Abstract
The CGG repeat in the 5′ untranslated region of the fragile X mental retardation 1 gene (FMR1) exhibits remarkable instability upon transmission from mothers with premutation alleles. A collaboration of 13 laboratories in eight countries was established to examine four issues concerning FMR1 CGG-repeat instability among females with premutation (∼55–200 repeats) and intermediate (∼46–60 repeats) alleles. Our central findings were as follows: (1) The smallest premutation alleles that expanded to a full mutation (>200 repeats) in one generation contained 59 repeats; sequence analysis of the 59-repeat alleles from these two females revealed no AGG interruptions within the FMR1 CGG repeat. (2) When we corrected for ascertainment and recalculated the risks of expansion to a full mutation, we found that the risks for premutation alleles with <100 repeats were lower than those previously published. (3) When we examined the possible influence of sex of offspring on transmission of a full mutation—by analysis of 567 prenatal fragile X studies of 448 mothers with premutation and full-mutation alleles—we found no significant differences in the proportion of full-mutation alleles in male or female fetuses. (4) When we examined 136 transmissions of intermediate alleles from 92 mothers with no family history of fragile X, we found that, in contrast to the instability observed in families with fragile X, most (99/136 [72.8%]) transmissions of intermediate alleles were stable. The unstable transmissions (37/136 [27.2%]) in these families included both expansions and contractions in repeat size. The instability increased with the larger intermediate alleles (19% for 49–54 repeats, 30.9% for 55–59, and 80% for 60–65 repeats). These studies should allow improved risk assessments for genetic counseling of women with premutation or intermediate-size alleles.
Introduction
The fragile X syndrome is one of a group of disorders caused by an expansion of a trinucleotide repeat. The fragile X mental retardation 1 gene (FMR1) (MIM 309550) was identified in 1991 and was found to include a CGG repeat within the first exon (Fu et al. 1991; Oberlé et al. 1991; Verkerk et al. 1991). In the general, unaffected population, the repeat is usually inherited stably, with repeat sizes varying from 6 to ∼50. In fragile X syndrome, as in the other repeat disorders, there are no distinct boundaries separating the different repeat-size categories. Individuals with intermediate-size alleles have ∼45 to ∼60 repeats, whereas males and females with premutation alleles have ∼55 to ∼200 repeats. The distinction between intermediate and premutation alleles is made by family history and repeat instability. Premutation alleles are known to be unstable and to expand to a full mutation (>200 repeats) in some family members. For intermediate alleles, the possibility of repeat instability is unknown; however, because of repeat size, the alleles are potential precursors of a full mutation in subsequent generations.
Affected males with fragile X full mutations typically exhibit a fragile site at Xq27.3 and have moderate-to-severe mental retardation. Other features frequently observed are large ears, prominent jaw, joint laxity, and macroorchidism. Additional abnormalities may include hyperactivity, attention deficits, and autismlike behavior. The prevalence of full-mutation males in the white population is ∼1/4,000 (Murray et al. 1996; Turner et al. 1996; Crawford et al. 2001). The disorder occurs in most ethnic and racial populations, but there is some evidence that the prevalence may vary from group to group (Crawford et al. 2002). In affected males with fully expanded repeats, the FMR1 promoter region is hypermethylated, resulting in transcriptional silencing of the gene (Oberlé et al. 1991; Sutcliffe et al. 1992). Thus, fragile X syndrome in males results from the absence of the fragile X mental retardation protein (FMRP) (Pieretti et al. 1991). Although nearly all affected males have full-mutation expansions, a few individuals have been reported with deletions in all, or in a portion of, the gene (Gedeon et al. 1992; Wöhrle et al. 1992; Gu et al. 1994; Meijer et al. 1994; Hirst et al. 1995; Quan et al. 1995; Parvari et al. 1999).
In families with fragile X, the CGG repeat is remarkably unstable and undergoes massive expansions to more than several hundred repeats in full-mutation alleles. Repeat instability is influenced by several factors. First, expansion to a full mutation occurs exclusively on transmission through premutation females. When a premutation allele is transmitted by a female, the repeat undergoes expansion in virtually every case, although it does not necessarily expand to a full mutation. In contrast, repeats transmitted from premutation males to their daughters may expand, contract, or remain unchanged (Nolin et al. 1996); however, with rare apparent exceptions, they do not expand to full-mutation alleles. Second, the instability of premutation alleles in females is correlated with the repeat size. Larger repeats carry greater risks of expansion than do smaller ones. Third, in the normal population, the CGG repeat is interrupted by AGG trinucleotides, most often at positions 10 and 20. In contrast, premutation alleles are distinguished by the absence of AGG or the presence of only one AGG interruption at the 5′ end of the repeat and long tracts of uninterrupted CGG repeats at the 3′ end (Eichler et al. 1994; Kunst and Warren 1994; Snow et al. 1994; Zhong et al. 1995).
The collaborative study presented here was undertaken to examine four issues regarding female transmissions of intermediate, premutation, and full-mutation alleles. As a first step, 13 laboratories shared pedigree information and DNA samples, to identify the smallest premutation alleles that expand to a full mutation in one generation. For premutation alleles, the precise number of repeats is difficult to determine, and size estimates from different laboratories may not be directly comparable. To ensure a uniform method for sizing, DNA samples were analyzed in parallel in a single laboratory, using a system designed to allow a careful determination of repeat size.
