Abstract
Bardet-Biedl syndrome (BBS) is a genetic disorder with the primary features of obesity, pigmentary retinopathy, polydactyly, renal malformations, mental retardation, and hypogenitalism. Patients with BBS are also at increased risk for diabetes mellitus, hypertension, and congenital heart disease. BBS is known to map to at least six loci: 11q13 (BBS1), 16q21 (BBS2), 3p13-p12 (BBS3), 15q22.3-q23 (BBS4), 2q31 (BBS5), and 20p12 (BBS6). Although these loci were all mapped on the basis of an autosomal recessive mode of inheritance, it has recently been suggested—on the basis of mutation analysis of the identified BBS2, BBS4, and BBS6 genes—that BBS displays a complex mode of inheritance in which, in some families, three mutations at two loci are necessary to manifest the disease phenotype. We recently identified BBS1, the gene most commonly involved in Bardet-Biedl syndrome. The identification of this gene allows for further evaluation of complex inheritance. In the present study we evaluate the involvement of the BBS1 gene in a cohort of 129 probands with BBS and report 10 novel BBS1 mutations. We demonstrate that a common BBS1 missense mutation accounts for ∼80% of all BBS1 mutations and is found on a similar genetic background across populations. We show that the BBS1 gene is highly conserved between mice and humans. Finally, we demonstrate that BBS1 is inherited in an autosomal recessive manner and is rarely, if ever, involved in complex inheritance.
Introduction
Bardet-Biedl syndrome (BBS [MIM 209900]) is a genetically heterogeneous disorder mapping to six known loci on chromosomes 11 (BBS1) (Leppert et al. 1994), 16 (BBS2) (Kwitek-Black et al. 1993), 3 (BBS3) (Sheffield et al. 1994), 15 (BBS4) (Carmi et al. 1995), 2 (BBS5) (Young et al. 1999), and 20 (BBS6) (Katsanis et al. 2000). The phenotype consists of central obesity, retinopathy, postaxial polydactyly, hypogonadism, renal anomalies, and mental retardation or developmental delay (Green et al. 1989; Beales et al. 1999). Other components of the phenotype include diabetes mellitus, hypertension, and congenital heart disease (Harnett et al. 1988; Elbedour et al. 1994). There has been substantial interest in understanding the molecular basis and biochemical pathways involved in Bardet-Biedl syndrome, because some components of the phenotype are common. Four genes involved in BBS have now been identified. Two groups working independently identified mutations in patients with BBS, in a gene previously shown to cause McKusick-Kaufman syndrome (the MKKS gene, now also known as “BBS6”) (Katsanis et al. 2000; Slavotinek et al. 2000). We used positional cloning to identify the genes causing BBS2 (Nishimura et al. 2001), BBS4 (Mykytyn et al. 2001), and, most recently, BBS1 (Mykytyn et al. 2002). The MKKS protein product shows similarity to type II chaperonins, and the BBS4 protein product shares homology with the O-linked acetylglucosamine transferase family of genes. The BBS1 and BBS2 protein products show no similarity to any proteins with known function.
Katsanis et al. (2001) recently hypothesized that in some cases the development of BBS requires the presence of three disease-causing mutations (two mutations at one locus and a third mutation at a second locus), a form of complex inheritance termed “triallelic inheritance.” The triallelic hypothesis is based on screening of a cohort of 163 families with BBS for sequence variations in both BBS2 and BBS6. This resulted in (1) the identification of three apparent mutant alleles in four pedigrees; (2) the detection of an unaffected individual in each of two pedigrees who carry two BBS2 mutations, but not a BBS6 mutation; and (3) the presence of homozygosity of anonymous polymorphic markers at one BBS locus, in combination with the presence of a single mutant allele at a second locus in some small families. In a subsequent article, Katsanis et al. (2002) screened the same BBS cohort, along with some additional families, for mutations in the BBS4 gene. They extended the triallelic hypothesis to include the possibility of tetra-allelic inheritance, on the basis of the identification of a patient who is homozygous for both BBS2 and BBS4 missense variants.
