and J.O.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshfieldclinic.org/research/genetics/
- Cooperative Human Linkage Center, The, http://gai.nci.nih.gov/CHLC/
- Digitalarkivet, http://digitalarkivet.uib.no/cgi-win/WebFront.exe?slag=vis&tekst=meldingar
- Entrez Genome, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi? (for NCBI Map Viewer, build 30)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CRLF1 cDNA [accession number NM_004750.2])
- GenLink, http://www.genlink.wustl.edu/
- Genome Database, The, http://www.gdb.org/
- LLNL Human Genome Center, http://greengenes.llnl.gov/genome/genome.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CISS [MIM 272430])
References
- Alexander WS, Rakar S, Robb L, Farley A, Willson TA, Zhang JG, Hartley L, Kikuchi Y, Kojima T, Nomura H, Hasegawa M, Maeda M, Fabri L, Jachno K, Nash A, Metcalf D, Nicola NA, Hilton DJ (1999) Suckling defect in mice lacking the soluble haemopoietin receptor NR6. Curr Biol 9:605–608 [DOI] [PubMed] [Google Scholar]
- Bonfield JK, Rada C, Staden R (1998) Automated detection of point mutations using fluorescent sequencing trace subtraction. Nucleic Acids Res 26:3404–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JR (1948) Outline for the study of scoliosis. American Academy of Orthopedic Surgeons Instructional Course Lectures 5:261–265 [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Davis S, Aldrich TH, Valenzuela DM, Wong VV, Furth ME, Squinto SP, Yancopoulos GD (1991) The receptor for ciliary neurotrophic factor. Science 253:59–63 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Vejsada R, Poueymirou WT, Acheson A, Suri C, Conover JC, Friedman B, McClain J, Pan L, Stahl N (1995) Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell 83:313–322 [DOI] [PubMed] [Google Scholar]
- Durham LK, Feingold E (1997) Genome scanning for segments shared identical by descent among distant relatives in isolated populations. Am J Hum Genet 61:830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson GC, Graber P, Losberger C, Herren S, Gretener D, Menoud LN, Wells TN, Kosco-Vilbois MH, Gauchat JF (1998) Cytokine-like factor-1, a novel soluble protein, shares homology with members of the cytokine type I receptor family. J Immunol 161:1371–1379 [PubMed] [Google Scholar]
- Elson GC, Lelievre E, Guillet C, Chevalier S, Plun-Favreau H, Froger J, Suard I, de Coignac AB, Delneste Y, Bonnefoy JY, Gauchat JF, Gascan H (2000) CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat Neurosci 3:867–872 [DOI] [PubMed] [Google Scholar]
- Gedde-Dahl T Jr (1973) Population structure in Norway: inbreeding, distance and kinship. Hereditas 73:211–232 [DOI] [PubMed] [Google Scholar]
- Lambert PD, Anderson KD, Sleeman MW, Wong V, Tan J, Hijarunguru A, Corcoran TL, Murray JD, Thabet KE, Yancopoulos GD, Wiegand SJ (2001) Ciliary neurotrophic factor activates leptin-like pathways and reduces body fat, without cachexia or rebound weight gain, even in leptin-resistant obesity. Proc Natl Acad Sci USA 98:4652–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser SS, Lo DC (2000) CNTF II, I presume. Nat Neurosci 3:851–852 [DOI] [PubMed] [Google Scholar]
- Maor G, Rochwerger M, Segev Y, Phillip M (2002) Leptin acts as a growth factor on the chondrocytes of skeletal growth centers. J Bone Miner Res 17:1034–1043 [DOI] [PubMed] [Google Scholar]
- McMemy DD, Neuhausser WM, Julius D (2002) Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature 416:52–58 [DOI] [PubMed] [Google Scholar]
- Peier AM, Moqrich A, Hergarden AC, Reeve AJ, Andersson DA, Story GM, Earley TJ, Dragoni I, McIntyre P, Bevan S, Patapoutian A (2002) A TRP channel that sense cold stimuli and menthol. Cell 108:705–715 [DOI] [PubMed] [Google Scholar]
- Sohar E, Shoenfeld Y, Udassin R, Magazanik A, Revach M (1978) Cold-induced profuse sweating on back and chest: a new genetic entity? Lancet 2:1073–1074 [DOI] [PubMed] [Google Scholar]
- Stockli KA, Lillien LE, Naher-Noe M, Breitfeld G, Hughes RA, Raff MC, Thoenen H, Sendtner M (1991) Regional distribution, developmental changes, and cellular localization of CNTF-mRNA and protein in the rat brain. J Cell Biol 115:447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Yokoji H, Misawa H, Hayashi M, Hu J, Deguchi T (1994) A null mutation in the human CNTF gene is not causally related to neurological diseases. Nat Genet 7:79–84 [DOI] [PubMed] [Google Scholar]
- Viana F, de la Pena E, Belmonte C (2002) Specificity of cold thermotransduction is determined by differential ionic channel expression. Nat Neurosci 5:254–260 [DOI] [PubMed] [Google Scholar]