Second, we calculated the risks of full-mutation expansion for all premutation alleles in females, after correction for ascertainment. The relationship between the repeat size in premutation females and full-mutation expansions demonstrates higher risks for individuals with larger repeat sizes (Fu et al. 1991; Heitz et al. 1992; Yu et al. 1992; Snow et al. 1993; Väisänen et al. 1994; Nolin et al. 1996; Ashley-Koch et al. 1998). However, because virtually all of the families in these earlier studies included affected males, premutation alleles with a higher degree of instability may be overrepresented. Furthermore, in many cases, DNA testing was not performed on all family members, and males with mental retardation were more likely to be tested than were individuals with normal intelligence. Thus, the previously published studies may have overestimated the risks of expansion of premutation repeats.
Third, we examined whether expansion to a full mutation is related to the sex of the offspring. Two reports have suggested that full-mutation expansions are more likely to occur in male than in female offspring (Rousseau et al. 1994; Loesch et al. 1995). As discussed above, pedigree studies of human disorders are frequently distorted, because some family members may be unavailable for analysis. For fragile X, unaffected females with a full mutation may be tested less frequently than unaffected males, resulting in an underestimate for females. A more recent study (Ashley-Koch et al. 1998) concluded that the findings of Rousseau et al. (1994) and Loesch et al. (1995) may be a result of ascertainment bias. To examine this possibility, we analyzed the prenatal studies from 7 of the 13 laboratories. By limiting the analysis to prenatal studies, we ensured an absence of ascertainment bias in the data collection.
Fourth, we analyzed the inheritance of intermediate-size repeats in females with no family history of fragile X. The progression of repeat expansions to premutation size in families with fragile X is likely to occur over many generations, with a gradual accumulation of repeat units. This is observed clearly in families identified through an individual with the full mutation. In the older generations, the propensity of the smaller premutation alleles to expand is 100%, because they lead to the full mutation. Analysis of intermediate and small premutation repeats in families without full-mutation alleles will provide useful information to predict the expansion risks of larger alleles.
Subjects and Methods
Thirteen laboratories reviewed pedigrees of families with fragile X, to identify premutation females with full-mutation offspring for whom DNA was available. DNA samples from the females with the smallest premutation alleles from each laboratory were sent to the New York State Institute for Basic Research in Developmental Disabilities for analysis. The study was approved by the institutional review board at the New York State Institute for Basic Research in Developmental Disabilities. The DNA samples were coded, and all identifiers were removed. The FMR1 CGG repeat region was amplified by PCR, using primers 1 and 3, and was analyzed in parallel by PAGE (Nolin et al. 1996). A second series of analyses were conducted on a subset of samples with the smallest premutation alleles. To ensure accurate sizing, a premutation male whose repeat region had been sequenced was included in the analysis.
For the transmission studies of intermediate-size and premutation alleles, as well as for the prenatal studies, each laboratory summarized its data and sent the information to the New York State Institute for Basic Research in Developmental Disabilities. The risks of expansion to full mutation among all premutation-size alleles were determined with and without correction for ascertainment, on the basis of family data from nine laboratories (The John F. Kennedy Institute; Wessex Regional Genetics Laboratory; Department of Human Genetics, Emory University; Abteilung Humangenetik, Universität Ulm; Laboratoire de Diagnostic Génétique, University Hospital, Strasbourg; Department of Obstetrics and Gynecology, Helsinki University Central Hospital; Department of Medical Genetics, University of Antwerp; Université de Laval; and Department of Human Genetics, New York State Institute for Basic Research in Developmental Disabilities). To correct for ascertainment bias, the proband in families with fragile X was excluded. In extended pedigrees, if all first- and second-degree relatives were examined irrespective of the phenotype, additional corrections were not made. If a sibship was examined only because of an affected individual, the affected individual was removed. For families in which more than one affected sibling was identified at the same time, all were removed from the analysis.
The prenatal studies were contributed by seven laboratories (Laboratoire de Diagnostic Génétique, Faculté de Médecine et CHRU; Department of Medical Genetics, University of Antwerp; Wessex Regional Genetics Laboratory; Abteilung Humangenetik, Universität Ulm; Department of Clinical Genetics, Oulu University Hospital; Department of Obstetrics and Gynecology, Helsinki University Central Hospital; and Department of Human Genetics, New York State Institute for Basic Research in Developmental Disabilities).
The studies of intermediate alleles were contributed by five laboratories (Laboratoire de Diagnostic Génétique, Faculté de Médecine et CHRU, Strasbourg; The John F. Kennedy Institute; Wessex Regional Genetics Laboratory; Department of Human Genetics, Emory University; and Department of Human Genetics, New York State Institute for Basic Research in Developmental Disabilities). Individuals with intermediate alleles were often referred for fragile X testing because of a family history of mental retardation. In two laboratories (Emory University and Wessex Regional Genetics Laboratory), some intermediate alleles were identified from screening populations with developmental disabilities.
