Abstract
In plants, changes in local auxin concentrations can trigger a range of developmental processes as distinct tissues respond differently to the same auxin stimulus. However, little is known about how auxin is interpreted by individual cell types. We performed a transcriptomic analysis of responses to auxin within four distinct tissues of the Arabidopsis thaliana root and demonstrate that different cell types show competence for discrete responses. The majority of auxin-responsive genes displayed a spatial bias in their induction or repression. The novel data set was used to examine how auxin influences tissue-specific transcriptional regulation of cell-identity markers. Additionally, the data were used in combination with spatial expression maps of the root to plot a transcriptomic auxin-response gradient across the apical and basal meristem. The readout revealed a strong correlation for thousands of genes between the relative response to auxin and expression along the longitudinal axis of the root. This data set and comparative analysis provide a transcriptome-level spatial breakdown of the response to auxin within an organ where this hormone mediates many aspects of development.
Keywords: Arabidopsis, development, root apical meristem, signaling gradient
Introduction
One of the main organizational cues in plant development is the signaling molecule auxin. A remarkable facet of auxin’s effect on plant development is the broad range of processes regulated by this simple compound (Vanneste and Friml, 2009). In meristems, auxin is a central modulator of growth and cellular differentiation (Bennett and Scheres, 2010). Auxin concentration gradients are maintained by active polar transport and have been proposed to give positional information that stages development and maturation in these growth centers (Friml et al, 2002; Benkova et al, 2003; Bhalerao and Bennett, 2003; Galinha et al, 2007; Ding and Friml, 2010; Dubrovsky et al, 2011).
Although auxin itself cannot be directly visualized, meristematic auxin gradients have been inferred from mass spectrometric measurement of tissue sections (in the Pinus sylvestris cambial meristem) and isolated cell types (of the Arabidopsis thaliana root apical meristem (RAM)) (Uggla et al, 1996; Petersson et al, 2009). A recently developed biosensor, DII-Venus (consisting of a fluorescent protein fusion of a labile component of the auxin perception and signaling machinery), has provided a new level of sensitivity in determining the distribution of auxin signaling activity in meristems (Vernoux et al, 2011). Readout of this sensor in the RAM suggests that there are cell type-specific aspects to auxin perception. In addition, it shows graded levels of auxin signaling intensity in the meristematic stele that are in line with a proximo-distal gradient of auxin itself (Supplementary Figure S1A; Brunoud et al, 2012). However, we lack an understanding of how cells interpret an auxin gradient in their broad transcriptional output (Overvoorde et al, 2010).
Localized auxin signaling output can be observed by visualizing the transcriptional response to auxin. DR5, a synthetic auxin-responsive promoter, driving a reporter gene (e.g., green fluorescent protein (GFP)) is often used as a proxy for the transcriptional auxin response (Supplementary Figure S1B; Ulmasov et al, 1997; Heisler et al, 2005). DR5 displays high expression in the tip of the RAM (specifically in the columella, QC and developing xylem), but its expression does not effectively match cell type-specific auxin measurements (Petersson et al, 2009) or fully complement DII-Venus levels in the RAM (Brunoud et al, 2012). Furthermore, the promoters of endogenous auxin-responsive genes, for example, SMALL AUXIN UP RNA (SAUR), AUXIN/INDOLE-3-ACETIC ACID INDUCED (Aux/IAA), BREVIS RADIX (BRX) or PLETHORA (PLT) genes, have been used to report the spatial influence of an auxin gradient on gene expression (Li et al, 1991; Galinha et al, 2007; Grieneisen et al, 2007; Santuari et al, 2011). However, these constructs give differing views of auxin-response distribution, with some showing an archetypal expression pattern similar to DR5 and others with a more graded expression in the proximal meristem. Hence, no single reporter provides a clear picture of how auxin gradients affect transcription throughout the root. Instead of singular auxin-induced reporters, a genome-wide assessment of auxin-responsive gene expression in relation to spatial expression could be used to visualize the hypothesized meristematic auxin-response gradient in silico. This global view can be used to assess the gradient’s influence on gene expression, both in the sense of its physical range and in the quantity of genes regulated. What is needed for such an analysis is a sensitive readout of the transcriptomic response to auxin in a particular tissue (e.g., the root) that can be superimposed on a spatial expression map of this tissue.
Another important issue in the study of auxin in plant development is how this simple molecule can elicit so many diverse responses in different cell types (Kieffer et al, 2010). Auxin distribution is dynamic and actively changes in response to environmental and developmental cues (Grunewald and Friml, 2010). Cells will encounter varying auxin levels throughout their lifespan and their response to auxin is determined by cellular context (i.e., cell identity and spatial domain). For instance, during the formation of lateral root primordia, an increase in auxin levels leads to cell proliferation specifically in distal xylem-pole (xp) pericycle cells (De Smet et al, 2008). In contrast, in the root epidermis, higher auxin levels do not induce cell division but rather inhibit cell expansion to mediate bending of the root tip during gravitropic growth (Swarup et al, 2005). Differences in the tissue-specific expression levels of the modular auxin perception and signal transduction machinery have been suggested to predispose cells to a particular response (Weijers et al, 2005; Kieffer et al, 2010; Rademacher et al, 2011; Vernoux et al, 2011; Hayashi, 2012) (Supplementary Figure S1C; Supplementary Table S1) and it is assumed that differences in the transcriptional response to auxin lie at the basis for many of the different observed physical responses. However, the importance of cellular context on the genome-wide transcriptional auxin response is undocumented. An assessment of the response to auxin at cellular resolution is needed to begin to sort out the influence of spatial context on the transcriptional auxin response.
The Arabidopsis seedling root apex is a highly amenable system for the examination of the role of auxin at a cellular resolution (Figure 1). The anatomical organization permits analysis of cell identity in the radial axis and developmental maturity in the longitudinal axis (Petricka and Benfey, 2008). Moreover, transcriptomic analyses of the individual cell types that make up this organ have provided a gene expression map of cell identities and high-resolution transcriptional data sets along the longitudinal developmental axis of the root tip (Birnbaum et al, 2003; Nawy et al, 2005; Lee et al, 2006; Levesque et al, 2006; Brady et al, 2007).
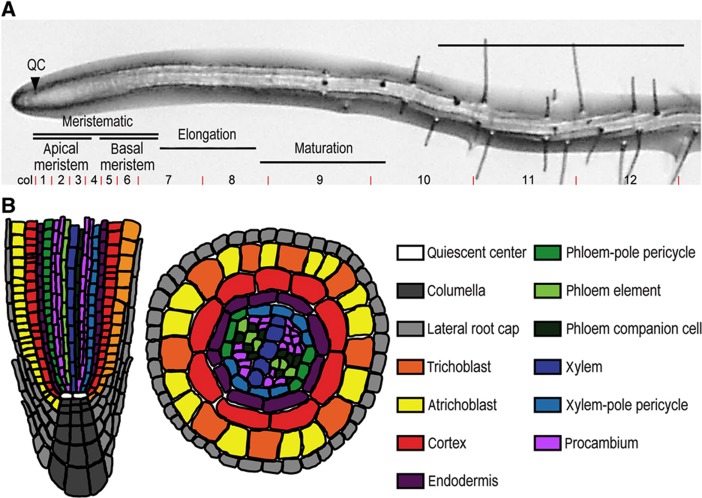
Figure 1.
The Arabidopsis thaliana root apex. (A) The apex of the seedling root can be divided into the meristematic zone (consisting of the apical and basal meristem), elongation zone and maturation zone; shown here in a 5-day post germination (dpg) seedling root tip. The longitudinal transcriptomic data sampling sections gathered by Birnbaum et al (2003) and Brady et al (2007) are indicated. QC, quiescent center, scale bar indicates 1 mm. (B) Schematic representation of the cell types in longitudinal and radial cross-sections of the Arabidopsis root apical meristem.
Here, we conduct a genome-wide, cell type-specific analysis of auxin-induced transcriptional changes in four distinct cell populations of the Arabidopsis root. This data set is used to (1) assess the relevance of cellular context on the transcriptional response to auxin and (2) test whether this comprehensive readout of auxin responses can delineate a genome-wide auxin-response gradient. The study uncovers both broad and tissue-specific auxin-responsive transcripts, and thus provides a resource to further examine the role of auxin in a cellular context and resolve how this important hormone guides plant development and growth. This sensitive readout of auxin responses together with the previous analysis of spatial gene expression in the root was used to generate, for the first time, a view of an inclusive auxin-response gradient in the RAM.
