Abstract
Helicobacter pylori is considered the most prevalent infectious agent among humans, and it causes gastric inflammation, gastroduodenal ulcers, and a risk of gastric cancer. We performed a genomewide linkage analysis among Senegalese siblings phenotyped for H. pylori–reactive serum immunoglobulin G. A multipoint LOD score of 3.1 was obtained at IFNGR1, the gene that encodes chain 1 of the interferon-γ (IFN-γ) receptor. Sequencing of IFNGR1 revealed −56C→T, H318P, and L450P variants, which were found to be associated with high antibody concentrations. The inclusion of these in the linkage analysis raised the LOD score to 4.2. The variants were more prevalent in Africans than in whites. Our findings indicate that IFN-γ signaling plays an essential role in human H. pylori infection, and they contribute to an explanation of the observations of high prevalences and relatively low pathogenicity of H. pylori in Africa. Moreover, they provide further support for the value of genomewide linkage studies in the analysis of susceptibility to infection and other complex genetic traits.
Introduction
Helicobacter pylori is considered the most common infectious agent among humans worldwide. In the United States and Europe, 25%–50% of the population are infected, and prevalences in developing countries reach 70%–90%, with almost all individuals acquiring the infection before the age of 10 years (Dunn et al. 1997). Approximately 10%–20% of infected individuals develop disease, such as gastritis or gastroduodenal ulcer, and have an increased risk of gastric cancer (Blaser 1998).
Several lines of evidence indicate a genetic influence on the susceptibility to H. pylori. Twin studies have shown a substantial genetic component (Malaty et al. 1994). Furthermore, evidence of ethnic differences was obtained by comparing North Americans of different ancestries (Graham et al. 1991). Finally, inbred mouse strains show varying degrees of susceptibility to experimental Helicobacter infection (Sawai et al. 1999).
H. pylori–reactive serum immunoglobulin G (IgG) concentrations, using whole bacterial lysates as antigen, have been found to be a reliable tool for defining the infection state of humans (Evans et al. 1989; Marchildon et al. 1999). Accordingly, they have been widely used in genetic and epidemiological studies (Evans et al. 1989; Graham et al. 1991; Malaty et al. 1994; Marchildon et al. 1999). Here, we present a genomewide linkage analysis of H. pylori infection, defined by reactive serum IgG, in Senegalese sibships.
Material and Methods
Study Group
The protocol was approved by the ethics committee of the Board of Physicians of Hamburg. Participants belonged to a study group analyzed for genetic factors controlling the intensity of infection with Schistosoma mansoni (Müller-Myhsok et al. 1997). They were recruited in the village of Ndombo, near the town of Richard Toll, in northern Senegal. The group comprised 10 families with 2 siblings, 11 with 3, 10 with 4, and 4 with 5.
Phenotyping
Serum levels of IgG antibodies to H. pylori were determined, as described elsewhere (Nilius et al. 2001), using a commercially available ELISA (Synelisa, Pharmacia, Upjohn, and Amersham Biosciences), which applies a composite of recombinant and whole-cell–lysate antigens. Serum levels of IgG antibody to phosphorylcholine were determined as reported elsewhere (Schenkein et al. 2001).
Linkage Analysis
The distribution of IgG values (U/ml) was substantially skewed to the left. Therefore, the values were log transformed to approximate normality. The genome screen was performed, as described elsewhere (Adams et al. 1998), using the human genome screening set, version 6, developed by J. L. Weber. The complete set consisted of 373 markers (86% tri- or tetranucleotide repeats), with an average heterozygosity of 76% and an average spacing of 10 cM (Invitrogen). Genotypes were determined using GeneScan and GenoTyper software (Applied Biosystems), yielding export files containing allele tables for each individual marker. Inheritance of alleles was verified by use of the PedCheck program (O'Connell and Weeks 1998). A quantitative-trait analysis was performed by applying a Haseman-Elston statistic, as implemented in GeneHunter2 (Kruglyak and Lander 1995). Allele frequencies were estimated in the study group, using the DownFreq program (Terwilliger 1995). Map order and distances of markers were obtained from the Marshfield map (Center for Medical Genetics, Marshfield Medical Research Foundation; Broman et al. 1998) and were verified by calculating the order with our own marker data, using the Crimap program (Lander and Green 1987), with a threshold for linkage of LOD = 3.0. A quantitative transmission/disequilibrium test was performed with the QTDT program as described elsewhere (Abecasis et al. 2000).