Sequence Analysis of the Premutation Allele from Females
Two stages of PCR were used to amplify the premutation allele from females prior to sequence analysis of the FMR1 CGG repeat. The first stage consisted of 25-μl reaction volumes, with 1× ThermoPol buffer (New England Biolabs), 2 mM MgSO4, 0.6 μM FRXPST283 forward primer (5′-AGG CGC TCA GCT CCG TTT CGG TTT CAC TTC-3′), 0.6 μM FRXPSF526 reverse primer (5′-AGC CCC GCA CTT CCA CCA CCA GCT CCT CCA-3′) (Levinson et al. 1994), 200 μM dATP, 200 μM dTTP, 200 μM dCTP, 100 μM dGTP, 100 μM 7 deaza-dGTP, 2 M betaine (Sigma), 4 U Deep Vent DNA Polymerase (New England Biolabs), and 100–200 ng DNA. The cycling was performed in an MJ Research PTC-100. Cycling conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 2 min, 62°C for 1 min, and 74°C for 2 min, followed by a 4°C hold. The products were separated on 3% NuSieve (BioWhittaker Molecular Applications) gel (0.5 μg/ml ethidium bromide, 1× TBE) for 6–8 h at 3.3 V/cm. Once the fragments were well resolved, the premutation band was excised, taking care to avoid the heteroduplex of the normal and premutation fragments. DNA was isolated from the gel slice, using a Qiaquick DNA gel extraction kit (Qiagen). The DNA was reamplified using 5 μl PCR product as a template in a 50-μl reaction with 1× native Pfu polymerase (Stratagene) plus buffer, 0.6 μM each forward and reverse primer, 200 μM dATP, 200 μM dTTP, 200 μM dCTP, 100 μM dGTP, 100 μM 7 deaza-dGTP, 10% glycerol, 1.5 M betaine (Sigma), and 2.5 U native Pfu polymerase (Stratagene). The cycling was performed in an MJ Research PTC-100, as follows: 98°C for 5 min, followed by 35 cycles of 98°C for 1 min, 65°C for 1 min, and 75°C for 2 min, followed by a 4°C hold. The PCR product was isolated using a Qiaquick PCR purification kit (Qiagen).
Sequencing Reaction
The PCR products were sequenced using the fmol DNA Cycle Sequencing kit (Promega), according to the manufacturer’s directions but with the following modifications: 5 μl PCR product was used as template, and the primers for sequencing were FRXPST283, for the forward reaction, and primer 2 (Brown et al. 1993), for the reverse direction. For the forward reaction, betaine was added (final concentration 1 M). The cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 30 s and 70°C for 30 s. For the reverse reaction, a final concentration of 1.75 M betaine was included in the reaction mixture. The cycling conditions were 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s, and 70°C for 1 min.
Results
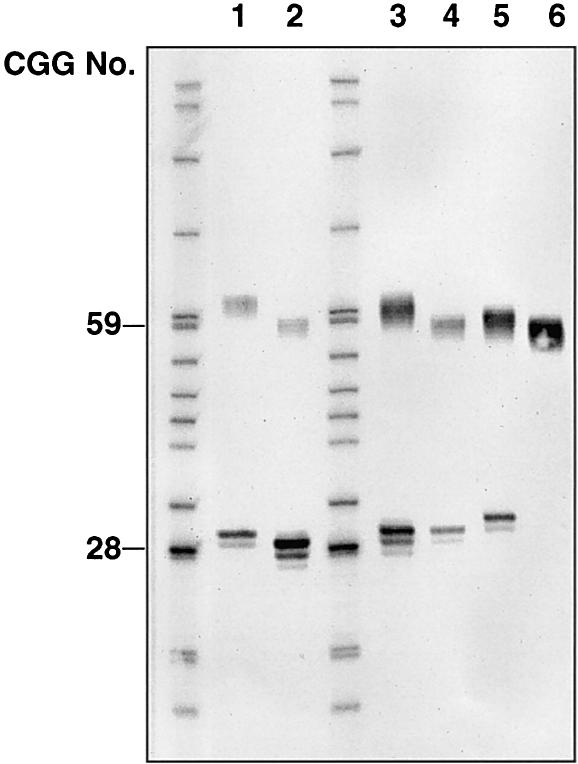
To identify the smallest premutation allele expanding to a full mutation in one generation, 13 laboratories reviewed fragile X pedigrees that included >1,500 females with premutation alleles. DNA samples were sent to one laboratory for analysis. A series of PCR analyses were performed to identify the smallest alleles among these samples.Figure 1 shows results of PCR analysis of DNA from five females with small premutation alleles. The two different samples (lanes 2 and 4) with 59 repeats were the smallest premutation alleles that expanded to a full mutation in one generation. To examine the repeat structure of the premutation alleles, sequence analysis of these alleles was performed. The studies revealed that neither premutation allele contained any AGG interruptions within the FMR1 repeat region (data not shown).
Figure 1.
Analysis of small premutation alleles from females with full-mutation offspring. Lane 1, female with 30 and 62 repeats. Lane 2, female with 29 and 59 repeats. Lane 3, female with 30 and 61 repeats. Lane 4, female with 30 and 59 repeats. Lane 5, female with 31 and 60 repeats. Lane 6, premutation male with 59 repeats. The two unmarked lanes are pBR322 digested with MspI as a size marker. FMR1 repeat-sizes for 28 and 59 CGGs are indicated (left).
Table 1 summarizes a total of 678 transmissions from 664 premutation females, after correction for ascertainment.Table 2 summarizes 1,338 transmissions from 936 mothers with premutation alleles, without correction for ascertainment. (Not all laboratories contributed to both sets of data.) The results confirmed the positive correlation between repeat size and risk of full-mutation expansion, as reported elsewhere (Fu et al. 1991; Heitz et al. 1991; Yu et al. 1992; Snow et al. 1993; Väisänen et al. 1994; Nolin et al. 1996; Ashley-Koch et al. 1998). After correction for ascertainment, however, the total percentage of full-mutation offspring in table 1 was lower than in table 2 or previous studies. The greatest reduction occurred in the repeat-size categories of 60–89. For these repeat sizes, the risks, after correction for ascertainment, were significantly smaller than those shown in table 2. For offspring of mothers with repeats >100, risks of full mutation were similar to those published elsewhere. Although, in most instances, the risks approached 100%, there were cases of expansion to larger premutation alleles or of contraction to smaller ones. In addition, five premutation alleles contracted to normal- or intermediate-size alleles. These were not included in the tables, since the analysis was limited to larger alleles. Three of the reversions from mother to child (82→33, 95→35, and 145→43) have been reported elsewhere (Brown et al. 1996). The fourth reversion contracted from 130 to 10 repeats (Holinski-Feder), and the fifth contracted from 70 to 54 repeats (Nolin and Brown). All of the reversions were transmitted to daughters. Four of the five daughters have a normal phenotype; the phenotype of the fifth is unknown, because the studies were performed for prenatal diagnosis. Stability of the reverted alleles cannot be determined at the present time, since none of the daughters have transmitted the allele to the next generation.