Results
Auxin-regulated gene-expression analysis in distinct cell types
To analyze the effect of auxin on separate spatial domains, transcriptional changes in response to auxin treatment were assayed by means of fluorescence activated cell sorting and microarray analysis of four distinct tissue-specific GFP-marker lines in Arabidopsis seedling roots. The assayed samples covered internal and external as well as proximal and distal cell populations; including marker lines for the stele, xp pericycle, epidermis and columella (Figure 2A). Roots were immersed in 5 μM indole-3-acetic acid (IAA) and treated for a total of 3 h (see Materials and methods). Expression of the markers used was stable within the treatment period (Supplementary Figure S2A). Analysis of the DII-Venus reporter under these treatment conditions showed that all tissues in the root responded to treatment within 30 min (Supplementary Figure S1A). For comparison, transcriptional responses to auxin were also assayed in intact (undigested) roots treated for 3 h.
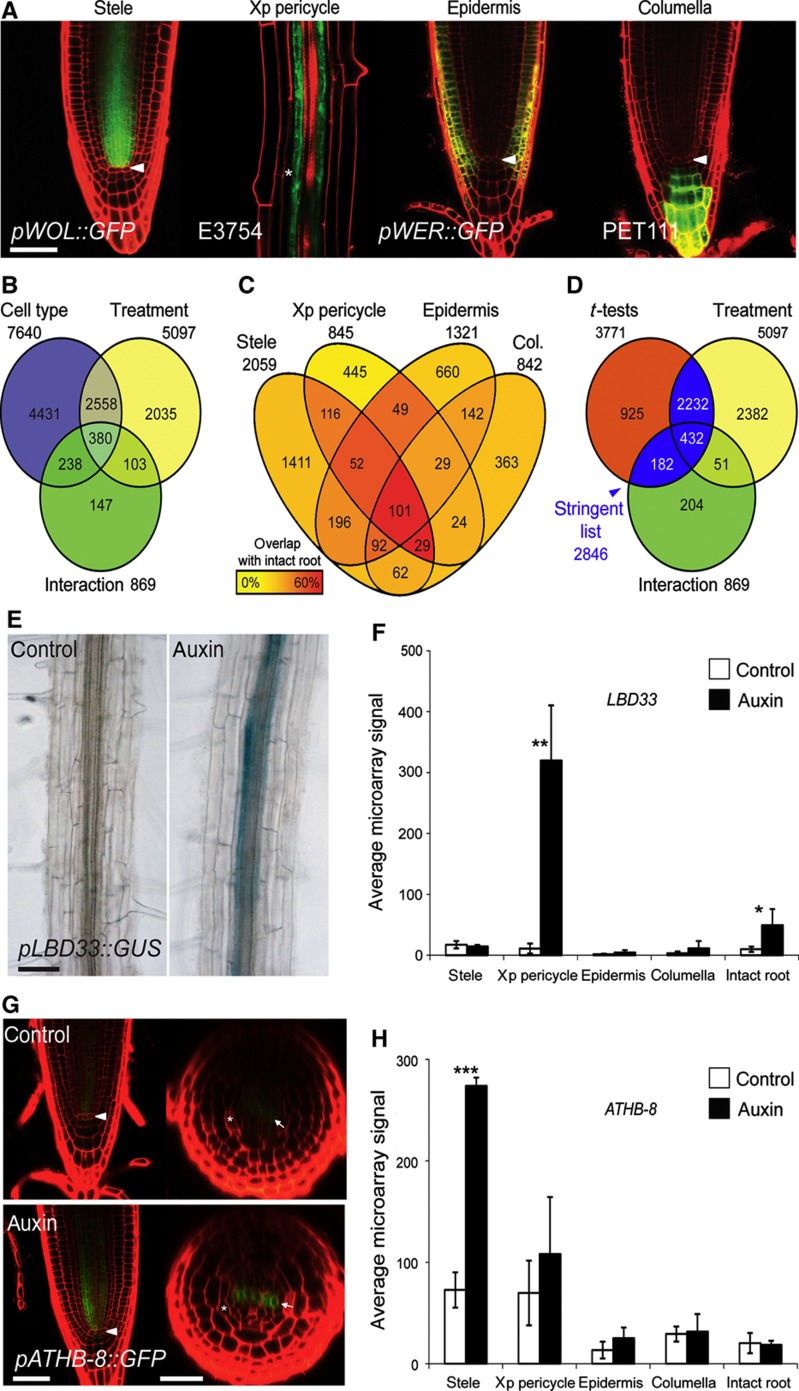
Figure 2.
Cell type-specific analysis of auxin responses. (A) Confocal micrographs of 5 dpg seedling roots of the four GFP-marker lines used for the fluorescence activated cell sorting of the stele (pWOL::GFP), xylem-pole pericycle (E3754), epidermis (and parts of the lateral root cap; pWER::GFP) and columella (PET111). Roots were incubated with propidium iodide to highlight cell boundaries (red); arrowheads indicate the QC, asterisk indicates the endodermis, scale bar indicates 50 μm. (B) A Venn diagram of the two-way ANOVA results showing the overlap between groups of genes found to be significant (P<0.01) for the factors cell type, treatment and interaction between cell type and treatment. (C) Venn diagram of the 3771 genes found to be significantly responding to auxin treatment in the four assayed tissues (t-test P<0.01, fold change >1.5). The yellow to red color code indicates percent overlap with auxin-responsive genes found in the analysis of intact roots. (D) Venn diagram of the overlap between lists of genes significantly regulated according to the ANOVA treatment or interaction factors (B) and the 3771 genes that pass any tissue-specific t-test (C). The blue-highlighted regions indicate genes in the ‘stringent’ list of 2846 auxin responders. (E) Micrograph of pLBD33::GUS reporter-gene line treated with auxin (5 μM IAA, 3 h), scale bar indicates 50 μm. (F) Histogram of microarray expression data for LBD33, showing significant induction specifically in the xylem-pole pericycle sample. Data are represented as mean±s.d.; n=3; t-test *P<0.05, **P<0.01. (G) Confocal analysis of pATHB-8::GFP reporter-gene line treated with auxin (1 μM 2,4-D, 16 h). Images were obtained with equal gain settings in the GFP channel. Arrowheads indicate the QC, arrows indicate the xylem pole, asterisks indicate endodermis and scale bars indicate 50 μm in longitudinal section and 25 μm in radial section. (H) Histogram of microarray expression data for ATHB-8, showing significant induction specifically in the stele sample. Data are represented as mean±s.d.; n=3, t-test ***P<0.001.
To establish that the tissue-specific expression profiles gathered here were consistent with the previously published root expression data (Birnbaum et al, 2003; Nawy et al, 2005; Lee et al, 2006; Levesque et al, 2006; Brady et al, 2007), we generated a list of cell type-specifically enriched (CTSE) genes using the public data and visualized their expression in our data set. This CTSE list was based on the expression profile template matching in a select data set of 13 non-overlapping, cell type-specific expression profiles of sorted GFP-marker lines (Supplementary Figure S2B; Supplementary Table S2). This procedure yielded a total of 3416 genes whose expression is enriched in one specific cell type (maturing xylem, developing xylem, xp pericycle, phloem-pole pericycle, phloem, phloem companion cell, quiescent center, endodermis, cortex, trichoblast, atrichoblast, lateral root cap or columella) or whose expression was enriched in two related cell types (xylem (developing and maturing xylem), pericycle (xp and phloem-pole pericycle), phloem (phloem and phloem companion cell), ground tissue (endodermis and cortex), epidermis (trichoblast and atrichoblast) or root cap (lateral root cap and columella)). The relative expression of the CTSE genes in the tissue-specific data generated in this study is differentially enriched in a manner that fits with the domains covered by the different markers used here (Supplementary Figure S2B). These results indicate a successful isolation of the transcriptomes of distinct cell types and show that the enrichment in specific tissues is consistent across the data sets.
