Abstract
Blepharophimosis syndrome (BPES), an autosomal dominant syndrome in which an eyelid malformation is associated (type I) or not (type II) with premature ovarian failure (POF), has recently been ascribed to mutations in FOXL2, a putative forkhead transcription factor gene. We previously reported 22 FOXL2 mutations and suggested a preliminary genotype-phenotype correlation. Here, we describe 21 new FOXL2 mutations (16 novel ones) through sequencing of open reading frame, 5′ untranslated region, putative core promoter, and fluorescence in situ hybridization analysis. Our study shows the existence of two mutational hotspots: 30% of FOXL2 mutations lead to polyalanine (poly-Ala) expansions, and 13% are a novel out-of-frame duplication. In addition, this is the first study to demonstrate intra- and interfamilial phenotypic variability (both BPES types caused by the same mutation). Furthermore, the present study allows a revision of the current genotype-phenotype correlation, since we found exceptions to it. We assume that for predicted proteins with a truncation before the poly-Ala tract, the risk for development of POF is high. For mutations leading to a truncated or extended protein containing an intact forkhead and poly-Ala tract, no predictions are possible, since some of these mutations lead to both types of BPES, even within the same family. Poly-Ala expansions may lead to BPES type II. For missense mutations, no correlations can be made yet. Microdeletions are associated with mental retardation. We conclude that molecular testing may be carefully used as a predictor for POF risk in a limited number of mutations.
Blepharophimosis-ptosis-epicanthus inversus syndrome (BPES [MIM 110100]) is an autosomal dominant genetic condition. In BPES type I, a complex eyelid malformation is associated with premature ovarian failure (POF), whereas in BPES type II, only the eyelid defect is observed (Zlotogora et al. 1983). Mutations in FOXL2 (GenBank accession number AF301906), a putative forkhead transcription factor gene located at 3q23 (MIM 605597), have recently been shown to cause both types of BPES (Crisponi et al. 2001). Murine Foxl2 mRNA was found to be highly expressed in embryonic mouse eyelids and in adult ovarian follicular cells (Crisponi et al. 2001). FOXL2 is the first human autosomal gene shown to be implicated in ovarian maintenance. It is a single-exon gene of 2.7 kb, and the predicted protein of 376 amino acids belongs to the large family of forkhead transcription factors, containing a 100-amino acid DNA-binding forkhead domain and a polyalanine (poly-Ala) tract of which the role has not yet been elucidated (Crisponi et al. 2001). Many members of the forkhead gene family are known to be involved in different developmental processes, and eight of them have already been shown to be implicated in human inherited developmental disorders (Carlsson and Mahlapuu 2002).
We described elsewhere the molecular characterization of a translocation in a patient with BPES and reported 22 FOXL2 mutations (De Baere et al. 2000, 2001). We and others have shown a preliminary genotype-phenotype correlation (Crisponi et al. 2001; De Baere et al. 2001): mutations resulting in a predicted truncated protein lead to type I BPES, whereas particular mutations leading to an extended protein cause type II BPES. Furthermore, we have assumed that FOXL2 haploinsufficiency may cause both types of BPES, suggested that there may be phenotypic overlap between the two types of BPES, and proposed that, in a fraction of patients with BPES, the genetic defect does not reside within the FOXL2 coding region and may be caused by a position effect (De Baere et al. 2001).
Since FOXL2 mutations have a pleiotropic effect, they have been expected to play a role in nonsyndromic POF (Crisponi et al. 2001; Prueitt and Zinn 2001). We analyzed FOXL2 in 100 patients with isolated POF but, so far, have not found causal mutations (De Baere et al. 2001, 2002). Recently, Harris et al. (2002) found two FOXL2 variants with a presumed pathogenic effect in 2 of 70 patients with nonsyndromic POF.
In the present report, we describe 21 new FOXL2 mutations, of which 16 are novel ones. Our study shows the existence of two mutational hotspots. In addition, we demonstrate, for the first time, that the assignment of an affected family to BPES type I or II is not always possible, since we observed intra- and interfamilial phenotypic variability. Furthermore, we revise the current genotype-phenotype correlation, since we found exceptions to it. We conclude that molecular testing may be carefully used as a predictor for POF risk in a limited number of mutations.
A total of 28 consenting probands—including affected individuals from two families with type I BPES, four families with type II, and one family in which both BPES types were present, plus 15 individuals with sporadic disease and patients from six families with BPES whose BPES type we were unable to assess—were screened for FOXL2 mutations. None of the probands have been included in previous FOXL2 mutation studies. Patients with BPES included in the present study presented with blepharophimosis, ptosis, epicanthus inversus, and telecanthus. POF (in BPES type I) was defined as cessation of menses for a duration of ⩾6 mo at age <40 years and an FSH concentration of >40 IU/liter. Patients with POF underwent a clinical assessment that included taking a complete medical and gynecological history and recording age at onset of menses, menstrual history, and age at menopause. A serum gonadotrophin assessment was performed. In six families and in most patients with sporadic disease, we were unable to assess the BPES type, mainly because of the prepubertal age and lack of endocrinological/gynecological data in affected females and because of an uninformative family history. All patients had an apparently normal karyotype. PCR amplification and sequencing of the FOXL2 coding region on genomic DNA was performed essentially as described by De Baere et al. (2001). For amplification and sequencing of the 5′ UTR and putative core promoter, we used the following primers: FOXL2-promF 5′-CGGCAGCAAACGGCAGAGCAA-3′; FOXL2-promR 5′-GGAGCTCAGCCTCTGGCCAT-3′; FOXL2-promseqF 5′-GGCAGGCCGGTCCAGGCT-3′; FOXL2-promseqR 5′-GCTCCACCGAGTTCCGCTT-3′. Sequencing was carried out on an ABI Prism 377 and 3100 sequencer (Applied Biosystems). For FISH analysis, probes (RP11 BAC RP11-54801, RPCI1 PACs 169-C10, 50-I6, 204-J22, 204-O7, and 300-F16; UK-MRC) were labeled by standard nick-translation, and FISH was performed as described elsewhere (De Baere et al. 1999). When mutations were found, we analyzed the segregation within the family for familial cases and in both parents for sporadic cases, to confirm the de novo nature of the mutation, whenever possible.