We recently used positional cloning to identify the BBS1 gene (Mykytyn et al. 2002), which is the gene thought, on the basis of linkage studies, to be most commonly involved in BBS. BBS1 mutations were found in each of six extended pedigrees mapping to the BBS1 locus. Each of the six families had either a homozygous mutation (three families) or compound heterozygous mutations. In addition, a screen for BBS1 mutations in 60 unrelated probands confirmed that this gene is commonly involved in BBS, and the screen resulted in the identification of a common missense mutation (M390R) that is involved in approximately one-third of all BBS cases in this patient cohort. In addition, we presented preliminary data indicating that BBS1 mutations do not participate in triallelic inheritance. This conclusion was based on the sequencing of the BBS2, BBS4, and MKKS genes in six families with BBS1 and in 10 unrelated individuals who were homozygous for the BBS1 M390R mutation, without finding any additional mutations. The identification of four BBS genes, including the gene most commonly involved in Bardet-Biedl syndrome, provides the opportunity to thoroughly examine whether BBS is inherited in a complex fashion. In this article, we present results from the evaluation of a large BBS cohort and report that we find no evidence for complex inheritance involving the BBS1 locus. In addition, we report the identification of 10 novel BBS1 mutations.
Patients and Methods
Patients and Families
Signed informed consent was obtained from each patient and family member, using protocols approved by the institutional review board at the University of Iowa and collaborating institutions. The diagnosis of BBS was based on clinical examination, using diagnostic criteria that consist of the presence of at least three of the cardinal features of BBS (obesity, polydactyly, renal anomalies, retinopathy, hypogonadism, and mental retardation). Retinopathy was diagnosed mainly by ophthalmoscopy. Electroretinography was also performed in some patients. Nearly all patients were of northern European ancestry.
Mutational and Genetic Analysis
Mutation detection was performed by direct sequencing of PCR amplification products. Primer sequences are available upon request from the corresponding author. In some cases, coding sequences of BBS1 were screened by SSCP, followed by direct DNA sequencing. Amplicons for SSCP analysis were designed to be ∼200 bp. For SSCP, PCR products were separated on native gels (7 ml of 50% glycerol, 3.5 ml 5× TBE, 8.8 ml 37.5:1 acrylamide:bis, and 50.7 ml ddH2O) for 3–4 h in 0.5× TBE at room temperature, with the temperature controlled by a cooling fan. The bands were visualized by silver staining (Bassam et al. 1991). Abnormal variants were sequenced and compared with a control sample (CEPH sample 1331-01) to detect any changes from that of the normal sequence.
PCR products for sequencing were amplified in a 25-μl reaction volume and visualized on 1.2% agarose gels. The corresponding bands were excised and purified using the QIAquick gel extraction kit (Qiagen). Plasmid DNA (150 ng in 4.5 μl) or 4.5 μl of purified PCR product was used as template for sequencing reactions. For plasmid, 5 pmol of primer and 2 μl of terminator sequencing mix (Applied Biosystems) were added for a final reaction volume of 10 μl. For PCR products, 10 pmol of primer and 1 μl of terminator sequencing mix were added for a final reaction volume of 10 μl. Cycling conditions were as specified by the manufacturer. Plasmid sequencing reactions were precipitated in the presence of linear acrylamide and were resuspended in 2 μl of loading buffer. PCR product sequencing reactions were plate precipitated in the presence of glycogen and isopropanol. The reactions were analyzed on an ABI 3700 sequencer.
Genotyping
We performed PCR amplification for the analysis of STRPs, using 40 ng of genomic DNA in 8.4-μl reaction volumes containing 1.25 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.8]; 500 mM KCl; 15 mM MgCl2; 0.01% gelatin [w/v]; 200 μM each of dATP, dCTP, dGTP, and dTTP; 2.5 pmol of each primer; and 0.2 U of Taq polymerase [Bioline]). Samples were subjected to 35 cycles of 94°C (50°C, 52°C, 55°C, or 57°C, as required) for 30 s and 72°C for 30s. Amplification products were separated on 6% polyacrylamide gels containing 7.7 M urea at 60 W for ∼2 h. Gels were silver stained, as described above.