For most auxin-responsive genes in our data set, transcript levels were affected in several cell types but often showed a relatively greater response to auxin in one or more of the tissues. Two separate criteria were used to define these different levels of response (see Materials and methods for a detailed description of the statistical analysis). First, a two-way analysis of variance (ANOVA) with the factors cell type and treatment was used to categorize auxin-regulated genes and the relation of responses between the different cell types. The ANOVA (P<0.01) yielded 7640 genes differentially expressed between the individual tissue samples; 5097 genes responded significantly to treatment across all tissues and formed a broad register of auxin-responsive genes in the root. In all, 869 genes showed a significant interaction between treatment and cell type, representing genes with the most dramatic spatial bias in regulation (Figure 2B; Supplementary Table S2). Second, Student’s t-tests were conducted on the individual tissue samples to classify the response within specific cell types (P<0.01; fold change>1.5). The number of significantly regulated genes in the stele, xp pericycle, epidermis and columella was 2059, 845, 1321 and 842, respectively (3771 unique genes; Figure 2C; Supplementary Table S2). In all, 1923 genes were found to be differentially regulated by auxin treatment in intact roots (t-test P<0.01, fold change>1.5; Supplementary Table S2). To generate a stringent list of auxin-responsive genes for the analysis of cell type-specific expression, we extracted the genes that passed the ANOVA for the treatment factor or interaction and also passed at least one of the four cell type-specific t-tests (Figure 2D), resulting in a total of 2846 auxin-responsive genes.
Measured auxin responses were corroborated at two levels. First, we observed the significant regulation of known auxin-responsive genes in the cell type-specific data set. This includes significant regulation of 22 members of the Aux/IAA family of auxin co-receptors (Calderon-Villalobos et al, 2010), 14 GH3 auxin conjugases (Hagen et al, 1991), 18 SAURs (Hagen and Guilfoyle, 2002) and 7 LATERAL ORGAN BOUNDARY DOMAIN CONTAINING PROTEIN (LBD) transcription factors (Shuai et al, 2002) (Supplementary Figure S2C–F; Supplementary Table S2). Several of the responsive LBD genes that are known to be involved in lateral root initiation (Okushima et al, 2007) displayed dramatic upregulation specifically in the xp pericycle, the tissue where lateral roots originate (Supplementary Figure S2E). These results indicate the robust induction of known auxin-responsive transcripts in the four cell types sampled in this work. Second, we confirmed that tissue-specific transcript level measurements matched auxin induction patterns in transcriptional reporter lines. This included xp pericycle-specific induction of pLBD33::GUS and pTMO6::GFP (TARGET OF MONOPTEROS 6 ) as well as stele-specific induction of pATHB-8::GFP (ARABIDOPSIS THALIANA HOMEOBOX GENE 8) and ubiquitous induction of pGH3.5::GFP and pIAA5::GUS (Figure 2E–H and Supplementary Figure S3) (Kang and Dengler, 2002; Lee et al, 2006; Okushima et al, 2007; Schlereth et al, 2010).
In a comparison of auxin responses in sorted cells and intact roots, genes that responded in a greater number of cell types (t-tests P<0.01, fold change>1.5) were more likely found responsive in intact roots (t-test P<0.01, fold change>1.5; Figure 2C). Moreover, genes previously associated with the gene-ontology (GO) term response to auxin stimulus are highly significantly overrepresented only in the group of 101 genes that respond in all 4 assayed tissues (20/101 genes; Fisher’s exact test P=3.51e−18). In Figure 2C, the heatmap overlaid on the Venn diagram shows the gain in sensitivity for detecting cell type-specific auxin responses compared with intact roots under the same treatment. Genes found to be regulated by auxin in only one tissue show a relatively small overlap with responses in the intact root (1411, 445, 600 and 363 genes in the stele, xp pericycle, epidermis and columella, respectively). Transcripts whose response was detected in higher numbers of tissues show a relatively larger overlap with those detected in the intact root. This suggests that many cell type-specific auxin responses may not be detected in analyses performed at the organ or organismal level because localized responses are diluted among otherwise non-responsive cells.
Functional analysis of cell type-specific auxin responses
Using the stringent list of (2846) auxin-responsive genes, expression patterns were ordered hierarchically by pairwise correlation. A heatmap of gene regulation patterns shows how almost all auxin-responsive genes exhibited some type of spatial bias in their regulation (Figure 3A). Although genes are most often regulated in the same direction (induced or repressed) in different cell types, the response is usually stronger in a subset of samples. These findings demonstrate a pervasive tissue-specific amplitude modulation of auxin responses, and suggest that most auxin-controlled genes have context-dependent aspects to their transcriptional regulation.
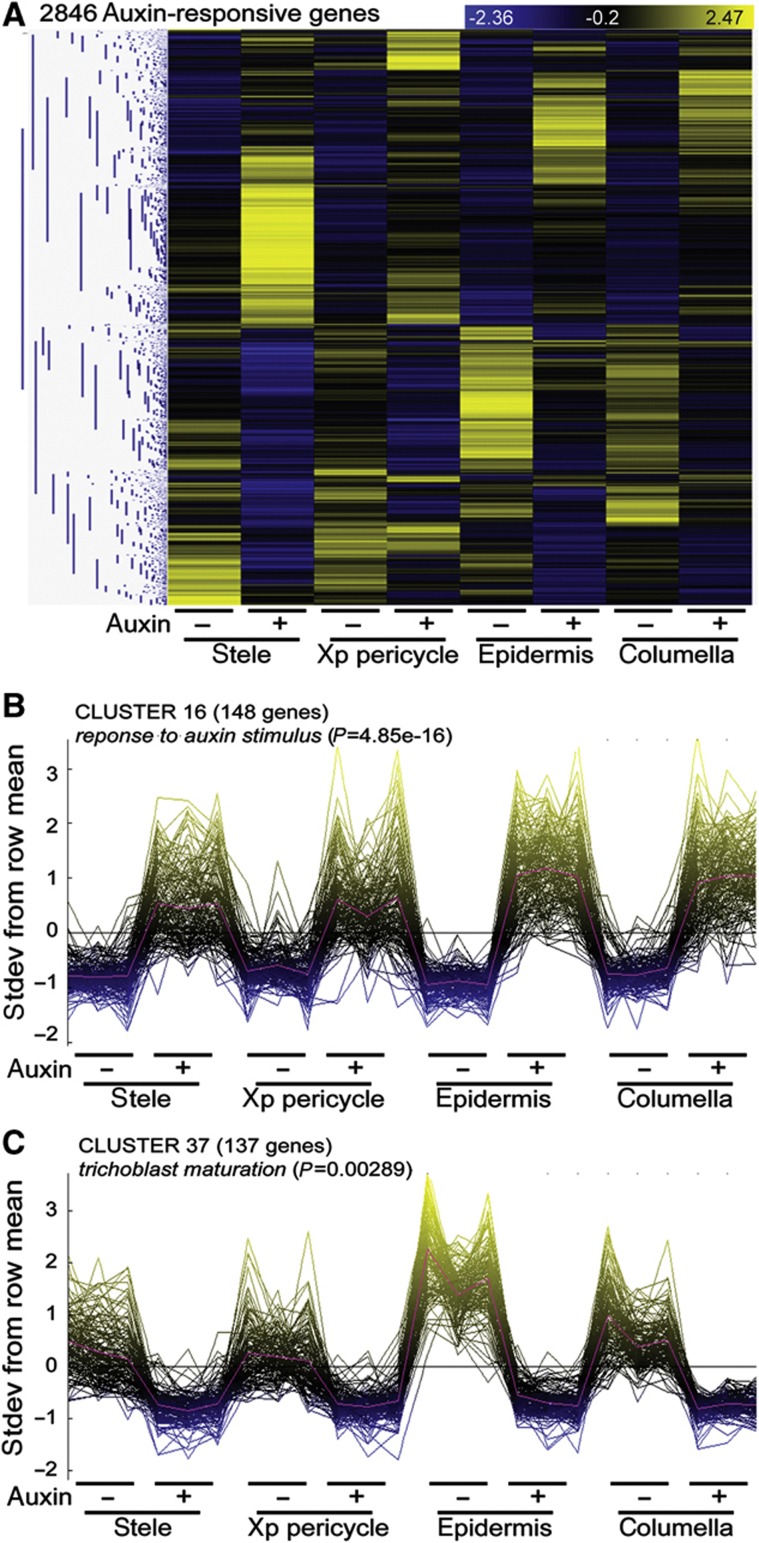
Figure 3.