New FOXL2 Mutations
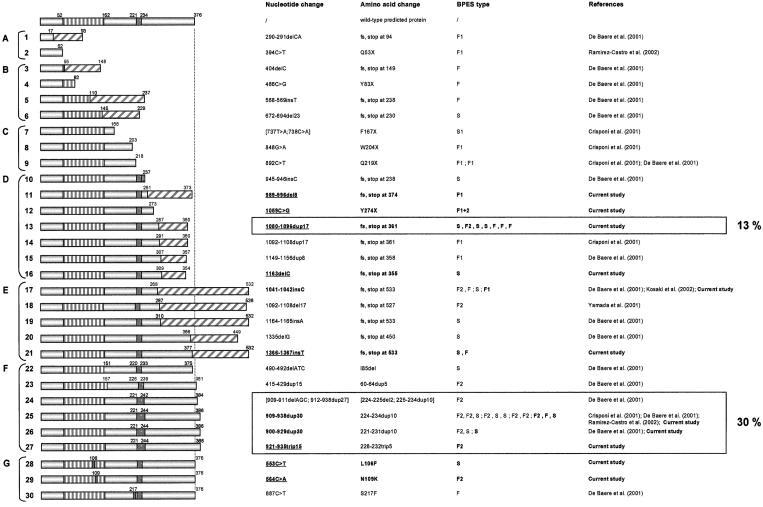
In the present study, we found 21 FOXL2 mutations in two families with BPES type I , four families with BPES type II, one family with BPES with intrafamilial phenotypic variability, nine patients with sporadic disease, and five families with BPES whose clinical type could not be determined. Specifically, 2 are missense mutations, 1 is a nonsense mutation, 12 are insertions or deletions leading to a frameshift, 5 are in-frame changes leading to poly-Ala expansions, and 1 is a microdeletion. Sixteen of them are novel. An overview of all FOXL2 mutations (53 in total, including those found in the current study) and variants described so far, as well as relevant clinical details, are given in table 1. None of the mutations found in the present study were present in 200 normal control chromosomes or in any of the unaffected family members whom we analyzed. We have subdivided the FOXL2 mutations in classes, according to their effect on the predicted protein (NCBI ORF finder). Groups A–D contain the predicted truncated proteins: without forkhead domain (group A), with partial forkhead domain (group B), with complete forkhead domain and without poly-Ala tract (group C), and with complete forkhead and poly-Ala domains (group D). Group E comprises frameshift mutations that lead to elongated proteins with complete forkhead and poly-Ala domains. Group F contains in-frame changes, and group G contains missense mutations. Group H comprises the cytogenetic rearrangements. Variants are included in group I. Figure 1 represents the predicted proteins of all FOXL2 mutations in the ORF reported so far. In figure 2, all relevant pedigrees are shown.
Table 1.
Overview of All FOXL2 Mutations and Variants Described So Far, as Well as Relevant Clinical Details[Note]
| Mutation Groupand Numbera | Effect | Position | Origin | BPESType | Clinical Information |
| A: | Truncation, no forkhead domain | ||||
| 1 | Fs, truncated to 93 aa | 290–291delCA | GBb | F1c | BPES and POF in 32-year-old female (De Baere et al. 2001) |
| 2 | Q53X | 394C→T | CO | F1 | Two-generation BPES type I pedigree; affected females have POF (Ramirez-Castro et al. 2002) |
| B: | Truncation, partial forkhead domain | ||||
| 3 | Fs, truncated to 148 aa | 404delC | BE | F | BPES in 8-year-old male proband, prepubertal sister, and father (De Baere et al. 2001) |
| 4 | Y83X | 486C→G | US | F | BPES in 3-year-old female and father (De Baere et al. 2001) |
| 5 | Fs, truncated to 237 aa | 568–569insT | US | F | BPES in father and 2 daughters (no endocrinological data) (De Baere et al. 2001) |
| 6 | Fs, truncated to 229 aa | 672–694del23 | US | S | BPES in prepubertal female (no endocrinological data) (De Baere et al. 2001) |
| C: | Truncation, complete forkhead domain, no poly-Ala tract | ||||
| 7 | F167X | [737T→A;738C→A] | Unknown | S1 | BPES in 21-year-old female showing growth retardation and delayed puberty at age 14 years. US: normal uterus and ovaries but no follicles. Low serum estradiol levels, elevated serum LHd and FSHe levels. Received hormonal treatment, resulting in breast development and regular menses. (Crisponi et al. 2001) |
| 8 | W204X | 848G→A | Unknown | F1 | Four-generation BPES type I pedigree (Crisponi et al. 2001) |
| 9 | Q219X | 892C→T | IT | F1 | Four-generation BPES type I pedigree (Amati et al. 1996; Crisponi et al. (2001) |
| CN | F1 | BPES, amblyopia, oligomenorrhea, and POF in affected females (De Baere et al. 2001) | |||
| D: | Truncation, complete forkhead domain and poly-Ala tract | ||||
| 10 | Fs, truncated to 237 aa | 945–946insC | BE | S | BPES in 22-year-old male, sporadic (De Baere et al. 2001) |
| 11 | Fs, truncated to 373 aa | 989–996del8 | AU | F1 | Three-generation family with BPES type I; index patient had menarche at age 26 years, menstrual cycles for 12 mo, amenorrhea at age 27 years (fig. 2, D11) (current study) |
| 12 | Y274X | 1059C→G | BE | F1+2 | Mother transmits BPES to daughter: oligomenorrhea at age 31 years, amenorrhea at age 32 years (low serum estradiol/progesterone and elevated serum LH and FSH levels; US: normal uterus/ovaries, secondary sex characteristics normal), ovum donation at age 33 years, resulting in one successful pregnancy (fig. 2, D12) (current study) |