Sequencing and SNP Analysis
All seven exons of IFNGR1 (MIM 107470) and 1,045 nt of the 5′-region of the gene were amplified from genomic DNA (table 1). Annealing temperatures were 54°C for all reactions, except those primed by Pr3/Pr4 and exon 5–sense/exon 5–antisense, for which they were 56°C and 55°C, respectively. PCR products were sequenced bidirectionally on an ABI 3100 sequencer (Applied Biosystems; EMBL and National Center for Biotechnology Information Nucleotide databases). SNPs were analyzed by dynamic allele-specific hybridization with fluorescence resonance energy transfer, in a LightCycler (Hoffmann–La Roche) (table 2). Annealing temperatures were 54°C for all reactions, except the one analyzing –56C→T, for which it was 58°C.
Table 1.
PCR Primer Pairs Used in Sequencing of the IFNGR1 Gene
| Exon | Sense (5′→3′) | Antisense (5′→3′) | Product Size(bp) |
| Pr1/2a | TGATTTGACACTGAATTGCTG | CCTGCTCACACCCTGCATGAC | 652 |
| Pr3/4a | TGATCTGGCTGATAATACCTC | ATCGGCTTGACCAAGAACTAC | 525 |
| 1 | GCACGACGCCGTGCTCACTGC | AACCACGGAGCCCCAGTCTCG | 473 |
| 2 | TATACATATCTGGGCAATGTG | GAAGGCTGATGAAAGAACACAG | 301 |
| 3 | ACTTTCTCTGGCGTCTCCATC | ATGCTCAACCTGTACTGACTC | 466 |
| 4 | GTCCTGCTTTAGAACAACCAG | AATGTATTCACATGGTCAGTG | 499 |
| 5 | CAGATGCATAGTATCGTGCTG | TCTAAGGAATGGAACTAATGC | 511 |
| 6 | ATTGCTATCTAAGACAGATAC | TGATTGATGGCAGGTGACATC | 436 |
| 7 | GGTGGTCCATTACTTCAGACC | ACAATTTCTGAGATCATAATC | 791 |
Pr1/2 and Pr3/4 are primer pairs for the proximal and distal parts of the 5′ region of IFNGR1. Primers were derived from the Homo sapiens chromosome 6 working draft sequence segment (NT_025741.8), published by the National Center for Biotechnology Information Annotation Project.
Table 2.
Primer Pairs of the IFNGR1 Variants with Anchor and Sensor Used in LightCycler Assay
| Variant | Sense (5′→3′)a | Antisense (5′→3′)b |
| −56C→T | GCACGACGCCGTGCTCACTGC | AACCACGGAGCCCCAGTCTCG |
| GACCAGCCCAGCACTGCCC | CCAGCCCCGGCCTTACGTCACTTCC | |
| H318P | GGTGGTCCATTACTTCAGACC | CTTCAGTAGTCACCACTTCTGTTA |
| CCAGGCATGCATACCGAAGAC | AAGAGCCGTTGTCTCCAGCAACAGT | |
| L450P | CCTCCTTTGGTTATGATAAACCACA | ACAATTTCTGAGATCATAATC |
| TCATCCACAAGTAGATCCACTAGC | AACCAATCAAGGACTCTTTACCGCT |
For each variant, the second nucleotide sequence represents the sensor.
For each variant, the second nucleotide sequence represents the anchor.
Results
Study Group and Phenotype
A total of 111 siblings aged 5 to 60 years, who were from 35 Senegalese nuclear families and comprised 143 sib pairs, were phenotyped for H. pylori infection, using serum IgG reactive to major H. pylori antigens. Sixty-three percent of the study group were positive, with no significant age dependence. Serum levels of anti–phosphorylcholine IgG, which is produced in reaction to ubiquitous dental-plaque bacteria (Schenkein et al. 2001), were measured as a control and were found not to be correlated with levels of anti–H. pylori IgG (r=0.12, P=.2).