We obtained oligonucleotide primers for the STRPs as MapPairs (Research Genetics). The custom primers required for this study were designed using the Primer3 program and were synthesized commercially (Research Genetics or Integrated DNA Technologies). For markers that proved difficult to amplify using the standard Taq polymerase, we substituted an equal amount of AmpliTaq (Applied Biosystems), along with an initial incubation of the PCR mixture at 94°C for 10 min.
Identification of BBS1 Homologues
To identify the mouse ortholog of BBS1, we used the human BBS1 DNA sequence to search the mouse genome subdivision of the Celera sequence database. A 500-Mb contig containing the entire Bbs1 coding sequence was identified and downloaded. The coding exons were then assembled into a contig and were conceptually translated. Similarity scores were calculated using the BLOSUM62 amino acid similarity matrix. We identified homologous bovine, zebrafish, and honeybee sequences by searching the translated EST database containing sequences from organisms other than human or mouse. Sequences were aligned using the ClustalW and Multiple Alignment programs (Baylor College of Medicine) and were formatted using the Boxshade program (EMBnet).
Results
Mutation Analysis of BBS1
Linkage analysis studies have suggested that BBS1 is the most common BBS locus, accounting for one-third to one-half of all BBS cases (Bruford et al. 1997). We reported elsewhere that screening of the BBS1 gene in 60 unrelated probands with BBS by SSCP analysis identified 22 individuals who had at least one copy of the M390R mutation, with 16 of these individuals being homozygous for this variant (Mykytyn et al. 2002). We have now screened a total of 129 unrelated individuals with BBS (including 69 previously unreported probands) for the M390R mutation, using SSCP analysis and/or direct sequencing. Of the 129 probands, 39 have at least one copy of the M390R mutation, and 27 demonstrate homozygosity for M390R, indicating that this mutation is involved in 30% of all BBS cases in our cohort. We sequenced the entire BBS1 gene in those individuals who were heterozygous for the M390R mutation, and we identified a second BBS1 mutation in 10 of 12 cases (table 1). These mutations are nonsense and deletion mutations, with the exception of one missense mutation (L518P) that was not found in 192 northern European control chromosomes. SSCP analysis of the entire BBS1 gene in 60 patients revealed 2 patients with 2 non-M390R mutations (table 1). These data indicate that, in at least 32% (41/129) of the probands in this cohort, the BBS phenotype is caused by mutations in BBS1, and the M390R mutation accounts for ∼80% of BBS1 disease-associated alleles in this population.
Table 1.
Mutations in Patients with BBS[Note]
| Exon | DNA Change | Protein Change |
| 1 | c.(−3)_37del | M1? |
| 4 | c.339T→G | Y113X |
| 4 | c.342delGa | V114fsX150 |
| 8 | c.599_604del | I200_T201del |
| 10 | c.851delA | Y284fsX288 |
| 11 | c.1040delT | M347fsX373 |
| 12 | c.1130_1134delb | C377_F378delfsX412 |
| 13 | c.1318C→T | R440X |
| 15 | c.1514_1515del | L505fsX556 |
| 15 | c.1553T→Cc | L518P |
Note.— Unless indicated, all mutations were found in combination with M390R.
Detected in combination with the L518P mutation.
Detected in the homozygous state.
Detected in 3 patients and in 0/96 control subjects.
Evaluation of Complex Inheritance Involving BBS1
We reported elsewhere the sequencing all of the known BBS genes (BBS2, BBS4, and MKKS) in probands from our six families with BBS1 and in 10 unrelated probands homozygous for the BBS1 M390R mutation, to search for additional mutations (Mykytyn et al. 2002). We did not identify any additional mutations in any of these individuals. To further evaluate the involvement of BBS1 in complex inheritance, we have now sequenced the BBS2, BBS4, and MKKS genes in a total of 43 unrelated probands, each having two BBS1 mutations. Although a few sequence variants were detected (table 2), they are all likely to be non–disease-causing polymorphisms, because they result in a conservative amino acid substitution, do not segregate in a manner consistent with disease causation, and/or are found in control individuals.