| 13 | Fs, truncated to 360 aa | 1080–1096dup17 | BE | S | BPES in 8-year-old male, sporadic, de novo (current study) |
| BE | F2 | Affected mother transmits BPES to son (stenosis of lacrimal ducts and intestinal malrotation) (fig. 2, D13) (current study) | |||
| BE | S | BPES in 5-year-old female, sporadic (current study) | |||
| NO | S | BPES in 1-year-old female, sporadic, de novo (current study) | |||
| GB | F | Two-generation BPES pedigree, type unknown (fig. 2, D13) (current study) | |||
| AU | F | BPES in family of unknown type (male-to-male transmission; fig. 2, D13) (current study) | |||
| DE | F | Two-generation BPES pedigree, type unknown (fig. 2, D13) (current study) | |||
| 14 | Fs, truncated to 360 aa | 1092–1108dup17 | Unknown | F1 | Four-generation BPES type I pedigree (Crisponi et al. 2001) |
| 15 | Fs, truncated to 357 aa | 1149–1156dup8 | GB | F1 | Four-generation BPES type I pedigree (Harrar et al. 1995; De Baere et al. 2001) (repeated in Bell et al. 2001) |
| 16 | Fs, truncated to 354 aa | 1163delC | BE | S | BPES in 5-year-old female, sporadic (current study) |
| E: | Elongation with complete forkhead domain and poly-Ala tract | ||||
| 17 | Fs, extended to 532 aa | 1041–1042insC | GB | F2 | BPES in mother and 2 daughters (De Baere et al. 2001) |
| GB | F | Three-generation family with BPES of unknown type (Harrar et al. 1995; De Baere et al. 2001) (repeated in Bell et al. 2001) | |||
| (dup1036C = 1041–1042insC) | JP | S | BPES in 12-year-old female, sporadic (Kosaki et al. (2002) | ||
| MX | F1 | II:4 (fig. 2, E17) with oligomenorrhea at age 17 years, secondary amenorrhea at age 29 years (low estradiol and progesterone, elevated FSH and LH, atrophic ovaries on US, primordial follicles on ovarian biopsy, and normal secondary sex characteristics); received ovum donation at age 34 years. Two other affected sisters have POF. II:14 (fig. 2, E17) oligomenorrhea at age 17 years, secondary amenorrhea at age 22 years (current study) | |||
| 18 | Fs, extended to 526 aa | 1092–1108del17 | JP | F2 | BPES in 5-year-old female, 38-year-old father, 71-year-old grandfather, and 74-year-old paternal grandaunt (who has 3 children) (Yamada et al. 2001) |
| 19 | Fs, extended to 532 aa | 1164–1165insA | BE | S | BPES in 3-year-old female (no endocrinological data) (De Baere et al. 2001) |
| 20 | Fs, extended to 449 aa | 1335delG | US | S | BPES in male patient (De Baere et al. 2001) |
| 21 | Fs, extended to 532 aa | 1366–1367insT | NL | S | BPES in 1-year-old female patient, de novo (current study) |
| DE | F | Three-generation BPES pedigree, unknown type (fig. 2, E21) (current study) | |||
| F: | In-frame mutation | ||||
| 22 | I85del, 375 aa | 490–492delATC | BE | S | BPES in 6-year-old female patient (no endocrinological data) (De Baere et al. (2001) |
| 23 | 60–64dup5 | 415–429dup15 | BE | F2 | Affected mother (oligomenorrhea and heavy menses) transmits BPES to son (De Baere et al. 2001) |
| 24 | [224–225del2;225–234dup10] | [909–911delAGC; 912–938dup27] | US | F2 | Four-generation BPES type II pedigree. Fertility problems in 2 affected females, of whom 1 gave premature birth to 2 babies who lived only a few days, and another had a pregnancy after fertility treatment, respectively (De Baere et al. 2001) |
| 25 | 224–234dup10 | 909–938dup30 | Unknown | F2 | Five-generation BPES type II pedigree, affected females transmit disease (Crisponi et al. 2001) |
| Unknown | F2 | Two-generation family with BPES type II, affected mother transmits BPES to daughter (Crisponi et al. 2001) | |||
| Unknown | S | BPES in male, sporadic, de novo (Crisponi et al. 2001) | |||
| US | F2 | Four-generation family with BPES type II (Small et al. 1995; De Baere et al. 2001) | |||
| IT | S | BPES in 5-year-old male patient (De Baere et al. 2001) | |||
| US | S | BPES in 17-year-old female patient (no endocrinological data) (De Baere et al. 2001) | |||
| CO | F2 | Three-generation BPES type II pedigree. Affected female transmits disease (Ramirez-Castro et al. 2002) | |||
| CO | F2 | Three-generation BPES type II pedigree. Affected female transmits disease (Ramirez-Castro et al. 2002) | |||
| DE | F2 | Three-generation BPES type II pedigree (fig. 2, F25) (current study) | |||