Categorizing cell type-specific auxin responses. (A) Spatial auxin-response patterns arranged by hierarchical clustering (pairwise Pearson’s correlation). 2846 genes significantly regulated in the ANOVA for treatment or for the interaction between treatment and cell type (P<0.01) and in at least one t-test of the four separate tissues assayed (P<0.01, fold change>1.5). The heatmap consists of row-normalized gene expression in rows and cell type±treatment in columns; blue (low) to yellow (high) color code indicates standard deviations from the row mean. (B) Dominant expression pattern 16 contains 148 genes with relatively uniform upregulation after auxin treatment; the GO term response to auxin stimulus is overrepresented in this list of genes (corrected Fisher’s exact test). (C) Dominant expression pattern 37 contains 137 genes that show high expression in the epidermis and are repressed by auxin treatment; the GO term trichoblast maturation is overrepresented in this list of genes (see Supplementary Figure S4 and Supplementary Table S3).
To dissect spatially distinct auxin responses, dominant expression patterns were extracted and used to group genes with similar responses (Supplementary Figure S4A; Orlando et al, 2009). These response clusters showed a significant overrepresentation of diverse GO terms (Supplementary Table S3). Extending the trend noted above for genes significantly regulated in all tissues, genes previously associated with the response to auxin stimulus as well as auxin mediated signaling and auxin homeostasis were mainly overrepresented in clusters containing genes with relatively uniform upregulation of expression; these included 10 Aux/IAAs and 4 GH3s (Figure 3B; Supplementary Figure S4B clusters 15 and 16; Supplementary Table S3). Four IAAs and GH3.3 were included in a cluster that showed relatively stronger induction in the stele and the induction of GH3.6/DWARF IN LIGHT 1 was strongest in the columella. Two genes previously associated with the response to auxin, LATE ELONGATING HYPOCOTYL and an uncharacterized homeodomain transcription factor (At1g74840), were found in a cluster of genes with strong downregulation in the pericycle. PIN-FORMED 7, NO VEIN and ACAULIS 5 are linked to the auxin-transport GO term found to be overrepresented in a cluster with relatively strong induction in the stele and pericycle. Genes associated with auxin biosynthesis were overrepresented in a cluster of uniformly downregulated genes. These enrichments show that, although most genes previously associated with the auxin response display broad induction, there are cell type-specific expression biases to the transcriptional regulation by auxin among genes that influence its own perception, metabolism and transport.
Several auxin-response clusters representing a localized spatial pattern of induction or repression showed overrepresentation of functions linked to growth processes known to be regulated by auxin. For example, clusters of genes that showed epidermis-specific downregulation by auxin (e.g., cluster 37) had statistically overrepresented GO terms for trichoblast maturation (Figure 3C; Supplementary Figure S4; Supplementary Table S3). These clusters of genes potentially identify a large component of the transcriptome influenced by auxin signaling in the epidermis to regulate development or responses to environmental cues. Genes associated with cell wall modification and cytoskeleton modification as well as transmembrane transport and peroxidase activity were also overrepresented in this cluster, pointing to processes that may mediate auxin’s specific effects on the epidermis.
Promoter analysis of the cell type-specific auxin-response clusters was conducted to look for overrepresentation of the canonical auxin-response element TGTCTC (Liu et al, 1994). Clusters 15 and 16, which show relatively uniform upregulation of gene expression across tissues (Supplementary Figure S4A), contain significantly more genes with this element in the 500-bp upstream of their transcription start site than expected by chance (hypergeometric distribution analysis; Supplementary Table S3). Additionally, the occurrence of the generic TGTCNC and the individual -A-, -C- and -G- variants was examined, with the finding that TGTCAC, TGTCCC and TGTCNC were also overrepresented in the promoters of the uniformly upregulated genes assigned to dominant expression patterns 15 and 16. Furthermore, TGTCAC was overrepresented in the promoters of genes assigned to pattern 34, which shows downregulation in all tissues that is strongest in the stele (Supplementary Figure S4A). None of these elements were significantly enriched in any other upregulated or downregulated clusters. These results suggest that direct targets of auxin signaling through the auxin-response promoter element are generally uniformly induced across tissues of the root, and that variants of the canonical element may also participate in auxin regulation of transcript levels.
Auxin effects on transcriptional cell identity
To explore the influence of auxin on cellular development in the root in more depth, the CTSE sets of cell-identity markers (Supplementary Figure S2A; Supplementary Table S2) were used to analyze the effect of auxin on tissue-enriched genes in our cell type-specific data set. The overlap between the stringent list of 2846 auxin-responsive genes and the 3416-gene CTSE list enabled us to assess whether auxin had a positive or inhibitory overall effect on transcriptional cell identity. Among the overlapping set, genes enriched specifically in the quiescent center and developing xylem are upregulated by auxin at a significantly higher proportion than expected by chance, whereas genes enriched in maturing xylem, cortex and trichoblasts are downregulated more frequently than expected (χ2-test P<0.01; Supplementary Figure S5A; Supplementary Table S4). Furthermore, auxin-responsive tissue-enriched gene clusters show cell type-specific auxin sensitivity. For example, auxin-responsive genes enriched in the developing xylem are predominantly induced in the stele and the majority of auxin-responsive genes enriched in trichoblasts are repressed in the epidermis (as judged by relative expression levels as well as the tissue-specific t-tests; Figure 4A, Supplementary Figure S5B).
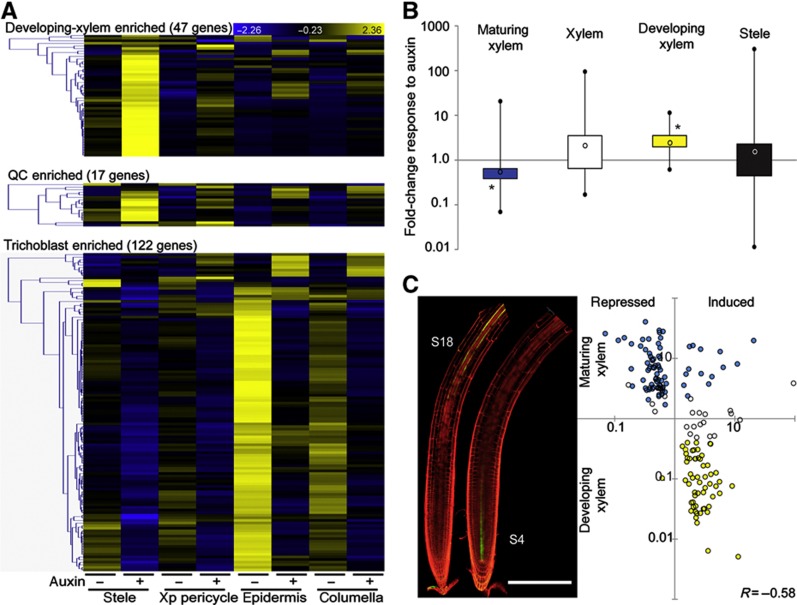
Figure 4.
Auxin-responsive cell-identity markers. (A) Cell type-specifically enriched auxin-responsive genes show spatially distinct auxin responses. Heatmaps display the overlap between lists of cell type-specifically enriched genes (Supplementary Figure S2B; Supplementary Table S2) and the stringent list of 2846 auxin-responsive genes; blue (low) to yellow (high) color code indicates standard deviations from the row mean. Auxin-responsive developing-xylem-enriched genes (top panel) and QC-enriched genes (middle panel) are predominantly upregulated, specifically in the stele; auxin-responsive trichoblast-enriched genes (lower panel) are predominantly downregulated and highly expressed in the epidermis before treatment. (B) Boxplot representation of the fold-change distribution of maturing-xylem- (blue), xylem- (white) and developing-xylem- (yellow) enriched genes that significantly respond to auxin treatment in the stele (t-test P<0.01, all auxin-responsive genes in the stele are represented by a black box). Black circles represent minimum and maximum values, black lines represent the first and fourth quartiles, boxes represent the second and third quartiles, open circle represents the median; *P<1e−10 χ2-test for ratio of induced-to-repressed genes. (C) The S18 and S4 marker lines for maturing and developing xylem, respectively (left panel), were used to plot the fold change in expression upon auxin treatment in the stele versus the expression ratio between maturing and developing xylem for the 157 auxin-responsive (maturing and/or developing) xylem-enriched genes. Pearson’s correlation R=−0.58, scale bar indicates 250 μm.