Table 2.
Other Sequence Variations in Patients with BBS
| Gene | Variation |
| BBS2 | I123V |
| BBS2 | A504V |
| BBS4 | K46R |
| BBS4 | I70V |
| BBS4 | T354I |
| MKKS | A8T |
| MKKS | R517C |
| MKKS | G532V |
We previously identified three large Bedouin kindreds with multiple affected individuals, each of which mapped to a different BBS locus (BBS2, BBS3, and BBS4) (Kwitek-Black et al. 1993; Sheffield et al. 1994; Carmi et al. 1995). Analysis of the inheritance pattern in each of these large pedigrees indicates autosomal recessive inheritance. Mutation analysis of the family with BBS2 indicates that all 11 affected individuals are homozygous for the BBS2 mutation. In addition, none of the 26 unaffected first-degree relatives have two copies of the mutation. All 14 affected members of the kindred with BBS3 are homozygous for the chromosome 3 disease haplotype, and none of the 36 unaffected first-degree relatives are homozygous for the disease haplotype. The BBS3 gene has yet to be identified, so we cannot confirm the haplotype data by mutation analysis. All eight affected individuals of the Bedouin family with BBS4 are homozygous for the disease-associated haplotype, as well as for the disease mutation. One of the 18 unaffected first-degree relatives in this family (a parent of affected individuals) was homozygous for the disease-associated haplotype. However, sequencing of the BBS4 gene reveals that this individual is heterozygous for the disease-causing mutation. In the three families combined, 80 unaffected first-degree relatives of patients with BBS were analyzed for the kindred-specific mutation or disease-associated haplotype, without detecting any unaffected homozygous individuals.
To determine whether BBS1 could contribute a third mutant allele in the three large Bedouin families with BBS, we sequenced the BBS1 gene in an affected proband from each kindred. No BBS1 sequence variants were identified. Finally, in six multiplex families in which affected individuals had two BBS1 mutations, none of 28 unaffected first-degree relatives were homozygous or compound heterozygous for BBS1 mutations, indicating complete disease penetrance.
Haplotype Analysis of the M390R Mutation
The identification of a single mutant BBS1 allele (M390R) involved in >30% of all BBS cases and 80% of all BBS1 disease-associated alleles, led us to ask whether this mutation was a recurrent one or whether these patients had inherited it from a common ancestor. We developed STRPs from an ∼450-kb region surrounding the BBS1 gene. Each genetic marker was genotyped in 14 unrelated control individuals of northern European descent to determine the approximate heterozygosity. Seventeen unrelated patients with BBS (also northern European) who were homozygous for the M390R mutation were also genotyped with these genetic markers. A marked reduction in the level of heterozygosity was observed in the BBS group when compared with the group of randomly selected individuals (table 3). This result suggests that the M390R mutation might be a relatively ancient mutation that arose on a single haplotype and is identical by descent in our patient population.
Table 3.
Heterozygosity of Markers Surrounding BBS1[Note]
|
Frequency ofM390R-AssociatedAllele |
Heterozygosity |
|||||
| Marker | Distance(kb) | No. ofObservedAlleles | Control | Patient | Control | Patient |
| 001107-AAT | 4 | .31 | .84 | .69 | .29 | |
| 001107-TAGA | 87.9 | 4 | .43 | .96 | .59 | .06 |
| 001107-TGA | 71.5 | 4 | .58 | 1.00 | .59 | .00 |
| 001107-TTTA | 2.5 | 2 | .81 | 1.00 | .31 | .00 |
| 001107-ATTT | 17.4 | 4 | .32 | .94 | .65 | .12 |
| 001107-CTATT | 56.3 | 7 | .19 | .66 | .74 | .47 |
| 001107-CTTTT | 32.5 | 4 | .43 | 1.00 | .56 | .00 |
| 019220-TAAAA | 52.0 | 4 | .25 | .97 | .59 | .06 |
| 019220-AG | 14.9 | 5 | .32 | .91 | .67 | .18 |
| 019220-AAT | 56.7 | 4 | .54 | .88 | .60 | .24 |
| 019220-TG | 54.8 | 4 | .75 | 1.00 | .40 | .00 |
| Average | .45 | .92 | .58 | .13 | ||
Note.— BBS1 is located between 001107-CTTTT and 019220-TAAAA.