Linkage Analysis
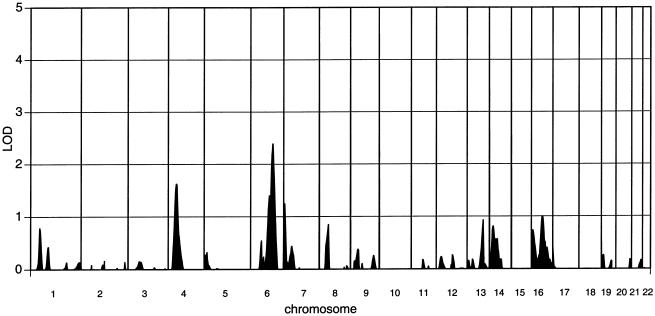
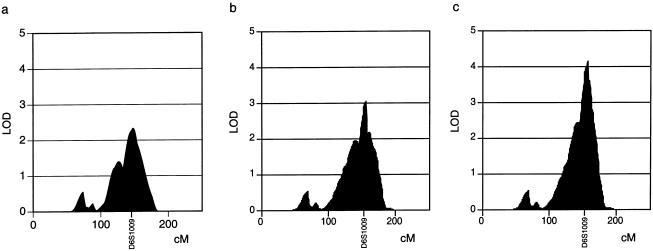
A genomewide linkage analysis yielded a peak that covered ∼90 cM on the long arm of chromosome 6, with a maximum multipoint LOD score of 2.3 at marker position D6S1009 (fig. 1). Because D6S1009 is located <1 cM from the IFNGR1 gene that encodes chain 1 of the interferon-γ (IFN-γ) receptor and because IFN-γ has been implicated in host defense against Helicobacter infection (Sawai et al. 1999; Kamradt et al. 2000), an additional marker, FA1, located in intron 6 of IFNGR1 (Altare et al. 1998), was included in the analysis. This raised the LOD score to 3.1 (fig. 2b).
Figure 1.
Genomewide linkage analysis of 143 sib pairs for the intensity of H. pylori infection, as defined by serum concentrations of H. pylori–reactive IgG.
Figure 2.
Multipoint LOD scores on chromosome 6q. a, Detailed profile of the multipoint LOD scores on chromosome 6q. b,Change in multipoint LOD scores when STR marker FA1, which is located in intron 6 of IFNGR1 (Altare et al. 1998), is included. c, Change in multipoint LOD scores when IFNGR1 variants –56T→C, H318P, and L450P are included as additional markers. The increase in LOD score can be explained by the combined markers becoming successively more informative, as indicated by an increment in information content from 0.73 to 0.84 and 0.94 (Kruglyak and Lander 1995).
Analysis of Polymorphisms
All seven exons and 1,045 nt of the 5′ flanking region of the IFNGR1 gene were sequenced in 10 siblings with extreme phenotypes from five families. Furthermore, exons 1 and 7 and the 5′ flanking region were sequenced in 20 additional siblings. Three variants (−611G→A, −527G→A, and –56C→T) were identified in the upstream region. Variant −611G→A was found in all but 4 of the 60 chromosomes studied, and −527G→A was found in only 6 of them; neither of these two variants showed evidence of an association with the phenotype, and they were not studied further. Variant −56C→T was found in approximately half of the chromosomes; 29 homozygous and 55 heterozygous carriers of the variant had higher levels of anti–H. pylori IgG than did the remaining 27 wild-type siblings (mean ± SD 1.33±0.44 log U/ml, 1.33±0.49 log U/ml, and 1.07±0.41 log U/ml, respectively).
In addition to the upstream variants, 1004A→C and 1400T→C exchanges were found in exon 7 of IFNGR1, resulting in replacements of histidine 318 and leucine 450 by proline residues (H318P and L450P, respectively). Among the 111 siblings studied, 14 carriers of H318P and 3 carriers of L450P were identified; both groups had higher anti–H. pylori IgG concentrations than the rest (for H318P, mean ± SD 1.52±0.52 log U/ml vs. 1.23±0.46 log U/ml; and for L450P, mean ± SD 1.51±0.90 log U/ml vs. 1.26±0.46 log U/ml, respectively).