| BE | F | Two-generation BPES pedigree (male-to-male transmission; fig. 2, F25) (current study) | |||
| FR | S | BPES in 10-year-old female, sporadic, de novo (current study) | |||
| 26 | 221–231dup10 | 900–929dup30 | US | F2 | BPES 25-year-old female proband and mother (De Baere et al. 2001) |
| BE | S | BPES in 23-year-old male patient (De Baere et al. 2001) | |||
| BE | S | BPES in 8-year-old male patient, sporadic (current study) | |||
| 27 | 228–232trip5 | 921–935trip15 | AU | F2 | Three-generation BPES type II pedigree (fig. 2, F27) (current study) |
| G: | Missense | ||||
| 28 | L106F | 553C→T | ZA | S | BPES in 1-year-old male patient, de novo (current study) |
| 29 | N109K | 564C→A | AU | F2 | Five-generation BPES type II pedigree (fig. 2, G29) (current study) |
| 30 | S217F | 887C→T | BE | F | BPES in father and 2 prepubertal daughters (no endocrinological data) (De Baere et al. 2001) |
| H: | Cytogenetic rearrangement | ||||
| 31 | t(3;21)(q23;q22.1) | 5′ to FOXL2 | US | S | BPES, no associated symptoms (Praphanphoj et al. 2000) |
| 32 | t(3;4)(q23;p15) | BPESC1 gene, 5′ to FOXL2 | JP | S | BPES, no associated symptoms (Fukushima et al. 1991; De Baere et al. 2000) |
| 33 | t(3;7)(q23;q32) | C3orf5 gene, 5′ to FOXL2 | IT | S | BPES, no associated symptoms (Boccone et al. 1994; Crisponi et al. 2001) |
| 34 | Microdeletion | FOXL2 and other genes | DK | F | BPES and mental retardation in father and prepubertal 7-year-old daughter (De Baere et al. 2001) |
| 35 | Microdeletion | FOXL2 and other genes | FI | S | BPES in 9-year-old male, mentally retarded, abnormally rotated ears, flat and broad nose bridge, scoliosis, high-arched palate, and umbilical hernia (current study) |
| I: | Variant ORFf | ||||
| F167F | 738C→T | Mix | NA | 12/140 (8.6%) POF, 4/46 (8.7%) XX male, 2/110 (1.8%) control chromg (De Baere et al. 2002) | |
| SI, NZ | NA | 5/140 (3.6%) POF chrom, 9/200 (4.5%) control chrom (Harris et al. 2002) | |||
| BE | S | BPES in 6-year-old female, sporadic (homozygous) (current study) | |||
| G174G | 759C→T | Mix | NA | 1/140 (0.7%) POF, 1/46 (2.2%) XX male, 0/110 control chrom (De Baere et al. 2002) | |
| A179G | 773C→G | SI, NZ | NA | 5/140 (3.6%) POF chrom, 9/200 (4.5%) control chrom (Harris et al. 2002) | |
| BE | S | BPES in 6-year-old female, sporadic (homozygous) (current study) | |||
| G187D | 797G→A | Unknown | NA | 1/46 (2.2%) XX male chrom; also present in unaffected family members (De Baere et al. 2002) | |
| 221–230del10 | 898–927del30 | SI | NA | 1/140 (0.7%) POF, 0/200 control chrom (Harris et al. 2002) | |
| Y258N | 1009T→A | NZ | NA | 1/140 (0.7%) POF, 0/200 control chrom (Harris et al. 2002) | |
| P285S | 1090C→T | Unknown | NA | 1/46 (2.2%) XX male chrom, 0/110 control chrom (De Baere et al. 2002) | |
| P286P | 1095T→G | Mix | NA | 3/140 (2.1%) POF, 3/46 (6.5%) XX male, 0/110 control chrom (De Baere et al. 2002) | |
| SI, NZ | NA | 3/140 (2.1%) POF chrom (Harris et al. 2002) | |||
| A301A | 1140G→T | Unknown | NA | 1/140 (0.7%) POF, 0/110 control chrom (De Baere et al. 2002) | |
| A369A | 1344G→T | Unknown | NA | 1/46 (2.2%) XX male, 0/110 control chrom (De Baere et al. 2002) | |
| Variant putative core promoter: | |||||
| Effect unknown | −317G→C | RU | S | Variant in typical patient with BPES, unaffected father and sister (current study) | |
| Effect unknown | −104G→A | DE | F | Variant in 5-year-old male patient with BPES, causal mutation found in ORF (current study) |
Note.— The mutations representing the two mutational hotspots demonstrated in the present study are boxed.
Numbering according to Crisponi et al. (2001).
AU = Australia; BE = Belgium; CN = China; CO = Colombia; FR = France; DE = Germany; DK = Denmark; FI = Finland; GB = United Kingdom; IT = Italy; JP = Japan; MX = Mexico; NL = The Netherlands; NO = Norway; NZ = New Zealand; RU = Russian Federation; SI = Slovenia; US = United States; ZA = South-Africa
fs = frameshift; US = ultrasound; F1 = familial, type I; F2 = familial, type II; F1+2 = familial, intrafamilial phenotypic variability; F = familial, type undetermined; S1 = sporadic, type I; S = sporadic; NA = not applicable.
LH = luteinizing hormone.
FSH = follicle-stimulating hormone.