When haplotypes were constructed for the 17 patients with BBS1 who demonstrated homozygosity for the M390R mutation and 7 patients heterozygous for the M390R mutation, nearly all of the patients were observed to have inherited a common haplotype (fig. 1A). A few differences were detected within the conserved haplotype that might have arisen because of a new mutation at the STRP locus. Only 3 of the 41 M390R-associated haplotypes that were examined differed at more than one marker within the region surrounding the BBS1 gene. This finding further indicates that the M390R mutation is an ancient mutation and that patients with this mutation have inherited it from a common ancestor.
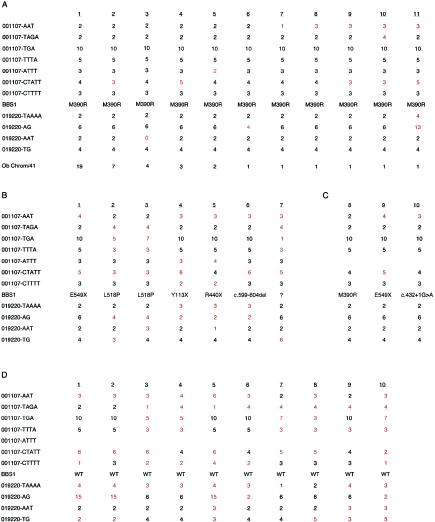
Figure 1.
Haplotype analysis of the BBS1 locus. A, Summary of the haplotype analysis of 41 chromosomes carrying the M390R mutation. The number of times each haplotype was observed is shown below each haplotype. Differences from the conserved haplotype are indicated in red. Of the 41 chromosomes, 19 correspond to a single haplotype, and 38/41 chromosomes differ at no more than one marker. B, Haplotype analysis of seven chromosomes carrying a non-M390R mutation in individuals of northern European descent. Differences from the M390R-associated haplotype are indicated in red. C, Haplotype analysis of the three disease-associated alleles identified in the Puerto Rican population. Differences from the M390R-associated haplotype are indicated in red. D, Ten non–disease-associated haplotypes from Puerto Rican BBS1 heterozygous individuals. Differences from the M390R-associated haplotype are indicated in red. WT = wild type.
We then examined the haplotypes of the chromosomes carrying a non-M390R mutation in the seven patients with BBS who had inherited the M390R mutation in the heterozygous state. The second mutation in these patients included the following: Y113X, R440X, L518P (two patients), E549X, c.599-604del, and one unknown. The haplotypes of the chromosomes not carrying M390R were found to exhibit extensive variation from each other and from the M390R haplotype (fig. 1B).
We previously used five multiplex Puerto Rican families to aid in the positional cloning of the BBS1 gene. In one family, the affected members were homozygous for the M390R mutation; in one family, they were homozygous for the E549X mutation; in two families, they were compound heterozygotes for the M390R mutation and the E549X mutation; and, in one family, they were compound heterozygotes for the E549X mutation and a c.432+1G→A splice site mutation (Mykytyn et al. 2002). The M390R mutation in these families is found on the same genetic background as in the northern European population. Interestingly, when haplotypes were constructed, it was observed that all three Puerto Rican mutations (M390R, E549X, and c.432+1G→A) appear to have occurred on a common genetic background (fig. 1C). Only the marker 001107-CTATT near the BBS1 gene was found to exhibit a difference among the three mutations, and that difference was a single repeat unit difference. The difference of a single repeat unit can be explained by the occurrence of a new mutation event at this locus on the common haplotype.