The variants were subjected to a quantitative transmission/disequilibrium test in 111 parent-child trios. L450P and H318P were too rare to be evaluated; −56C→T was positively associated with high anti–H. pylori IgG concentrations (P=.03). When –56C→T, H318P, and L450P were included in the linkage analysis, the maximum LOD score increased to 4.2 (fig. 2c).
Frequencies in Unselected Individuals
The frequencies of −56C→T, H318P, and L450P were assessed in chromosomes of 100 unrelated individuals from the Senegalese study population and from 100 unrelated members of a German volunteer group (table 3). H318P and L450P were found in the Senegalese sample only, and −56C→T appeared to be more frequent among the Senegalese than among German sample.
Table 3.
Frequencies of IFNGR1 Variants in Unselected Individuals[Note]
|
% Affected Chromosomes in |
||
| Variant | African Subjects | White Subjects |
| −56C→T | 47 | 36 |
| 1004A→C (H318P) | 6 | 0 |
| 1400T→C (L450P) | 1 | 0 |
Note.— For the three variants combined, the difference between African and white chromosomes is significant (P<.02).
Discussion
As the key mediators of the T helper-1 (Th1) lymphocyte response, IFN-γ and its receptor are part of an important signaling system in the immune response to pathogens (Shtrichman and Samuel 2001). With regard to H. pylori, clinical studies have shown that the human immune response in the gastric mucosa is dominated by Th1 cells, which produce IFN-γ (D'Elios et al. 1997). In addition, studies in the mouse model of H. felis revealed that IFN-γ is involved in the pathogenesis of gastritis and in immunoprotection (Sawai et al. 1999; Kamradt et al. 2000).
Here, we show that high levels of circulating anti–H. pylori IgG are linked and associated with promoter and structural polymorphisms of IFNGR1. Anti–H. pylori IgG levels had elsewhere been shown to be a reliable indicator of H. pylori infection (Evans et al. 1989; Marchildon et al. 1999). Since the concentrations of antibodies against H. pylori may also reflect antibody production per se, which could be expected to be influenced by IFN-γ signaling, they were compared with levels of an antibody to ubiquitous dental-plaque bacteria, but no correlation was found. Therefore, we believe that it is not antibody production but the course of H. pylori infection that is reflected by the phenotype of anti–H. pylori IgG used here and that was found to be influenced by IFNGR1 polymorphisms.
We have not yet performed functional studies; therefore, we can only speculate about functional implications. The −56C→T variant affects a major transcriptional initiation site of IFNGR1 (Merlin et al. 1997), which suggests that it influences transcription. H318P is located in the intracellular domain of IFNGR1, between the binding site for janus kinase (JAK) 1 at amino acid positions 266–269 and a tyrosine residue at position 440, which is phosphorylated in the process of signal transduction by JAK1 in conjunction with JAK2 (Bach et al. 1997). L450P lies in close proximity to the phosphorylation site. Both variants introduce a proline residue, which may cause substantial structural alterations by changing the direction of the polypeptide backbone. Thus, they might affect the accessibility of Y440 to phosphorylation by JAKs and, thereby, influence IFNGR1 signal transduction.
It is also unclear whether −56C→T and the other variants cause a gain or loss of IFNGR1 function. Circumstantial evidence argues in favor of a loss: Clinical and epidemiological findings in humans, as well as experimental studies in mice, have indicated that serum antibody levels reflect Helicobacter infection and not immunoprotection (Blanchard et al. 1999); in addition, studies in the mouse model have shown that IFN-γ contributes to protection and clearance of the infection (Sawai et al. 1999; Kamradt et al. 2000). Therefore, since –56C→T was associated with high levels of anti–H. pylori antibodies, it may be concluded that the variant was associated with high susceptibility to H. pylori infection, which suggests that it causes a loss of IFNGR1 function. This presumption requires experimental validation.