ORF = open reading frame.
chrom = chromosomes.
Figure 1.
The predicted protein translations for FOXL2 mutations detected in this and previous studies are shown at the left and are assembled into groups. Groups A–D are predicted truncated proteins: A, without forkhead domain; B, with partial forkhead; C, with complete forkhead and without poly-Ala tract; D, with complete forkhead and poly-Ala domains. Group E comprises frameshift mutations leading to elongated proteins with complete forkhead and poly-Ala domains; group F, in-frame mutations; and group G, missense mutations. Vertical stripes indicate the forkhead domain, dark gray shows the poly-Ala tract, and diagonal stripes represent novel amino acids caused by a frameshift mutation. In the first column, the corresponding nucleotide changes are represented. The numbering is according to Crisponi et al. (2001). Mutations found in the present study are indicated in bold, and novel mutations are underlined. In the second column, the respective amino acid changes are shown. The two mutational hotspots demonstrated in the present study are boxed, and their percentages are shown at the right: 30% of the mutations lead to poly-Ala expansions, and 13% are a novel duplication 1080–1096dup17. Abbreviations: fs = frameshift; stop = stop codon. In the third column, the BPES type is shown (F1 = familial, type I; F2 = familial, type II; F1+2 = familial, occurrence of both BPES types in the same family; F = familial, type undetermined; S1 = sporadic, type I; and S = sporadic).
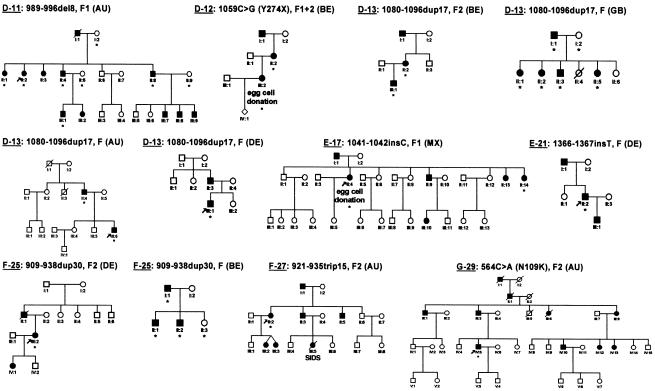
Figure 2.
Pedigrees and respective FOXL2 mutations of patients with familial BPES, represented in accordance with the subdivision and numbering of figure 1. For patients with sporadic disease, no pedigree information is shown. Affected individuals are indicated by filled symbols; individuals tested are indicated by an asterisk below the symbol. SIDS = sudden infant death syndrome. Abbreviations: AU = Australia; BE = Belgium; DE = Germany; GB = United Kingdom; MX = Mexico.
The first series of mutations found in the present study belongs to the group of the predicted truncated proteins with complete forkhead and poly-Ala domains (group D) (table 1; fig. 1, translations 10–16). Of particular interest is the novel and unique mutation 1059C→G (Y274X) found in a family in which a mother affected by BPES type II transmitted the disease to her daughter, affected by BPES type I. The latter received ovum donation at age 33 years, resulting in one successful pregnancy (fig. 2, D12). This is the first reported case in which the occurrence of both types of BPES in the same family caused by the same mutation is clearly documented (intrafamilial phenotypic variability).
The second series of new mutations belongs to the group of predicted elongated proteins with complete forkhead and poly-Ala domains (group E) (table 1; fig. 1, protein translations 17–21). It is interesting that the recurrent insertion 1041–1042insC, which was previously found in one family with type II BPES, one family of unknown type, and a patient with sporadic disease (De Baere et al. 2001; Kosaki et al. 2002), was found here in a family with type I BPES, in which one of the affected females received ovum donation (fig. 2, E17). This is the first mutation shown to lead to both BPES types in different families (interfamilial phenotypic variability).
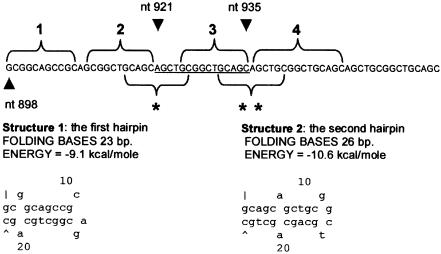
The third series of new mutations belongs to the group of in-frame changes (group F) (table 1; fig. 1, protein translations 22–27). It is notable that we found, in a family with BPES type II, a novel in-frame 15-bp triplication 921–935trip15, leading to a poly-Ala expansion 228–232trip5 (fig. 2, F-27). This is the first triplication observed in the poly-Ala tract of FOXL2. It may be explained by a replication error, further suggesting that DNA hairpin–induced polymerase slippage is the most probable mechanism underlying in-frame changes in the poly-Ala tract of FOXL2 (hypothesis outlined in fig. 3), as we suggested elsewhere (De Baere et al. 2001).
Figure 3.
Explanation of the mechanism of the novel triplication 921–935trip15 by a replication error. Predictions of DNA hairpins are based on MFOLD (Walter et al. 1994). When the DNA polymerase is stalling because of the GC richness of the region, the sequences 1 and 2 of the newly synthesized strand form a DNA hairpin because they are self-complementary (structure 1). This allows the sequence 3 to hybridize with the template of the region 2, since they have exactly the same sequence, and this leads to a duplication of the underlined sequence (nt 921–935). Once this extra sequence is present (4), there may be a rearrangement of the hairpin, which leads to the formation of another one involving the sequences (single and double asterisk). The formation of the second hairpin is favorable, because it is more stable (by an energy excess of 1.5 kcal/mole; see structure 2). Finally, the 3′ end of the sequence 4 hybridizes with the template of sequence 2, which results in the triplication after escaping repair. Further, notice that sequences 1–4 form a very stable, almost perfect, hairpin (−28 kcal/mole).