Since all three mutations in the Puerto Rican patients with BBS appeared to have occurred on a similar haplotype, we wished to determine whether or not this particular haplotype was prevalent in this population. A total of 10 unrelated carriers were available for study from the Puerto Rican families with BBS. The disease-associated haplotype could be determined for all 10 samples. Only 1 of 10 non–disease-associated haplotypes displayed similarity to the disease-associated haplotype in this population (fig. 1D). Thus, it appears that the disease-associated haplotype is not commonly found in this population.
Evolutionary Conservation of BBS1
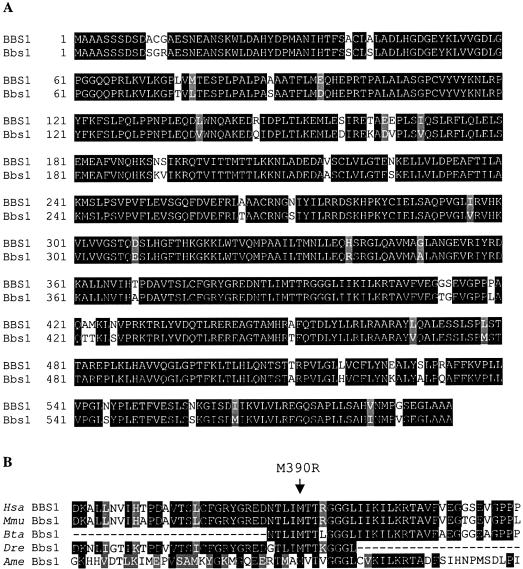
We were interested in determining the conservation of BBS1 across species. We were able to determine the mouse sequence for BBS1 by sequence alignment of the human cDNA sequence against the Celera mouse genome database. This procedure identified a 500-Mb contig containing the entire coding sequence of mouse Bbs1. We were then able to assemble the individual mouse exons to generate the full-length coding sequence. Translating the mouse cDNA yields a sequence of 593 amino acids, which is also the length of human BBS1. Comparing the human and mouse protein sequence reveals that they are 92% identical and 96% similar (fig. 2A).
Figure 2.
Evolutionary conservation of BBS1. A, Alignment of human BBS1 amino acid sequence with mouse Bbs1. Identical amino acids are shaded in black and conserved amino acids are shaded in gray. B, Alignment of human (Hsa) BBS1 peptide sequence and mouse (Mmu), bovine (Bta), zebrafish (Dre), and honeybee (Ame) peptide sequences surrounding the methionine at position 390 (indicated by an arrow).
To ask how well conserved the methionine at position 390 is across more-diverse species, we performed a BLAST search with the BBS1 amino acid sequence against a translated EST database containing sequences from organisms other than human or mouse. These analyses demonstrate that the methionine at position 390 is conserved across many species, including mouse, cow, zebrafish, and honeybee (fig. 2B), providing additional support that M390R is a disease-causing mutation. BLAST analysis indicates the leucine at position 518 is conserved in rat, although in mouse there is a nonconservative substitution of histidine (fig. 2A). The substitution of a proline seen in three patients likely has a deleterious effect on the structure of the protein.
Discussion
The recent identification of the BBS1 gene provides the opportunity to further evaluate the hypothesis that BBS displays complex inheritance. Examination of this hypothesis could facilitate the understanding of complex inheritance in other disorders, as well as aid in the understanding of the biochemical pathways involved in BBS-associated phenotypes. In addition, determination of the frequency of complex inheritance of BBS is requisite in providing proper recurrence-risk estimates to family members at risk of BBS . The complex inheritance hypothesis predicts each of the following: (1) When examining a gene commonly involved in BBS, one should be able to identify a subset of affected individuals that have only a single mutant allele in that gene. (2) In individuals with two mutations in one gene, one should be able to identify an additional mutation in one of the other BBS genes. (3) Some first-degree relatives with two mutant alleles at one locus should be clinically unaffected, because their genotype at the second locus is normal.