Table 1.
Transmission of FMR1 Premutation Repeats from Females, with Correction for Ascertainment
|
No. of Offspring with |
||||
| Repeat Sizeof MaternalAllele | No. of Mothers | Premutation | Full Mutation | % Expandedto FullMutation |
| 55–59 | 21 | 26 | 1 | 3.7 |
| 60–69 | 80 | 107 | 6 | 5.3 |
| 70–79 | 76 | 62 | 28 | 31.1 |
| 80–89 | 133 | 59 | 81 | 57.8 |
| 90–99 | 118 | 22 | 89 | 80.1 |
| 100–109 | 84 | 0 | 70 | 100 |
| 110–119 | 72 | 1 | 53 | 98.1 |
| 120–129 | 33 | 1 | 35 | 97.2 |
| 130–139 | 23 | 1 | 17 | 94.4 |
| 140–149 | 5 | 0 | 1 | 100 |
| 150–159 | 6 | 0 | 2 | 100 |
| 160–169 | 9 | 0 | 10 | 100 |
| 170–199 | 4 |
0 |
6 |
100 |
| Total | 664 | 279 | 399 | 58.8 |
Table 2.
Transmission of FMR1 Premutation Repeats from Females, without Correction for Ascertainment
|
No. of Offspring with |
||||
| Repeat Sizeof MaternalAllele | No. ofMothers | Premutation | Full Mutation | % Expandedto FullMutation |
| 55–59 | 27 | 35 | 2 | 5.4 |
| 60–69 | 108 | 124 | 29 | 19.0 |
| 70–79 | 90 | 65 | 64 | 49.6 |
| 80–89 | 192 | 76 | 206 | 73.0 |
| 90–99 | 187 | 34 | 224 | 86.8 |
| 100–109 | 116 | 4 | 151 | 97.4 |
| 110–119 | 98 | 4 | 128 | 97.0 |
| 120–129 | 36 | 3 | 61 | 95.3 |
| 130–139 | 38 | 0 | 55 | 100 |
| 140–149 | 17 | 0 | 31 | 100 |
| 150–159 | 10 | 0 | 12 | 100 |
| 160–169 | 10 | 0 | 12 | 100 |
| 170–199 | 7 |
0 |
11 |
100 |
| Total | 936 | 352 | 986 | 73.7 |
To examine sex bias in full-mutation transmissions, prenatal studies from seven laboratories were summarized (table 3). A total of 567 offspring from 448 mothers with premutation and full-mutation alleles were analyzed. There were more males than females, consistent with the normal excess of males to females in births. No differences were observed in the proportion of full-mutation male and female offspring born to mothers with premutation alleles. Among the full-mutation fetuses of full-mutation mothers, a small excess of full-mutation males was observed, but the difference was not statistically significant (χ21=1.35; P=.25).
Table 3.
Prenatal FMR1 Repeat Transmissions from Mothers with Premutation or Full Mutation Alleles
|
Fetus |
|||||
| Mother | Sex | Normal | Premutation | Full Mutation | Total |
| Premutation | Male | 115 | 15 | 96 | 226 |
| Premutation | Female | 98 | 17 | 90 | 205 |
| Full mutation | Male | 37 | 0 | 39 | 76 |
| Full mutation | Female | 33 |
0 |
27 |
60 |
| Total | 283 | 32 | 252 | 567 | |
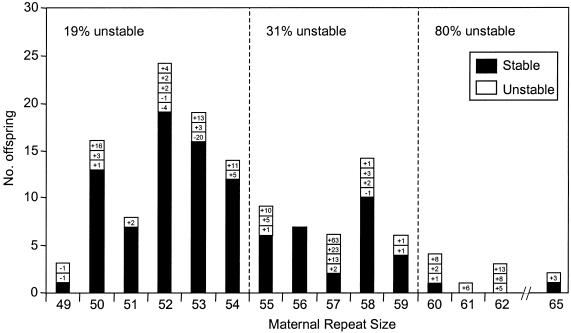
In figure 2 and table 4, we have summarized the FMR1 CGG repeats in 136 offspring of 92 mothers with intermediate alleles with 49–65 repeats and with no family history of fragile X. The intermediate alleles were limited to the larger sizes, since there is virtually no risk of expansion to a full mutation for alleles with fewer repeats. The repeat sizes of the offspring from each mother are detailed in the Appendix (table A1). Several trends could be observed among the transmissions. First, most (72.8%) of the intermediate alleles were stably inherited. A change was observed in 27.2% (37/136) of the offspring, and it included 31 expansions and 6 contractions. In contrast, transmissions of premutation alleles from females in families with fragile X are always associated with instabilities. Second, larger repeat sizes were more likely to be unstable than were smaller sizes (χ2=17.19; P<.0002). Specifically, 19% (16/84) of the mothers with 49–54 repeats had changes in their offspring, and the same was true of 30.9% (13/42) of those with 55–59 repeats and of 80% (8/10) of those with 60–65 repeats. Third, most of the contractions from mother to child occurred among the smallest intermediate sizes.