In the xylem, separate expression profiles for developing and maturing cell populations permitted analysis of auxin responses in relation to expression along the longitudinal maturation gradient within a specific cell lineage (tissue-specific marker lines S4 and S18, respectively; Lee et al, 2006; Figure 4B and C; Supplementary Table S4). Analysis showed that auxin promotes the expression of developing-xylem genes and represses the expression of genes enriched in maturing xylem. Fifty-six of fifty-seven auxin-regulated developing-xylem-enriched genes in the stele sample were induced by auxin (χ2-test P=5.66e−11) and 63 out of 77 auxin-regulated maturing-xylem-enriched genes were repressed (χ2-test P=7.59e−11). In planta, the expression of developing-xylem identity marker pTMO5::GFP (Lee et al, 2006; Schlereth et al, 2010) intensifies and expands from the apical meristem further into the basal (shootward) meristem upon auxin treatment (Supplementary Figure S5B–D), corroborating the transcriptomic data and showing that this increase in expression seen in the stele sample takes place exclusively within the xylem lineage. Notably, the auxin sensitivity of xylem-enriched genes (i.e., the degree of induction or repression by auxin as measured by fold change) was significantly correlated with the ratio of expression between the developing and maturing xylem. Genes that show a higher relative expression in the developing xylem tend to be more strongly induced, and genes that show a higher relative expression in the maturing xylem tend to be more strongly repressed (Figure 4C; Pearson’s correlation R=−0.58, Z-score=7.13 non-parametric randomization test for significance). A transcript’s auxin sensitivity is therefore a reliable predictor of longitudinal expression in the RAM for xylem-enriched genes.
Longitudinal expression correlates with genome-wide auxin responses
We next addressed the link between the global response to auxin and spatial expression along the longitudinal axis of the entire root tip. There are two transcriptomic data sets of gene expression in the longitudinal dimension of the Arabidopsis seedling root: one of a 13-slice sampling of two individual roots (Brady et al, 2007) and another of a three-section sampling comprising the meristematic, elongation and maturation zones gathered in pooled replicates (Birnbaum et al, 2003; Figure 1A).
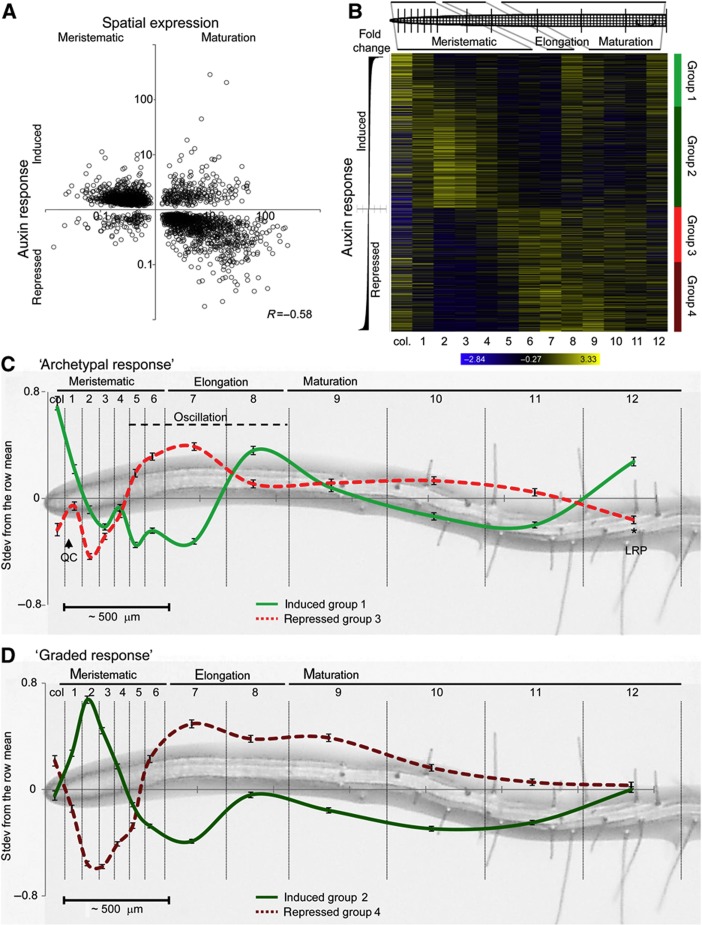
To quantify the relationship between auxin response and spatial expression along the longitudinal root axis, we examined the overlap of the 6850 genes with differential expression between the meristematic and maturation zone (t-test P<0.01; Supplementary Table S5; Birnbaum et al, 2003) and our extensive list of 5097 auxin-responsive genes according to the ANOVA treatment factor. The two lists yielded an intersection of 2437 genes, for which fold change of the auxin response (averaged over four tissues) was plotted against the fold change in expression between the meristematic and maturation zone (Figure 5A). The correlation between these two independent data sets is highly significant (Pearson’s correlation R=−0.58, Z-score=28.06 non-parametric randomization test for significance) and indicates that, for thousands of genes, auxin-sensitivity predicts longitudinal expression.
Figure 5.
Visualization of a global auxin-response gradient in the root meristem. (A) The fold-change response to auxin treatment (ANOVA treatment P<0.01, 5097 genes) was plotted versus the expression ratio between the meristematic zone and maturation zone (t-test P<0.01, 6850 genes; Supplementary Table S5; Birnbaum et al, 2003) for the genes that are both significantly responsive to auxin and significantly differentially expressed between meristematic and maturation zones (2437-gene intersect). Pearson’s correlation R=−0.58. (B) Heatmap of the spatial expression of auxin-responsive genes (ANOVA treatment P<0.01, 5097 genes) in the 13-slice longitudinal data set (root1; Brady et al, 2007). Genes were ordered by fold-change response to auxin treatment; blue (low) to yellow (high) color code indicates standard deviations from the row mean. Upregulated and downregulated genes were further subdivided into groups 1–4 based on relative induction or repression and broad differences in longitudinal expression (red and green color coding). (C, D) Average normalized longitudinal expression patterns of auxin-responsive genes, ±s.e.m. (groups 1–4 in B). The relative spatial separation of the 13-slice data set (Brady et al, 2007) is represented on the x axis and the standard deviations from the row mean on the y axis. (C) Longitudinal expression of archetypal auxin-responsive genes (groups 1 and 3 in B), consisting of the top 1000 induced and the first 1000 repressed genes. The quiescent center (QC), oscillation zone and first lateral root primordium (LRP) are indicated. (D) Longitudinal expression of graded auxin-responsive genes (groups 2 and 4 in B), consisting of the remaining 1842 induced and 1255 repressed genes.
To visualize the relation between transcriptional auxin sensitivity and the regulation of expression along the longitudinal axis of the root, genes were ordered by fold change in expression after auxin treatment and plotted in a heatmap of the 13-slice data set (Figure 5B; Brady et al, 2007). This representation again revealed a link between relative responsiveness to auxin and spatial expression along the longitudinal axis of the root. The upregulated genes displayed a longitudinal expression gradient with a meristematic maximum and slope linked to the intensity of the auxin response; downregulated genes showed complementary expression with a minimum in the apical end of the meristem (Figure 5B). These meristematic response gradients were also seen in the replicate root sampled in the 13-slice data set (Supplementary Figure S6A). Secondary expression peaks were observed in the elongation and maturation zones of both sampled roots; however, these regions vary between root 1 and root 2 (Figure 5B; Supplementary Figure S6A).