With respect to each of the above criteria, we find no evidence for complex inheritance in our study. First, we have identified 41 families too small for individual linkage analysis, in which the affected patients have at least one BBS1 mutation. In all but two cases, sequencing of the coding region and flanking splice sites identified the second mutant allele. In the two instances in which we identified only a single mutant BBS1 allele, the patients did not have mutations in the BBS2, BBS4, or MKKS genes. These data are consistent with our previous study in which the sequencing of the BBS1 gene in 12 families that contained two BBS2, BBS4, or MKKS mutations failed to identify BBS1 mutations (Mykytyn et al. 2002). These data lead to the conclusion that a mutation in BBS1 does not contribute to complex inheritance involving the other BBS loci.
Second, complete sequencing of the coding sequence and flanking spice sites in the BBS2, BBS4, and MKKS genes in 43 unrelated patients with BBS with two mutant BBS1 alleles failed to identify additional mutant alleles. These data indicate that homozygous or compound heterozygous BBS1 mutations are sufficient for disease penetrance.
Third, in the examination of 80 unaffected first-degree relatives of patients with BBS in three large kindreds, we found no evidence of two mutant alleles (i.e., no evidence of reduced penetrance). Furthermore, no evidence of reduced penetrance was found in numerous parents and unaffected siblings of patients from small kindreds with BBS1. These combined data make it unlikely that complex inheritance is commonly involved in BBS, and they indicate that, in the vast majority of cases, BBS is inherited in an autosomal recessive pattern. We conclude that families who have two mutations in BBS1 should be assigned recurrence risks according to a traditional autosomal recessive Mendelian inheritance model.
Our data confirm that BBS1 is the most common BBS locus and that a single mutation accounts for the majority of BBS1 cases. Our method of mutation screening was to screen all patients first for the M390R mutation. In those patients who had only one copy of the M390R variant, the full coding region and consensus splice sites were sequenced. Although this approach may not detect all mutations, the distribution of M390R mutations among homozygotes and heterozyotes indicates that we are detecting most BBS1 mutations in our cohort by this approach. This is evident by the fact that Hardy-Weinberg equilibrium is present in those patients with at least one copy of the M390R mutations (p=0.8 based on observed homozygote frequency of p2=0.64, with expected heterozygote frequency of 2pq=0.32 and observed heterozygote frequency of 2pq=0.29, where p is the allele frequency of M390R and q is the allele frequency of all non-M390R mutations).
We also report haplotype analysis of the common BBS1 M390R mutation. Our data suggest that the M390R mutation is an ancient mutation, because it resides on a single haplotype both within and across populations. Interestingly, when we examine the mutation-associated haplotypes of our Puerto Rican patients, we find that they all share a similar haplotype, regardless of the mutation. It is unclear why multiple mutations would arise on a similar haplotype. It is possible that the disease haplotype was common in the Puerto Rican population when the mutations first arose or that the disease-associated haplotype is more susceptible to mutation.
A wide range of inter- and intrafamilial clinical variation in families with BBS has been documented (Riise et al. 1997, 2002). It is likely, on the basis of these clinical observations, that there are other genes that modify the effects of mutations in BBS genes. These modifier genes may be the BBS genes themselves or other genes that code for proteins that interact with the BBS proteins directly or indirectly. The present study, in which we did not find evidence to support complex inheritance, does not exclude the possibility that sequence alterations or expression differences at one BBS locus might influence the phenotype caused by recessive mutations at another BBS locus. The study of such gene interactions will be greatly facilitated by the development of BBS animal models. The recent identification of four BBS genes will enable the development of these models.