Figure 2.
Maternal transmission of intermediate alleles. The maternal repeat-sizes are shown on the X-axis, and the number of offspring on the Y-axis. The offspring with stably inherited alleles are shown in black. Each unstable transmission is shown as a white rectangle with the change in repeat number (+ or −) indicated.
Table 4.
Unstable Intermediate Alleles
|
Offspring Alleles |
||||
| Repeat Sizeof MaternalAlleles | No. Unstable/Total (%) | No.Contracted | No.Expanded | Repeat-SizeChanges |
| 49–54 | 16/84 (19.0) | 5 | 11 | −20 to +16 |
| 55–59 | 13/42 (30.9) | 1 | 12 | −1 to +63 |
| 60–65 | 8/10 (80.0) |
0 |
8 |
+1 to +13 |
| Total | 37/136 (27.2) | 6 | 31 | |
The intermediate allele transmissions to more than one sibling in a family are summarized in table 5. The families fall into three categories. In 17 families, all intermediate alleles were stably transmitted. In nine families, a mix of stable and unstable transmissions was observed, and in 5 families, all transmissions were unstable. Thus, the patterns of instability varied among families and were not necessarily related to maternal repeat size.
Table 5.
Intermediate-Allele Stability within Sibships
|
No. of Sibships |
||||
| Repeat Sizein Mother | Stablea | Unstablea | Mixedb | Total (%)a |
| 49–54 | 12 (26) | 2 (4) | 6 (10/7) | 20 (47) |
| 55–59 | 5 (13) | 2 (6) | 2 (2/2) | 9 (23) |
| 60–65 | 0 | 1 (3) | 1 (1/1) | 2 (5) |
Data in parentheses are number of transmissions.
Data in parentheses are number of stable/unstable transmissions among mixed sibships.
Discussion
The progression of trinucleotide repeats from a normal to an affected size is not well understood and remains an area of intense investigation. A retrospective examination of families with intermediate and premutation FMR1 alleles provides information about the repeat sizes that undergo rapid expansion to full mutations, information that can be valuable for genetic counseling. In addition, such studies provide insight into the mutational process. In the present study, the smallest alleles to undergo expansion to full mutation in one generation contain 59 CGG repeats with no AGG interruptions. These findings underscore the probable role of AGG interruptions in ensuring repeat stability from one generation to another. The study does not, however, rule out a slight possibility of a smaller premutation allele expanding to a full mutation in one generation, but it does suggest that this would be a rare event. To allow size comparisons among laboratories, a lymphoblastoid cell line from a female with 59 repeats has been established and is available from the American Type Culture Collection (accession number CRL-2704).
In the present study, full-mutation expansions in offspring of women with premutation-size alleles were reexamined to obtain accurate risk estimates for use in genetic counseling. Risk estimates can be determined by retrospective studies, as performed here, or by prospective studies. The former usually has the advantage of a large sample size but may have biases due to ascertainment. For example, in our study, correction for ascertainment was made because full-mutation males are more likely to be tested than are full-mutation females or premutation individuals. Prospective studies have the advantage of random sampling but may have smaller sample sizes. Table 6 compares the full-mutation expansions in the present study with those in two prospective and one retrospective study without correction for ascertainment. (For the purposes of comparison, all figures are expressed as percent expansion to full mutation.) In the two prospective studies (Pesso et al. 2000; Toledano-Alhadef et al. 2001), nearly 25,000 pregnant women in Israel were screened for fragile X carrier status and were offered prenatal diagnosis when found to carry premutation alleles. The results of the prenatal testing are summarized in table 6, where the small sample sizes for risk estimation are obvious. A comparison of the different studies suggests that, because of the correction for ascertainment, the estimates derived in the present study may better represent the true risk of expansion to full mutation.
Table 6.
Comparison of Full-Mutation Expansion from Maternal Premutation Allele
|
% Full Mutation Expansion (No. of Offspring with Full-MutationExpansions/Total No. of Pregnancies) |
||||
| MaternalRepeat Size | Pesso et al. 2000 | Toledano-Alhadef et al. 2001 | Nolin et al. 1996 | Present Study |
| 55–59 | 0 (0/11) | 0 (0/22) | 13 (3/22) | 4 (1/27) |
| 60–69 | 12 (1/8) | 10 (2/20) | 21(7/34) | 5 (6/113) |
| 70–79 | 50 (1/2) | 17 (1/6) | 58 (59/102) | 31 (28/90) |
| 80–89 | 50 (1/2) | … | 73 (78/107) | 58 (81/140) |
| 90–99 | 100 (1/1) | … | 94 (83/88) | 80 (89/111) |
| 100–200 | 75 (3/4) | … | 99 (177/179) | 98 (194/197) |
Among the premutation alleles transmitted from mother to child, five in the normal or intermediate range reverted to a smaller size. Reports of three other such reversions have been published (Snow et al. 1993; van den Ouweland et al. 1994; Vits et al. 1994). In all eight cases, the reversion has occurred only in transmission from mother to daughter. The apparent sex bias in these samples is intriguing but needs to be examined in a larger data set.
The intermediate-repeat alleles ascertained through individuals with no family history of the fragile X syndrome represent an interesting group of alleles that may be susceptible to repeat instability. These alleles were identified either through screening programs or because of a family history of mental retardation unrelated to fragile X. For the latter group, it is essential that clinicians obtain a careful history and perform testing of individuals with mental retardation whenever possible.