Within the groups of induced and repressed genes, there were also notable differences in longitudinal expression patterns that were associated with relative sensitivity to auxin treatment. Using induction versus repression and relative fold change of the response to auxin treatment to subdivide response strength, the 5097 auxin-responsive genes could be broadly subdivided into four categories: group (1) strongly auxin-induced genes with high expression in the apex that quickly diminishes in the apical meristem and displays variable secondary peaks in the elongation and maturation zones; group (2) moderately to weakly auxin-induced genes with high expression in the apical end of the meristem, a graded decline toward the basal end of the meristem and lacking prominent secondary peaks; group (3) weakly auxin-repressed genes with the inverse spatial expression pattern of group 1; and group (4) moderately to strongly auxin-repressed genes that complement the expression of group 2 (Figure 5B–D). Thus, expression patterns along the longitudinal axis are also linked to the degree of induction or repression by auxin.
In a validation of the overall trends in the data, the relation between auxin response and longitudinal expression could be recapitulated using independent auxin-response data sets collected in the root, in this study and by others (Vanneste et al, 2005; Bargmann and Birnbaum, 2009). The correlation for the various lists of genes regulated by auxin signaling was quantified by cross-referencing with the meristematic versus maturation data set (Supplementary Figure S6B–K; Supplementary Table S5; Birnbaum et al, 2003). First, correlation was also evident with auxin responses measured in individual tissues and with auxin responses in the intact root as well as the stringent list of 2846 auxin-responsive genes (Supplementary Figure S6B–G; Supplementary Table S5). In addition, correlation was observed between longitudinal expression and previously published data of auxin responses in proximal root tissues above the primary meristem (excluding the RAM; Vanneste et al, 2005), indicating that this correlation is not restricted to responses in the root apex (Supplementary Figure S6H). However, no strong correlation between auxin response and spatial expression was evident when response data was generated from whole seedlings (Okushima et al, 2007), suggesting that responses outside the root do not correlate with expression in the root tip (Supplementary Figure S6I). Finally, an inverse correlation could be observed between longitudinal expression and the transcriptional response to transient expression of gain-of-function Aux/IAA repressors measured in root epidermal protoplasts (Bargmann and Birnbaum, 2009). Here, genes repressed by the expression of Aux/IAA19mII repressor (Tiwari et al, 2001) displayed relatively high meristematic expression, whereas genes induced by the expression of a gain-of-function Aux/IAA repressor showed low expression in the meristem (Supplementary Figure S6J). Consequently, as a correlation between auxin signaling and longitudinal expression is recapitulated by the manipulation of the canonical auxin signal transduction pathway in dissociated cells, the observed correlation can be attributed to a direct cellular response to auxin signaling.
Discussion
Auxin affects cell type-specific development
This study demonstrates the influence of cellular context on genome-wide transcriptional responses to auxin treatment and reveals a broad range of tissue specificity in these responses. A relatively small proportion of transcripts show uniform regulation across tissues (Figure 3A; Supplementary Figure S4A), while the majority displays a spatial bias toward one or more of the tissues analyzed. Examining the auxin responsiveness of CTSE genes, it appears that auxin treatment, in general, does not promote all cells toward a common developmental state. Instead, auxin can promote or inhibit cell character by enhancing or repressing the expression level of cell-specific markers differentially in the separate tissue samples analyzed here (Figure 4A; Supplementary Figure S5A and B).
A significant repressive effect of auxin on trichoblast-enriched gene expression was observed in our data set, particularly in the epidermis sample (Figure 4A; Supplementary Figure S5A). The repression was not seen for genes enriched in atrichoblasts or for genes enriched throughout the epidermis (Supplementary Figure S5A). This result was independently recapitulated by statistical overrepresentation of genes previously associated with the GO term trichoblast development in dominant expression-pattern clusters that show downregulation specifically in the epidermis (Figure 3C; Supplementary Figure S4; Supplementary Table S3). These findings are in line with a previous study of root-hair defects in aux1 auxin-importer mutants (Jones et al, 2009), where increased transcriptional auxin signaling in trichoblasts was associated with defects in root-hair development. The categorization of auxin-regulated trichoblast-enriched genes presented here can be used to further investigate the mechanisms by which auxin may influence root-hair development.
Analysis of radial patterning in the stele of the RAM has indicated cross-talk between auxin and cytokinin modulates a PIN-driven high-auxin domain in the xylem that mediates cell specification (Bishopp et al, 2011). Consistent with high auxin levels in the xylem lineage, a subset of highly auxin-induced genes (including Aux/IAA6, 8, 19 and 29) shows high basal expression throughout the xylem (enriched in both the developing and maturing xylem; Supplementary Figure S1C; Figure 4B and C; Supplementary Table S4). The auxin-responsive, developing- or maturing-xylem-enriched transcriptomes can be used to investigate xylem specification by looking for potential auxin-responsive regulators of development.
Aside from cell-fate specification, auxin also has a role in xylem differentiation and maturation. Analysis of the tissue-specific auxin responses in relation to xylem development demonstrates how auxin may regulate lineage-specific differentiation through moderating activation and repression of genes associated with juvenile and maturing transcriptional states, respectively. Auxin significantly promotes developing-xylem identity and inhibits the expression of maturing-xylem genes in the stele (Figure 4; Supplementary Figure S5). Xylem development has previously been proposed to be directly regulated by local auxin levels in the cambial meristem of wood-forming tissues (Bhalerao and Bennett, 2003; Yoshida et al, 2009). In this tissue, the radial developmental gradient (characterized by sequential division, expansion and secondary cell wall deposition) parallels an auxin concentration gradient, as measured by mass-spectrometric analysis of IAA in cryo-sections (Uggla et al, 1996). However, a transcriptional link between the perception of an auxin concentration gradient and the regulation of cellular maturation was not found (Nilsson et al, 2008). In the root, we did find that sensitivity to auxin treatment directly correlates with the slope of expression of xylem-enriched genes along the longitudinal developmental gradient (Figure 4C). This correlation provides evidence that an endogenous auxin gradient directly influences the global transcriptional state of cells along this dimension to regulate maturation.
Mapping the longitudinal auxin-response transcriptome
The availability of transcriptomic data sets along the longitudinal axis of the Arabidopsis seedling root (Birnbaum et al, 2003; Brady et al, 2007) allowed us to plot the spatial expression of comprehensive sets of auxin-responsive genes and to identify a transcriptional auxin-response gradient within this tissue as a whole. The analysis in effect uses the entire auxin-responsive transcriptome as a reporter for the endogenous auxin response; as opposed to the use of singular auxin-induced promoters. Moreover, we could also observe the longitudinal expression of auxin-repressed genes in this context and factor in the relative sensitivity of auxin-responsive genes.
The auxin response can be seen to be bipartite; consisting of genes with an archetypal expression, including high expression in the root tip and secondary shootward peaks (group 1 and complementary group 3), and genes with a graded meristematic expression pattern (group 2 and group 4; Figure 5B–D). The archetypal response resembles the expression of the DR5 reporter (Supplementary Figure S1B), for which auxiliary expression has also been observed in more shootward portions of the root, similarly to the secondary peaks of group 1 (Figure 5C). For DR5, these shootward peaks of expression have been shown to correspond to the regions of pre-branch site specification and lateral root primordia (De Smet et al, 2007; Dubrovsky et al, 2008; Moreno-Risueno et al, 2010). The graded response, however, more closely matches measured auxin concentrations (Petersson et al, 2009) and slopes along with the cellular-maturation gradient in the apical and basal meristem (Figures 1A and 5B–D). One speculation is that the archetypal response is directly under control of the auxin signal transduction machinery, including the negative feedback regulation, which could explain why these genes do not mirror the measured concentration gradient. The graded response may be under non-canonical or indirect regulation, conceivably through auxin-responsive master-regulator transcription factors that do reflect the developmental- and auxin-concentration gradient in the meristem (such as the PLTs; Galinha et al, 2007).