The fourth series of mutations belongs to the group of missense changes (group G) (table 1; fig. 1, protein translations 28–30). In a male patient with sporadic BPES and a male patient from a family with BPES type II (fig. 2, G-29), we found a transition 553C→T (L106F) and a transversion 564C→A (N109K), respectively. Both missense mutations are novel, unique, and the first ones reported in the forkhead domain. The absence of L106F in the unaffected parents, the drastic amino acid change (asparagine to lysine) in N109K, and the absence of the mutations in 200 control chromosomes are suggestive of a pathogenic effect of both mutations. In addition, leucine at position 106 and asparagine at position 109 are both conserved in 24 forkhead domains of human genes (Crisponi et al. 2001) and in four orthologous genes (mouse, rat, goat, and pufferfish) (Cocquet et al. 2002).
Furthermore, we identified a microdeletion (group H) in a male patient with sporadic BPES, mental retardation, and other associated symptoms. FISH analysis, using probes from the BPES region at 3q23, showed a heterozygous deletion in 100% of the mitoses studied (50 per probe) (data not shown). The microdeletion, estimated to be at least 200 kb, contains several known genes: FOXL2, C3orf5 (Crisponi et al. 2001), BPESC1 (De Baere et al. 2000), RBP1, RBP2, β′-COP (De Baere et al. 1999), and other as-yet-unidentified genes. Furthermore, it contains the human counterpart of an 11.7-kb deletion, identified in the polled intersex (PIS) goat and known to contain elements controlling the expression of several genes of that region (Schibler et al. 2000; Pailhoux et al. 2001). The fine mapping of the extent of this and other microdeletions will be the subject of further studies. Polymorphic and unclassified variants (group I) have been shown in table 1.
Finally, we were unable to find a causal mutation in one family with BPES of unknown type and in six sporadic BPES cases using the current strategy. This may be explained by a misdiagnosis in some cases, by a total gene deletion, or by a defect residing outside the transcription unit (position effect), which may be missed through the current screening method. Further attempts are under way to elucidate the molecular defects in these patients.
Spectrum of FOXL2 Mutations
Forty-seven percent (25/53) of all FOXL2 mutations reported so far in the ORF are changes leading to a frameshift. Thirty-four percent (18/53) are in-frame changes, of which the majority lead to poly-Ala expansions (16/53). Thirteen percent (7/53) are nonsense mutations, and 6% (3/53) are missense mutations, of which two are located in the forkhead domain (fig. 1). In addition, two microdeletions containing FOXL2 and other genes were found in patients with BPES, as were three balanced translocations, which were located <200 kb 5′ of FOXL2 and exerted a position effect (table 1).
Polyalanine Expansions
The present study further confirms the existence of a mutational hotspot in the region coding for the poly-Ala domain, since 30% of all mutations in the ORF lead to poly-Ala expansions, resulting mainly in BPES type II (fig. 1; table 1). There are at least six other genes in which stable poly-Ala expansions exceeding a critical number have been shown to cause human disease: mutations of HOXD13 in synpolydactyly (MIM 142989), of RUNX2 in cleidocranial dysplasia (MIM 600211), of PABP2 in oculopharyngeal muscular dystrophy (MIM 164300), of ZIC2 in holoprosencephaly (MIM 603073), of HOXA13 in hand-foot-genital syndrome (MIM 142959), and of ARX in X-linked mental retardation (MIM 300382) (Stromme et al. 2002). Although the mutational mechanisms may differ, the net result (addition of 1–10 extra alanine residues) is the same. So far, the role of the poly-Ala tract in FOXL2 has not yet been elucidated. However, the number of alanine residues (i.e., 14) is strictly conserved among the human, mouse, rat, and goat, suggesting the existence of strong functional or structural constraints (Cocquet et al. 2002). In several transcription factors, alanine-rich regions, which form α-helices, were found to be responsible for repression of target genes (Han and Manley 1993). Indeed, computer-assisted secondary structure predictions (nnPredict-UCSF) suggest that the poly-Ala tract of FOXL2 consists of an α-helical region (data not shown). The poly-Ala expansions observed in FOXL2 either may directly interfere with protein-protein interactions or may distort an essential secondary structure. To elucidate the true effect of the poly-Ala expansion on FOXL2, further studies are required.
Other Recurrent Mutations
Apart from poly-Ala expansions, we found a novel duplication 1080–1096dup17 in seven families (13% of all mutations in the ORF reported so far). This duplication occurs de novo in patients with sporadic disease and in unrelated families of heterogeneous ethnic origins (table 1), suggesting an independent origin and representing a second mutational hotspot. Other recurrent mutations are 892C→T (Q219X), found in families of Italian and Chinese origin with BPES type I (table 1). The insertion 1041–1042insC was found in four unrelated families, including a patient with sporadic disease (table 1). Finally, an insertion in the stop codon 1366–1367insT was found twice, de novo in a patient with sporadic disease and in a family (table 1). These data suggest that the last three mutations also have an independent origin.
Intra- and Interfamilial Phenotypic Variability
Elsewhere, we described menstrual abnormalities and reduced female fertility in two families with BPES type II, suggesting phenotypic overlap between both BPES types (De Baere et al. 2001). Bell et al. (2001) described a family with BPES type I in which the first generations of affected females are infertile and three affected young women in the youngest generation have normal pelvic ultrasound and hormone levels, suggestive of phenotypic variability. However, in this family, early age of the affected women may preclude accurate prediction of their ovarian phenotypic outcome, on the basis of these test results, since the onset of POF usually occurs at a later age.