Our combined studies demonstrate that, as a group, the intermediate alleles exhibit a level of instability different from those in families with fragile X. Whereas fragile X premutation alleles always undergo repeat instability when transmitted by females, nearly 73% of the intermediate alleles were stably transmitted. For the unstable ones, some of the intermediate alleles that expanded on transmission in our study are likely to be premutation alleles that will continue to expand to a full mutation within several generations. Sixteen percent of the unstable intermediate transmissions, however, were contractions. This finding is in marked contrast to the repeat instability observed in families with fragile X, where contractions to smaller repeats from females are rare. Nevertheless, the pattern of a greater instability with larger repeats was similar for premutation alleles from families with fragile X and for intermediate alleles. Sullivan et al. (2002) also observed a positive correlation of repeat size and instability among intermediate alleles, although the percentage of unstable maternal transmissions was lower than that observed here. Interestingly, they observed a higher degree of instability in paternal than maternal transmissions of intermediate alleles.
How should families be counseled when an intermediate allele (50–60 repeats) is identified? Ideally, PCR analysis of other family members should be performed, to examine the allele instability within a family. Even so, not all alleles in this range will be transmitted unstably, and few transmissions may be available to define instability. In table 5, a summary of allele stability within sibships indicates that some families undergo only stable or unstable transmissions, whereas other families may undergo a mixture of both. Although the sample size is small, there is no clear association between repeat size and allele instability. The differences are likely due, at least in part, to differences in AGG interruptions within the FMR1 repeat. This study underscores the element of uncertainty that is likely to remain for each family.
When counseling pregnant females with premutation and intermediate alleles, clinicians must consider the risks of repeat instability in allele transmission from mother to child. For premutation alleles, there is a clear risk for expansion to a full mutation. For intermediate alleles, the risks of expansion cannot be easily determined because of our inability to differentiate stable intermediate alleles from unstable premutation alleles in the same size range. At present, this distinction can be made only by observing transmissions in future generations. Nevertheless, clinicians must provide risk estimates to assist families in making decisions about prenatal diagnosis. For this reason, we have examined the risk of full-mutation expansions among all females with 55–59 repeats. For several reasons, the actual risk for this group may be lower than the estimates shown in table 1. First, the 3.7% estimate may not be accurate, because it is based on only one full mutation in 27 transmissions. Second, and more important, only females from families known to be segregating fragile X were included. As a result, stable alleles in this size category were excluded. Third, in identifying the smallest premutation alleles that expand to a full mutation in one generation, the offspring of ∼1,500 premutation females were examined for full-mutation alleles. We identified only two premutation alleles with 59 repeats that expanded to full mutation in one generation, suggesting that expansions from this size category occur very infrequently. If we assume that 31% of offspring from women who have intermediate alleles with this repeat size inherited an unstable allele from their mothers, then the overall risk of expansion would be 1.1% (3.7% from table 1 [31%]). Thus, the risk of a full-mutation expansion in one generation from an allele of 55–59 repeats is 3.7%–1.1%. In considering whether to seek prenatal diagnosis, prospective parents and their physicians must carefully weigh the risks of chorionic villus sampling or amniocentesis against the risks of expansion. However, the concerns of parents must be regarded as an important factor, and prenatal diagnosis for reassurance may be warranted in some cases.
An additional issue for mothers with premutation alleles is the possibility of transmitting a premutation-size repeat. Recent findings of a progressive neurological disorder in a portion of older premutation males (Tassone et al. 2000; Hagerman and Hagerman 2002) and findings of premature ovarian failure among premutation females (Sherman, 2000) raise additional issues for families and genetic counselors to consider.
To summarize, our collaborative study has examined repeat instability among intermediate and premutation alleles transmitted by females and has concluded that 59 repeats is the smallest premutation allele known to expand to a full mutation in one generation. The risks of expansion for intermediate and premutation alleles summarized here should provide valuable information both for genetic counseling and for insight into the mutational pathway.
Acknowledgments
We thank members of the many families with fragile X for their participation and support. We also thank Dr. Carl Dobkin for reviewing the manuscript and for helpful discussion. This work was supported in part by the New York State Office of Mental Retardation and Developmental Disabilities and by National Institutes of Health grant HD29909 (to S.L.S. and A.S.).
Appendix A
Table A1.