Nevertheless, the reduced frequencies of the IFNGR1 variants found among whites would further explain the observation that, in some African countries, the occurrences of peptic ulcer and gastric cancer are remarkably low when compared with the high H. pylori prevalences that are found (Holcombe 1992; Mitchell et al. 2002). This observation prompted studies in the murine model, revealing that the Th1 response and gastritis caused by H. felis were ameliorated by helminth coinfection, which were interpreted to show that the pathology due to H. pylori may in Africa be mitigated because of frequent helminth co-infections (Fox et al. 2000). In conjunction with the experimental findings that IFN-γ signaling contributes to both clearance and pathology of Helicobacter infection (Sawai et al. 1999; Kamradt et al. 2000), our data provide some evidence suggesting that IFNGR1 variants might also contribute to reduced clearance and mitigated gastric pathology of H. pylori in African populations.
Previous studies on the human genetics of infection have revealed a crucial role for IFN-γ signaling in the immune defense, specifically against mycobacteria (Dupuis et al. 2000). Severe and atypical childhood mycobacterial diseases, classified as Mendelian susceptibility to mycobacteria (McKusick 1998), were found to be associated with rare mutations of IFNGR1 and other components of the IFN-γ signaling pathway, which strongly inhibit signal transduction. The variant −56C→T is common and presumably causes milder forms of functional impairment. Such polymorphisms have already been postulated to exist and to be involved in the susceptibility to classical mycobacterial diseases, which, like H. pylori infections, affect hundreds of millions of humans worldwide (Dupuis et al. 2000). That additional variants of the IFN-γ signaling pathway were identified by a genomewide linkage analysis provides further proof of principle for the value of genomewide approaches in the analysis of susceptibility to infection and other complex genetic traits.
Acknowledgments
This study was financed by institutional funds of the Bernhard Nocht Institute. The authors are indebted to the people of Ndombo, for their willingness to cooperate, and to the health authorities of Richard Toll and Dakar, for support. They also thank Bruno Gryseels and his team, for logistic help, and appreciate the expert technical assistance of Birgit Muntau and Gerd Ruge. This work is part of the M.D. thesis of T.T.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/ (for order and genetic distances of microsatellite markers)
- EMBL Nucelotide Sequence Database, http://www.ebi.ac.uk/embl/ (for human IFNGR1 promoter −527 [accession number AJ490332] and exon 7 1004A→C [accession number AJ490331])
- National Center for Biotechnology Information Nucleotide Database, http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=Nucleotide (for human IFNGR1 primer sequences [working draft sequence NT_025741.8])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IFNGR1 [MIM 107470])
References
- Abecasis GR, Cardon LR, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams LJ, Mitchell PB, Fielder SL, Rosso A, Donald JA, Schofield PR (1998) A susceptibility locus for bipolar affective disorder on chromosome 4q35. Am J Hum Genet 62:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altare F, Jouanguy E, Lamhamedi-Cherradi S, Fondanèche MC, Fizame C, Ribiérre F, Merlin G, Dembic Z, Schreiber R, Lisowska-Grospierre B, Fischer A, Seboun E, Casanova JL (1998) A causative relationship between mutant IFNgR1 alleles and impaired cellular response to IFNγ in a compound heterozygous child. Am J Hum Genet 62:723–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach EA, Aguet M, Schreiber RD (1997) The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol 15:563–591 [DOI] [PubMed] [Google Scholar]
- Blanchard TG, Nedrud JG, Reardon ES, Czinn SJ (1999) Qualitative and quantitative analysis of the local and systemic antibody response in mice and humans with Helicobacter immunity and infection. J Infect Dis 179:725–728 [DOI] [PubMed] [Google Scholar]