In the present study, we show, for the first time, families with clear intra- and interfamilial phenotypic variability caused by the same mutation (number 12 and 17, respectively, in table 1 and fig. 1). Phenotypic variability is an important item for genetic counseling, because parents considering reproduction would like to know how severely affected a child will be. Variability may be caused by a combination of the effects of “modifier” genes and the environment (modulating the severity of POF or the age at onset). Haploinsufficient conditions are especially sensitive to such effects (Veitia 2002).
Revised Genotype-Phenotype Correlation
To add novel insights to the genotype-phenotype correlation demonstrated by us and others (Crisponi et al. 2001; De Baere et al. 2001), we revised genotypes and phenotypes of this and previous studies. The establishment of a reliable genotype-phenotype correlation provides challenges for genetic counseling, since molecular testing may allow prediction of the development of POF.
We observe that mutations leading to predicted truncated proteins without forkhead domain (group A), or with complete forkhead domain but without poly-Ala tract (group C), lead almost certainly to BPES type I. Mutations of group A are supposed to lead to haploinsufficiency of FOXL2, since DNA-binding activity is abolished because of disruption of the forkhead domain. Although, in general, nonsense- or frameshift-mutated transcripts are degraded by nonsense-mediated decay (NMD), intronless genes may be insensitive to NMD (as reported for the melanocortin 4-receptor gene; Brocke et al. 2002). The intronless FOXL2 gene might therefore be NMD insensitive, giving rise to stable mutated transcripts and expressed mutated proteins. For none of the mutations that lead to predicted proteins with partial forkhead domain (group B) can a correlation be made, because of lack of informative families.
Nonsense mutation 1059C→G (Y274X) (fig. 1, translation 12), which belongs to the group of predicted truncated proteins with complete forkhead and poly-Ala domains (group D), leads to intrafamilial phenotypic variability, whereas 1080–1096dup17 (fig. 1, translation 13) occurs in patients with BPES of unknown type and, surprisingly, also in a pedigree with BPES type II. Taken together, these two truncating mutations of group D lead to a BPES phenotype that would not have been expected on the basis of the previous genotype-phenotype correlation, not accounting for interindividual variability.
Another mutation 1041–1042insC (fig. 1, translation 17), which belongs to the group of predicted elongated proteins with complete forkhead and poly-Ala domain (group E) leads, surprisingly, to BPES type I and to BPES type II in different families. This is the third mutation that is not in line with the previously suggested genotype-phenotype correlation and is the second mutation found to lead to both types of BPES, although not in the same family (interfamilial phenotypic variability).
In-frame mutations 909–938dup30, 900–929dup30, and 921–935trip15 (fig. 1, translation 25, 1–26, and 1–27, respectively), which belong to group F and lead to poly-Ala expansions, occur in families with BPES of unknown type and in families with BPES type II. So far, we can conclude that poly-Ala expansions most likely lead to BPES type II and may allow childbearing.
Missense mutation 564C→A (N109K) of group G leads to BPES type II (fig. 1, 29), whereas, for the other ones, no correlations can be made. The microdeletions in group H occur in patients with sporadic or familial BPES, with mental retardation as a major associated feature.
In conclusion, we assume that for predicted proteins with a truncation before the poly-Ala tract, the risk for development of POF is high. For mutations that lead to a predicted truncated or extended protein containing an intact forkhead and poly-Ala tract, no clear-cut predictions are possible so far, since such mutations may lead to the two types of BPES (even within the same family). Moreover, our data strongly suggest that poly-Ala expansions lead to BPES type II. For missense mutations, no predictions can be made yet, because of the small number and lack of informative families. Microdeletions lead to a contiguous gene syndrome, including mental retardation, but in most cases, that is already diagnosed before molecular analyses. Finally, we conclude that molecular testing may be carefully used as a predictor for POF risk in a limited number of mutations.