Intermediate Allele Transmissions
| Repeat Size of Allele in | |
| Mother | Offspringa |
| 49 | 48, 48, 49 |
| 50 | 50 |
| 50 | 50 |
| 50 | 50 |
| 50 | 50 |
| 50 | 50, 50 |
| 50 | 50, 50 |
| 50 | 50 |
| 50 | 50 |
| 50 | 50, 66 |
| 50 | 53 |
| 50 | 51 |
| 50 | 50, 50 |
| 51 | 51 |
| 51 | 51, 51, 51, 53 |
| 51 | 51 |
| 51 | 51, 51 |
| 52 | 48 |
| 52 | 52 |
| 52 | 52 |
| 52 | 52 |
| 52 | 52 |
| 52 | 52 |
| 52 | 54 |
| 52 | 52 |
| 52 | 52, 52 |
| 52 | 52, 52, 54 |
| 52 | 51, 56 |
| 52 | 52 |
| 52 | 52 |
| 52 | 52, 52 |
| 52 | 52 |
| 52 | 52, 52, 52, 52 |
| 53 | 53, 53, 56 |
| 53 | 53 |
| 53 | 53 |
| 53 | 33, 53 |
| 53 | 53 |
| 53 | 53 |
| 53 | 53, 53 |
| 53 | 66 |
| 53 | 53 |
| 53 | 53, 53 |
| 53 | 53 |
| 53 | 53 |
| 53 | 53, 53 |
| 54 | 54 |
| 54 | 54 |
| 54 | 54 |
| 54 | 54 |
| 54 | 54, 54 |
| 54 | 59, 65 |
| 54 | 54 |
| 54 | 54 |
| 54 | 54, 54 |
| 54 | 54 |
| 54 | 54 |
| 55 | 65 |
| 55 | 55 |
| 55 | 55 |
| 55 | 56 |
| 55 | 55, 55, 55 |
| 55 | 55 |
| 55 | 60 |
| 56 | 56 |
| 56 | 56 |
| 56 | 56, 56, 56, 56 |
| 56 | 56 |
| 57 | 59 |
| 57 | 57 |
| 57 | 57 |
| 57 | 70, 80, 120 |
| 58 | 59, 60, 61 |
| 58 | 58, 58 |
| 58 | 58 |
| 58 | 58, 57 |
| 58 | 58 |
| 58 | 58 |
| 58 | 58 |
| 58 | 58 |
| 58 | 58, 58 |
| 59 | 59 |
| 59 | 59, 59 |
| 59 | 59, 60 |
| 59 | 60 |
| 60 | 68 |
| 60 | 62 |
| 60 | 60, 61 |
| 61 | 67 |
| 62 | 67, 70, 75 |
| 65 | 68 |
| 65 | 65 |
Unstable transmissions are underlined.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- American Type Culture Collection, http://www.atc.org (for lymphoblastoid cell line [accession number CRL-2704])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for fragile X syndrome [MIM 309550])
References
- Ashley-Koch AE, Robinson H, Glicksman AE, Nolin SL, Schwartz CE, Brown WT, Turner G, Sherman SL (1998) Examination of factors associated with instability of the FMR1 CGG repeat. Am J Hum Genet 63:776–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WT, Houck GE, Jeziorowska A, Levinson F, Ding X, Dobkin C, Zhong N, Henderson J, Brooks SS, Jenkins EC (1993) Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. JAMA 270:1569–1575 [PubMed] [Google Scholar]
- Brown WT, Houck GE. Ding X, Zhong N, Nolin S, Glicksman A, Dobkin C, Jenkins EC (1996) Reverse mutations in the fragile X syndrome. Am J Med Genet 64:287–292 [DOI] [PubMed] [Google Scholar]
- Crawford DC, Acuna JM, Sherman SL (2001) FMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med 3:359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Meadows KL, Newman JL, Taft LF, Scott E, Leslie M, Shubec L, Holmgreen P, Yeargin-Allsopp M, Boyle C, Sherman SL (2002) Prevalence of the fragile X syndrome in African-Americans. Am J Med Genet 110:226–233 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Holden JJ, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL (1994) Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet 8:88–94 [DOI] [PubMed] [Google Scholar]
- Fu Y-H, Kuhl DPA, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG Jr, Warren ST (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67:1047–1058 [DOI] [PubMed] [Google Scholar]
- Gedeon AK, Baker E, Robinson H, Partington MW, Gross B, Manca A, Korn B, Poustka A, Yu S, Sutherland GR, Mulley JC (1992) Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet 1:341–344 [DOI] [PubMed] [Google Scholar]
- Gu Y, Lugenbeel KA, Vockley JG, Grody WW, Nelson D (1994) A de novo deletion in FMR1 in a patient with developmental delay. Hum Mol Genet 3:1705–1706 [DOI] [PubMed] [Google Scholar]
- Hagerman RJ, Hagerman PJ (2002) The fragile X premutation: into the phenotypic fold. Curr Opin Genet Dev 12:278–283 [DOI] [PubMed] [Google Scholar]
- Heitz D, Rousseau F, Devys D, Saccone S, Abderrahim H, Le Paslier D, Cohen D, Vincent A, Toniolo D, Della Valle G, Honhson S, Schlessinger D, Oberlé I, Mandel JL (1991) Isolation of sequences that span the fragile X and identification of a fragile X–related CpG island. Science 251:1236–1239 [DOI] [PubMed] [Google Scholar]
- Hirst M, Grewal P, Flannery A, Slatter R, Maher E, Barton D Fryns JP, Davies K (1995) Two new cases of FMR1 deletion associated with mental impairment. Am J Hum Genet 56:67–74 [PMC free article] [PubMed] [Google Scholar]
- Kunst CB, Warren ST (1994) Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell 77:853–861 [DOI] [PubMed] [Google Scholar]
- Levinson G, Maddalena A, Palmer FT, Harton GL, Bick DP, Howard-Peebles PN, Black SH, Schulman JD (1994) Improved sizing of fragile X CCG repeats by nested polymerase chain reaction. Am J Med Genet 51:527–534 [DOI] [PubMed] [Google Scholar]
- Loesch DZ, Huggins R, Petrovic V, Slater H (1995) Expansion of the CGG repeat in fragile X in the FMR1 gene depends on the sex of the offspring. Am J Hum Genet 57:1408–1413 [PMC free article] [PubMed] [Google Scholar]
- Meijer H, de Graaff E, Merckx DML, Jongbloed RJE, de Die-Smulders CEM, Engelen JJM, Fryns JP, Curfs PMG, Oostra BA (1994) A deletion of 1.6 kb proximal to the CGG repeat of the FMR1 gene causes the clinical phenotype of the fragile X syndrome. Hum Mol Genet 3:615–620 [DOI] [PubMed] [Google Scholar]