Acknowledgments
We are grateful to the patients and their families for their participation in this study. We thank Jean Andorf for technical assistance and Denise Aguiar Crouch for administrative assistance. This work was supported by National Institutes of Health grants P50-HL-55006 (to V.C.S.), R01-EY-11298 (to E.M.S. and V.C.S.), and NCRR-RCMI 2G12RR03050 (to A.S.C.) and by the following organizations: Foundation Fighting Blindness (support to S.G.J., E.M.S. and V.C.S.), Carver Endowment for Molecular Ophthalmology (support to E.M.S. and V.C.S.), Research to Prevent Blindness, New York (support for Department of Ophthalmology, University of Iowa, and Department of Ophthalmology, University of Tennessee-Memphis, and a Senior Scientific Investigator Award [to S.G.J.]). V.C.S. is an associate investigator of the Howard Hughes Medical Institute, and E.M.S. is an investigator of the Howard Hughes Medical Institute.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Baylor College of Medicine Search Launcher, http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html (for ClustalW 1.8 and Multiple Alignment Programs)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Boxshade 3.21, http://www.ch.embnet.org/software/BOX_form.html (for printing of multiple-alignment files)
- GenBank, http://www.ncbi.nih.gov/Genbank, (for BBS1 cDNA [accession number AF503941] BBS2 cDNA [accession number NM_031885], BBS4 cDNA [accession number NM_033028], MKKS (BBS6) cDNA [accession number NM_018848])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBS [MIM 209900]) [PubMed]
References
- Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83 [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA (1999) New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 36:437–446 [PMC free article] [PubMed] [Google Scholar]
- Bruford EA, Riise R, Teague PW, Porter K, Thomson KL, Moore AT, Jay M, Warburg M, Schinzel A, Tommerup N, Tornqvist K, Rosenberg T, Patton M, Mansfield DC, Wright AF (1997) Linkage mapping in 29 Bardet-Biedl syndrome families confirms loci in chromosomal regions 11q13, 15q22.3-q23, and 16q21. Genomics 41:93–99 [DOI] [PubMed] [Google Scholar]
- Carmi R, Rokhlina T, Kwitek-Black AE, Elbedour K, Nishimura D, Stone EM, Sheffield VC (1995) Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Hum Mol Genet 4:9–13 [DOI] [PubMed] [Google Scholar]
- Elbedour K, Zucker N, Zalzstein E, Barki Y, Carmi R (1994) Cardiac abnormalities in the Bardet-Biedl syndrome: echocardiographic studies of 22 patients. Am J Med Genet 52:164–169 [DOI] [PubMed] [Google Scholar]
- Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O’Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet-Biedl syndrome, a form of Lawrence-Moon-Biedl syndrome. N Engl J Med 321:1002–1009 [DOI] [PubMed] [Google Scholar]
- Harnett JD, Green JS, Cramer BC, Johnson G, Chafe L, McManamon P, Farid NR, Pryse-Phillips W, Parfrey PS (1988) The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N Engl J Med 319:615–618 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR (2001) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293:2256–2259 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, Scambler PJ, Beales PL, Lupski JR (2002) BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet 71:22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwitek-Black, AE, Carmi R, Duyk GM, Buetow KH, Elbedour K, Parvari R, Yandava CN, et al (1993) Linkage of Bardet-Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 5:392–396 [DOI] [PubMed] [Google Scholar]
- Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA (1994) Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet 7:108–112 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen H, Beck J, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Lüleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31:435–438 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 [DOI] [PubMed] [Google Scholar]
- Riise R, Andreasson S, Borgastrom MK, Wright AF, Tommerup N, Rosenberg T, Tornqvist K (1997) Intrafamilial variation of the phenotype in Bardet-Biedl syndrome. Br J Ophthalmol 81:378–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riise R, Tornqvist K, Wright AF, Mykytyn K, Sheffield VC (2002) The phenotype in Norwegian patients with Bardet-Biedl syndrome with mutations in the BBS4 gene. Arch Ophthalmol 120:1364–1367 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Carmi R, Kwitek-Black A, Rokhlina T, Nishimura D, Duyk GM, Elbedour K, Sunden SL, Stone EM (1994) Identification of a Bardet-Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 3:1331–1335 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 [DOI] [PubMed] [Google Scholar]
- Young TL, Penney L, Woods MO, Parfrey PS, Green JS, Hefferton D, Davidson WS (1999) A fifth locus for Bardet-Biedl syndrome maps to 2q31. Am J Hum Genet 64:900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]