It is important to note that there is likely a cell lineage-specific aspect to the interpretation (or maintenance) of auxin gradients. This can be observed with the auxin signaling reporters DR5::3xVenus and DII-Venus (Supplementary Figure S1; Santuari et al, 2011; Brunoud et al, 2012). The correlation seen between the auxin response and the whole-root longitudinal data (Birnbaum et al, 2003; Brady et al, 2007), therefore, represents a global response gradient that may consist of several distinct cell type-specific gradients. The availability of longitudinally separated markers of the same cell lineage in the xylem (Figure 4C) makes possible the visualization of a response gradient in this tissue specifically, that is consistent with mass spectrometric auxin measurements in the stele (Petersson et al, 2009). It will be interesting to see whether similar gradients can be seen in other cell types, as more specific marker lines become available; especially in the epidermis where DR5::3xVenus and DII-Venus reporters potentially indicate an inverted gradient (Supplementary Figure S1A and B).
Overall, the significant correlation between the transcriptomic auxin response and spatial expression within the root suggests that auxin sensitivity together with spatial gradients of auxin distribution is a determinant in the spatial expression of thousands of genes.
Cellular competence for a unique auxin response
Cells perceive auxin as though selectively processed by a set of filters that accompany a given cell identity and represent the auxin-sensing and -response machinery active in the cell. Our data present the transcriptional output of several such innate response-machinery filters, providing an important view of how auxin is perceived by individual cell types.
The canonical auxin perception and signal transduction pathway (composed of the TRANSPORT INHIBITOR RESPONSE 1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) receptors, the Aux/IAA co-receptor/transcriptional repressors and the AUXIN RESPONSE FACTOR (ARF) transcription factors (Dharmasiri et al, 2005; Okushima et al, 2005; Overvoorde et al, 2005)) is encoded by large gene families that show divergent cell type-specific expression patterns (Supplementary Figure S1; Rademacher et al, 2011). Promoter-swap and misexpression studies using specific ARFs and gain-of-function Aux/IAAs have demonstrated that individual components of this modular auxin-response pathway can bestow specific responses in different tissues of the root and embryo (Knox et al, 2003; Rademacher et al, 2012). The cellular TIR1/AFB-Aux/IAA-ARF composition is thought to represent an ‘auxin code’ that is the principal determinant of the specificity of the output.
However, additional regulatory interactors of the TIR1/AFB-Aux/IAA-ARF pathway, for example, the TOPLESS transcriptional co-repressors, the MYB77 transcription factor and the miR165 microRNA (Shin et al, 2007; Szemenyei et al, 2008), may also impart tissue specificity. Both TMO5 and TMO6 are targets of the same auxin-response factor (ARF5/MONOPTEROS; Schlereth et al, 2010), yet show highly divergent patterns of induction. TMO5 is expressed and induced specifically in the xylem, whereas TMO6 is seen to have high expression in the phloem and procambium and is induced in the xp pericycle at sites of initiating lateral roots (Supplementary Figures S3 and S5). The discrepancy between the induction of these direct auxin-response targets reveal an intriguing aspect of cell type-specific regulation of transcriptional auxin responses and show that additional factors, aside from ARF5, must be involved in the activation of their expression in different cell types.
Interestingly, the ARF binding site (the auxin-response element TGTCTC) and several variants thereof (TGTCNC) are only found to be overrepresented among genes with a relatively uniform upregulation (Supplementary Figure S4; Supplementary Table S3). This could indicate that, if the more spatially distinct responses and auxin-regulated gene repression are in part directly mediated by binding of particular ARF isoforms, a more complex DNA binding-site recognition may be involved in the target specificity of different ARF isoforms. Alternatively, the spatially distinct responses could be composed more of indirect target genes that are regulated by secondary activators or repressors. Finally, signal transduction outside of the TIR1/AFB-Aux/IAA-ARF auxin-response pathway may also account for cell type-specific responses.
Data availability
The raw microarray data generated in this study have been deposited in the Gene Expression Omnibus in the form of.cel files and are available online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35580.
Materials and methods
Plant materials and treatment
All Arabidopsis thaliana plant lines used in this study are listed in Supplementary information. Seed was sterilized by 5 min incubation with 96% ethanol followed by 20 min incubation with 50% household bleach and rinsing with sterile water and stratified for 2 days at 4°C in the dark. Seedlings were grown hydroponically on nylon mesh in phytatrays (Sigma) with growth medium (2.2 g/l Murashige and Skoog Salts (Sigma-Aldrich), 1% (w/v) sucrose, 0.5 g/l MES hydrate (Sigma-Aldrich), pH 5.7 with KOH), or plated on square petri dishes (Fisher Scientific) with growth medium plus 1% (w/v) agar (as in Bargmann and Birnbaum, 2010). Phytatrays and plates were placed in an Advanced Intellus environmental controller (Percival) set to 35 μmol/m2 s−1 and 22°C with an 18 h-light/6 h-dark regime. For the cell sorting and microarray experiment, 1-week-old (5 dpg) seedlings were treated with 5 μM IAA (Sigma-Aldrich) or mock treated with solvent alone for a total of 3 h (2 h in phytatray and 1 h during the protoplast and sorting procedure). Duration of treatment was chosen to obtain a relatively early yet robust representation of responses to auxin in the root; before morphological effects, such as cell division, could be observed but late enough to include secondary/indirect target genes. A 10-mM IAA stock was dissolved in ethanol and stored at −20°C. 2,4-Dichlorophenoxyacetic acid (2,4-D; Sigma-Aldrich) treatments (used for auxin-responsive GFP reporter lines) were performed by transferring seedlings to plates supplemented with 1 μM 2,4-D from a 10-mM stock dissolved in ethanol and stored at −20°C. The pGH3.5::GFP reporter line was generated by cloning 3546, bp upstream of the GH3.5 start codon (using primers Fwd 5′-cagtttaattatactccatttattcgtca-3′ and Rev 5′-ggtttaagagaaagagagaagtctgagaaaatg-3′) in front of the GFP open reading frame in pMDC107 (Curtis and Grossniklaus, 2003) using Gateway recombination via pENTR-D-TOPO (Invitrogen). The resulting vector was used to transform Col-0 Arabidopsis with Agrobacterium tumefaciens (GV3101). The pIAA5::GUS reporter line was generated by cloning 913 bp upstream of the IAA5 start codon (using primers Fwd 5′-cacctatcacaaagtcttgttgtgttattca-3′ and Rev 5′-ctttgatgtttttgattgaaagtattg-3′) in front of the uidA open reading frame in pMDC163 (Curtis and Grossniklaus, 2003) using Gateway recombination via pENTR-D-TOPO (Invitrogen). The resulting vector was used to transform Col-0 Arabidopsis with A. tumefaciens (GV3101).
Generation of protoplasts, flow cytometry and fluorescence activated cell sorting
Protoplast isolation was performed as described previously (Bargmann and Birnbaum, 2010). Roots from one phytatray containing ∼1500 one-week-old seedlings were harvested (after a 2-h treatment with 5 μM IAA or solvent alone) and placed into a gently shaking 50 ml tube with 15 ml protoplasting solution (supplemented with 5 μM IAA or solvent alone) for 45 min. Protoplasting solution was prepared with 1.25% (w/v) cellulase (Yakult), 0.3% (w/v) macerozyme (Yakult), 0.4 M mannitol, 20 mM MES, 20 mM KCl, 0.1% (w/v) BSA, 10 mM CaCl2, 5 mM β-mercapto ethanol, pH adjusted to 5.7 with Tris/HCl pH 7.5. The protoplast solution was filtered through 40-μm cell strainer (BD Falcon, USA), transferred to 15 ml conical tubes and centrifuged for 5 min at 500 g. In all, 14 ml of the supernatant was aspirated and pellets were resuspended.
Protoplast suspensions were cytometrically analyzed and sorted using FACSAria (BD Biosciences) equipped with a 488-nm laser and fitted with a 100-μm nozzle to measure fluorescent emission at 530/30 and 610/20 nm for GFP and red-spectrum autofluorescence, respectively. Positive events were identified based on their red-to-green fluorescence ratio, sorted directly into 350 μl RNA extraction buffer and stored at −80°C.