- Murray A, Youings S, Dennis N, Latsky L, Linehan P, McKechnie N, Macpherson J, Pound M, Jacobs P (1996) Population screening at the FRAXA and FRAXE loci: molecular analyses of boys with learning difficulties and their mothers. Hum Mol Genet 5:727–735 [DOI] [PubMed] [Google Scholar]
- Nolin SL, Lewis FA 3rd, Ye LL, Houck GE Jr, Glicksman AE, Limprasert P, Li SY, Zhong N, Ashley AE, Feingold E, Sherman SL, Brown WT (1996) Familial transmission of the FMR1 CGG repeat. Am J Hum Genet 59:1252–1261 [PMC free article] [PubMed] [Google Scholar]
- Oberlé I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boué J, Bertheas MF, Mandel JL (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252:1097–1102 [DOI] [PubMed] [Google Scholar]
- Pesso R, Berkenstadt M, Cuckle H, Gak E, Pelet L, Frydman M, Barkai G (2000) Screening for fragile X syndrome in women of reproductive age. Prenat Diagn 20:611–614 [DOI] [PubMed] [Google Scholar]
- Quan F, Zonana J, Gunter K, Peterson KL, Magenis RE, Popovich BW (1995) An atypical case of fragile X syndrome caused by a deletion that includes the FMR1 gene. Am J Hum Genet 56:1042–1051 [PMC free article] [PubMed] [Google Scholar]
- Parvari R, Mumm S, Galil A, Manor E, Bar-David Y, Carmi R (1999) Deletion of 8.5 Mb, including the FMR1 gene, in a male with the fragile X syndrome phenotype and overgrowth. Am J Med Genet 83:302–307 [DOI] [PubMed] [Google Scholar]
- Pieretti, M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66:817–822 [DOI] [PubMed] [Google Scholar]
- Rousseau F, Heitz D, Tarleton J, Macpherson J, Malmgren H, Dahl N, Barnicoat A Mathew C, Mornet E, Tejada I, Maddalena A, Spiegel R, Schinzel A, Marcos JAG, Schorderet DF, Schaap T, Maccioni l, Russo S, Jacobs PA, Schwartz C, Mandel JL, Sherman S (1994) Higher rate of transition from fragile X premutations into full mutation in males than in females suggest post-conceptional expansion of the CGG repeats. Am J Hum Genet Suppl 55:A240 [Google Scholar]
- Sherman SL (2000) Premature ovarian failure in the fragile X syndrome. Am J Med Genet 97:189–194 [DOI] [PubMed] [Google Scholar]
- Snow K, Doud LK, Hagerman R, Pergolizzi RG, Erster SH, Thibodeau SN (1993) Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet 53:1217–1228 [PMC free article] [PubMed] [Google Scholar]
- Snow K, Tester DJ, Kruckeberg KE, Schaid DJ, Thibodeau SN (1994) Sequence analysis of the fragile X trinucleotide repeat: implications for the origin of the fragile X mutation. Hum Mol Genet 3:1543–1551 [DOI] [PubMed] [Google Scholar]
- Sullivan AK, Crawford DC, Scott EH, Leslie ML, Sherman SL (2002) Paternally transmitted FMR1 alleles are less stable than maternally transmitted alleles in the common and intermediate size range. Am J Hum Genet 70:1532–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST (1992) DNA methylation represses FRM-1 transcription in fragile X syndrome. Hum Mol Genet 1:397–400 [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ (2000) Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet 66:6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano-Alhadef H, Basel-Vanagaite L, Magal N, Davidov B, Ehrlich S, Drasinover V, Taub E, Halpern GJ, Ginott N, Shohat M (2001) Fragile-X carrier screening and the prevalence of premutation and full-mutation carriers in Israel. Am J Hum Genet 69:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Webb T, Robinson H (1996) Prevalence of fragile X syndrome. Am J Med Genet 64:196–197 [DOI] [PubMed] [Google Scholar]
- Van den Ouweland AMW, Deelen WH, Kunst CB, Uzielli ML, Nelson DL, Warren ST, Oostra BA, Halley DJJ (1994) Loss of mutation at the FMR1 locus through multiple exchanges between maternal X chromosomes. Hum Mol Genet 3:1823–1827 [DOI] [PubMed] [Google Scholar]
- Väisänen ML, Kähkönen M, Leisti J (1994) Diagnosis of fragile X syndrome by direct mutation analysis. Hum Genet 93:143–147 [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP (1991) Identification of a gene FMR-1 containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed] [Google Scholar]
- Vits L, De Boulle K, Reyniers E, Handig I, Darby JK, Oostra B, Willems PJ (1994) Apparent regression of the CGG repeat in FMR1 to an allele of normal size. Hum Genet 94:523–526 [DOI] [PubMed] [Google Scholar]
- Wöhrle D, Kotzot D, Hirst MC, Manca A, Korn B, Schmidt A, Barbi G, Rott HD, Poustka A, Davies KE, Steinbach P (1992) A microdeletion of less than 250 kb, including the proximal part of the FMR1 gene and the fragile-X site, in a male with the clinical phenotype of fragile-X syndrome. Am J Hum Genet 51:299–306 [PMC free article] [PubMed] [Google Scholar]
- Yu S, Mulley J, Loesch D, Turner G, Donelly A, Gedeon A, Hillen D, Kremer E, Lynch M, Pritchard M, Sutherland GR, Richards RI (1992) Fragile-X syndrome: unique genetics of the heritable unstable element. Am J Hum Genet 50:968–980 [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Yang W, Dobkin C, Brown WT (1995) Fragile X gene instability: anchoring AGGs and linked microsatellites. Am J Hum Genet 57:351–361 [PMC free article] [PubMed] [Google Scholar]