- Blaser MJ (1998) Helicobacter pylori and gastric diseases. BMJ 316:1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elios MM, Manghetti M, De Carli M, Costa F, Baldari CT, Burroni D, Telford JL, Romagnani S, Del Prete G (1997) T helper 1 effector cells specific for Helicobacter pylori in the gastric antrum of patients with peptic ulcer disease. J Immunol 158:962–967 [PubMed] [Google Scholar]
- Dunn BE, Cohen H, Blaser MJ (1997) Helicobacter pylori. Clin Microbiol Rev 10:720–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis S, Doffinger R, Picard C, Fieschi C, Altare F, Jouanguy E, Abel L, Casanova JL (2000) Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol Rev 178:129–137 [DOI] [PubMed] [Google Scholar]
- Evans DJ Jr, Evans DG, Graham DY, Klein PD (1989) A sensitive and specific serologic test for detection of Campylobacter pylori infection. Gastroenterology 96:1004–1008 [DOI] [PubMed] [Google Scholar]
- Fox JG, Beck P, Dangler CA, Whary MT, Wang TC, Shi HN, Nagler-Anderson C (2000) Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces Helicobacter-induced gastric atrophy. Nat Med 6:536–542 [DOI] [PubMed] [Google Scholar]
- Graham DY, Malaty HM, Evans DG, Evans DJ Jr, Klein PD, Adam E (1991) Epidemiology of Helicobacter pylori in an asymptomatic population in the United States: effect of age, race, and socioeconomic status. Gastroenterology 100:1495–1501 [DOI] [PubMed] [Google Scholar]
- Holcombe C (1992) Helicobacter pylori: the African enigma. Gut 33:429–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamradt AE, Greiner M, Ghiara P, Kaufmann SH (2000) Helicobacter pylori infection in wild-type and cytokine-deficient C57BL/6 and BALB/c mouse mutants. Microbes Infect 2:593–597 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaty HM, Engstrand L, Pedersen NL, Graham DY (1994) Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med 120:982–986 [DOI] [PubMed] [Google Scholar]
- Marchildon P, Balaban DH, Sue M, Charles C, Doobay R, Passaretti N, Peacock J, Marshall BJ, Peura DA (1999) Usefulness of serological IgG antibody determinations for confirming eradication of Helicobacter pylori infection. Am J Gastroenterol 94:2105–2108 [DOI] [PubMed] [Google Scholar]
- McKusick VA (1998) Mendelian inheritance in man: catalog of human genes and genetic disorders. Johns Hopkins University Press, Baltimore [Google Scholar]
- Merlin G, van der Leede BJ, McKune K, Knezevic N, Bannwarth W, Romquin N, Viegas-Pequignot E, Kiefer H, Aguet M, Dembic Z (1997) The gene for the ligand binding chain of the human interferon gamma receptor. Immunogenetics 45:413–421 [DOI] [PubMed] [Google Scholar]
- Mitchell HM, Ally R, Wadee A, Wiseman M, Segal I (2002) Major differences in the IgG subclass response to Helicobacter pylori in the first and third worlds. Scand J Gastroenterol 37:517–522 [DOI] [PubMed] [Google Scholar]
- Müller-Myhsok B, Stelma FF, Guisse-Sow F, Muntau B, Thye T, Burchard GD, Gryseels B, Horstmann RD (1997) Further evidence suggesting the presence of a locus, on human chromosome 5q31-q33, influencing the intensity of infection with Schistosoma mansoni. Am J Hum Genet 61:452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius M, Wex T, Muller-Dietz G, Leodolter A, Schilling D, Malfertheiner P (2001) Comparative evaluation of two hemagglutination tests for the detection of anti–Helicobacter pylori antibodies. Diagn Microbiol Infect Dis 41:221–223 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai N, Kita M, Kodama T, Tanahashi T, Yamaoka Y, Tagawa Y, Iwakura Y, Imanishi J (1999) Role of gamma interferon in Helicobacter pylori–induced gastric inflammatory responses in a mouse model. Infect Immun 67:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkein HA, Berry CR, Purkall D, Burmeister JA, Brooks CN, Tew JG (2001) Phosphorylcholine-dependent cross-reactivity between dental plaque bacteria and oxidized low-density lipoproteins. Infect Immun 69:6612–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtrichman R, Samuel CE (2001) The role of gamma interferon in antimicrobial immunity. Curr Opin Microbiol 4:251–259 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]