RNA extraction and microarray hybridization
RNA was extracted from 20 000 sorted cells per replicate using an RNeasy Micro Kit with RNase-free DNase Set according to the manufacturer’s instructions (QIAGEN). RNA was quantified with a Bioanalyzer (Agilent Technologies) and reverse-transcribed, amplified and labeled with WT-Ovation Pico RNA Amplification System and FL-Ovation cDNA Biotin Module V2 (NuGEN). The labeled cDNA was hybridized, washed and stained on an ATH-121501 Arabidopsis full genome microarray using a Hybridization Control Kit, a GeneChip Hybridization, Wash, and Stain Kit, a GeneChip Fluidics Station 450 and a GeneChip Scanner (Affymetrix). Three independent biological replicates were collected for all treatments. The raw data files generated by others and used in this analysis were obtained from the Benfey lab or from http://www.ncbi.nlm.nih.gov/geo/.
Data analysis
Data normalization, analysis and visualization were performed using freely available code and R-based software (listed in Supplementary information). Raw microarray data were MAS5.0 normalized with a scaling factor of 250 and log transformed before homoscedastic statistical analysis (Student’s t-test and two-way ANOVA; Flexarray). Ambiguous probesets (Supplementary Table S2) were removed from further analysis. False discovery rates (FDR) were calculated based on the P-value distribution (Q-value). For the comparison between statistical tests in separate tissues, equal cutoffs were set at P<0.01. FDR at this cutoff and q-values for the individual probesets are reported in Supplementary Table S2. Additionally, a fold-change cutoff of >1.5 was set for the tissue-specific and intact root t-tests. To generate the stringent list of 2846 auxin responders, genes had to pass the ANOVA for treatment or interaction (P<0.01) and at least one of the tissue-specific t-tests (P<0.01, fold change>1.5).
For co-expression analysis, hierarchical clustering was performed on the stringent list of auxin responders with pairwise Pearson’s correlation using gene expression lists of replicate sample averages that were row normalized (Multiple Experiment Viewer). Branch length distribution of the HCL tree and the figure of merit (FOM) of iterative K-means clustering runs were used to gauge the expected number of clusters (Multiple Experiment Viewer). A Fuzzy K-means clustering search for dominant expression patterns was executed employing the R script by Orlando and co-workers for the manipulation of large-scale Arabidopsis microarray data sets (Orlando et al, 2009). Clusters containing <10 genes were omitted from further analysis. GO-term overrepresentation was analyzed using VirtualPlant with TAIR10 gene annotations. Promoter element enrichment was based on the absence or presence of motifs in the 500-bp upstream of the transcription start sites (hypergeometric distribution test with FDR correction q<0.01, TAIR10 annotation).
Template matching for the isolation of CTSE gene sets was performed using the Pavlidis template matching algorithm (Multiple Experiment Viewer) on previously generated transcriptomic data from 13 non-overlapping tissue marker lines (Supplementary Table S2) with a similarity cutoff of R>0.8.
Microscopy
Confocal microscopy was performed with SP5 (Leica) and LSM710 (Zeis) microscopes and software. Cell walls were stained by 10 min incubation in 10 μg/ml propidium iodide (dissolved in water). GUS reporter gene lines were stained in 50 mM phosphate buffer pH 7, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, 0.05% (v/v) Triton X, 1 mM X-Gluc, for 24 h at 37°C. The staining reaction was stopped and seedlings were fixed and cleared with ethanol and mounted in water. Staining was visualized with an Axioskop (Zeiss) microscope.
Supplementary Material
Supplementary Information
Expression of the auxin signaling components in different cell types and longitudinal sections of the Arabidopsis root
Cell type-specific auxin responses in the Arabidopsis root
Dominant expression patterns in cell type-specific auxin responses
chi squared tests for ratio's of induced-to-repressed expression of cell type-specifically enriched genes
Analysis of publicly available microarray data
Acknowledgments
We would like to thank Hidehiro Fukaki (Kobe University) for the pLBD33::GUS reporter. This work was supported by grants from the NSF (DBI-0519984) and NIH (R01-GM078270) to KDB, the NIH (R01-GM086632) to DCB, the NIH (GM43644), Howard Hughes Medical Institute and Gordon and Betty Moore Foundation to ME, the EMBO (LTF) to IE, the Vaadia-BARD (FI-431-10) to ES, and the Research Foundation of Flanders to SV.
Author contributions: BORB helped conceptualize, design, perform and write this work, SV helped conceptualize and write this work, GK helped perform the clustering analysis, TN helped perform the confocal microscopy, IE helped perform the promoter and correlation analyses, ES and GC helped perform reporter-gene analysis, JF, DCB and ME helped conceptualize this work. KDB helped conceptualize, design and write this work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bargmann BO, Birnbaum KD (2009) Positive fluorescent selection permits precise, rapid, and in-depth overexpression analysis in plant protoplasts. Plant Physiol 149: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Birnbaum KD (2010) Fluorescence activated cell sorting of plant protoplasts. J Vis Exp 36: e1673, ; DOI: 10.3791/1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bennett T, Scheres B (2010) Root development-two meristems for the price of one? Curr Top Dev Biol 91: 67–102 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Bennett MJ (2003) The case for morphogens in plants. Nat Cell Biol 5: 939–943 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Help H, El-Showk S, Weijers D, Scheres B, Friml J, Benkova E, Mahonen AP, Helariutta Y (2011) A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr Biol 21: 917–926 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M (2010) Auxin perception--structural insights. Cold Spring Harb Perspect Biol 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, Inze D, Bennett MJ, Beeckman T (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134: 681–690 [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, Wang JY, Meuli N, Vanneste S, Friml J, Hilson P, Jurgens G, Ingram GC, Inze D, Benfey PN, Beeckman T (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J (2010) Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA 107: 12046–12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Napsucialy-Mendivil S, Duclercq J, Cheng Y, Shishkova S, Ivanchenko MG, Friml J, Murphy AS, Benkova E (2011) Auxin minimum defines a developmental window for lateral root initiation. New Phytol 191: 970–983 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, Palme K (2002) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Grunewald W, Friml J (2010) The march of the PINs: developmental plasticity by dynamic polar targeting in plant cells. EMBO J 29: 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hagen G, Martin G, Li Y, Guilfoyle TJ (1991) Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol 17: 567–579 [DOI] [PubMed] [Google Scholar]
- Hayashi K (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HM, Grierson CS (2009) Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Dengler N (2002) Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216: 212–219 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Neve J, Kepinski S (2010) Defining auxin response contexts in plant development. Curr Opin Plant Biol 13: 12–20 [DOI] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, Scheres B, Benfey PN (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hagen G, Guilfoyle TJ (1991) An auxin-responsive promoter is differentially induced by auxin gradients during tropisms. Plant Cell 3: 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Karlberg A, Antti H, Lopez-Vernaza M, Mellerowicz E, Perrot-Rechenmann C, Sandberg G, Bhalerao RP (2008) Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 20: 843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Brady SM, Koch JD, Dinneny JR, Benfey PN (2009) Manipulating large-scale Arabidopsis microarray expression data: identifying dominant expression patterns and biological process enrichment. Methods Mol Biol 553: 57–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Benfey PN (2008) Root layers: complex regulation of developmental patterning. Curr Opin Genet Dev 18: 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E, Moller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D (2011) A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606 [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222 [DOI] [PubMed] [Google Scholar]
- Santuari L, Scacchi E, Rodriguez-Villalon A, Salinas P, Dohmann EM, Brunoud G, Vernoux T, Smith RS, Hardtke CS (2011) Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr Biol 21: 1918–1923 [DOI] [PubMed] [Google Scholar]
- Schlereth A, Moller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jurgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007) The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B (1996) Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA 93: 9282–9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, Inze D, Fukaki H, Beeckman T (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, Guedon Y, Armitage L, Picard F, Guyomarc’h S, Cellier C, Parry G, Koumproglou R, Doonan JH, Estelle M, Godin C et al. (2011) The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Systems Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Iwamoto K, Demura T, Fukuda H (2009) Comprehensive analysis of the regulatory roles of auxin in early transdifferentiation into xylem cells. Plant Mol Biol 70: 457–469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Expression of the auxin signaling components in different cell types and longitudinal sections of the Arabidopsis root
Cell type-specific auxin responses in the Arabidopsis root
Dominant expression patterns in cell type-specific auxin responses
chi squared tests for ratio's of induced-to-repressed expression of cell type-specifically enriched genes
Analysis of publicly available microarray data
Data Availability Statement
The raw microarray data generated in this study have been deposited in the Gene Expression Omnibus in the form of.cel files and are available online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35580.