Acknowledgments
This work was supported by Fonds voor Wetenschappelijk Onderzoek–Vlaanderen grant KAN 315-201.99N (to E.D.B. and L.M.) and by a Bijzonder Onderzoeksfonds grant from the Ghent University (to E.D.B.). We are most grateful to the families, clinicians, and researchers who participated in the present study.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for FOXL2 [accession number AF301906])
- MFOLD, http://bioweb.pasteur.fr/seqanal/interfaces/mfoldsimple.html
- NCBI ORF finder, http://www.ncbi.nlm.nih.gov/gorf/orfig.cgi
- nnPredict-UCSF, http://www.cmpharm.ucsf.edu/cgi-bin/nnpredict.pl
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BPES [MIM 110100], FOXL2 [MIM 605597], SPD [MIM 142989], CCD [MIM 600211], OPMD [MIM 164300], HPE [MIM 603073], HFGS [MIM 142959], and X-L MR [MIM 300382])
- UK-Human Genome Mapping Project, http://www.hgmp.mrc.ac.uk/
References
- Amati P, Gasparini P, Zlotogora J, Zelante L, Chomel JC, Kitzis A, Kaplan J, Bonneau D (1996) A gene for premature ovarian failure associated with eyelid malformation maps to chromosome 3q22-q23. Am J Hum Genet 58:1089–1092 [PMC free article] [PubMed] [Google Scholar]
- Bell R, Murday VA, Patton MA, Jeffery S (2001) Two families with blepharophimosis/ptosis/epicanthus inversus syndrome have mutations in the putative forkhead transcription factor FOXL2. Genet Test 5:335–338 [DOI] [PubMed] [Google Scholar]
- Boccone L, Meloni A, Falchi AM, Usai V, Cao A (1994) Blepharophimosis, ptosis, epicanthus inversus syndrome, a new case associated with de novo balanced autosomal translocation [46,XY,t(3;7)(q23;q32)]. Am J Med Genet 51:258–259 [DOI] [PubMed] [Google Scholar]
- Brocke KS, Neu-Yilik G, Gehring N, Hentze MW, Kulozik AE (2002) The human intronless melanocortin 4–receptor gene is NMD insensitive. Hum Mol Genet 11:331–335 [DOI] [PubMed] [Google Scholar]
- Carlsson P, Mahlapuu M (2002) Forkhead transcription factors: key players in development and metabolism. Dev Biol 250:1–23 [DOI] [PubMed] [Google Scholar]
- Cocquet J, Pailhoux J, Jaubert F, Servel N, Xia X, Pannetier M, De Baere E, Messiaen L, Cotinot C, Fellous F, Veitia R (2002). Evolution and expression of FOXL2. J Med Genet 39:916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27:159–166 [DOI] [PubMed] [Google Scholar]
- De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, Gillerot Y, et al. (2001) Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype-phenotype correlation. Hum Mol Genet 10:1591–1600 [DOI] [PubMed] [Google Scholar]
- De Baere E, Fukushima Y, Small K, Udar N, Van Camp G, Verhoeven K, Palotie A, De Paepe A, Messiaen L (2000) Identification of BPESC1, a novel gene disrupted by a balanced chromosomal translocation, t(3;4)(q23;p15.2), in a patient with BPES. Genomics 68:296–304 [DOI] [PubMed] [Google Scholar]
- De Baere E, Lemercier B, Christin-Maitre S, Durval D, Messiaen L, Fellous M, Veitia R (2002) FOXL2 mutation screening in a large panel of POF patients and XX males. J Med Genet 39:e43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baere E, Van Roy N, Speleman F, Fukushima Y, De Paepe A, Messiaen L (1999) Closing in on the BPES gene on 3q23: mapping of a de novo reciprocal translocation t(3;4)(q23;p15.2) breakpoint within a 45-kb cosmid and mapping of three candidate genes, RBP1, RBP2, and β′-COP, distal to the breakpoint. Genomics 57:70–78 [DOI] [PubMed] [Google Scholar]
- Fukushima Y, Wakui K, Nishida T, Ueoka Y (1991) Blepharophimosis sequence and de novo balanced autosomal translocation [46,XY,t(3;4)(q23;p15.2)]: possible assignment of the trait to 3q23. Am J Med Genet 40:485–487 [DOI] [PubMed] [Google Scholar]
- Han K, Manley JL (1993) Functional domains of the drosophila engrailed protein. EMBO J 12:2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrar HS, Jeffery S, Patton MA (1995) Linkage analysis in blepharophimosis-ptosis syndrome confirms localisation to 3q21–24. J Med Genet 32:774–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SE, Chand AL, Winship IM, Gersak K, Aittomaki K, Shelling AN (2002) Identification of novel mutations in FOXL2 associated with premature ovarian failure. Mol Hum Reprod 8:729–733 [DOI] [PubMed] [Google Scholar]
- Kosaki K, Ogata T, Kosaki R, Sato S, Matsuo N (2002) A novel mutation in the FOXL2 gene in a patient with blepharophimosis syndrome: differential role of the polyalanine tract in the development of the ovary and the eyelid. Ophthalmic Genet 23:43–47 [DOI] [PubMed] [Google Scholar]
- Pailhoux E, Vigier B, Chaffaux S, Servel N, Taourit S, Furet JP, Fellous M, Grosclaude F, Cribiu EP, Cotinot C, Vaiman D (2001) A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat Genet 29:453–458 [DOI] [PubMed] [Google Scholar]
- Praphanphoj V, Goodman BK, Thomas GH, Niel KM, Toomes C, Dixon MJ, Geraghty MT (2000) Molecular cytogenetic evaluation in a patient with a translocation (3;21) associated with blepharophimosis, ptosis, epicanthus inversus syndrome (BPES). Genomics 65:67–69 [DOI] [PubMed] [Google Scholar]
- Prueitt RL, Zinn AR (2001) A fork in the road to fertility. Nat Genet 27:132–134 [DOI] [PubMed] [Google Scholar]
- Ramirez-Castro JL, Pineda-Trujillo N, Valencia AV, Muneton CM, Botero O, Trujillo O, Vasquez G, Mora BE, Durango N, Bedoya G, Ruiz-Linares A (2002) Mutations in FOXL2 underlying BPES (types 1 and 2) in Colombian families. Am J Med Genet 113:47–51 [DOI] [PubMed] [Google Scholar]
- Schibler L, Cribiu EP, Oustry-Vaiman A, Furet JP, Vaiman D (2000) Fine mapping suggests that the goat polled intersex syndrome and the human blepharophimosis ptosis epicanthus syndrome map to a 100-kb homologous region. Genome Res 10:311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KW, Stalvey M, Fisher L, Mullen L, Dickel C, Beadles K, Reimer R, Lessner A, Lewis K, Pericak-Vance MA (1995) Blepharophimosis syndrome is linked to chromosome 3q. Hum Mol Genet 4:443–448 [DOI] [PubMed] [Google Scholar]
- Stromme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SG, Fryns JP, Sutherland GR, Mulley JC, Gecz J (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445 [DOI] [PubMed] [Google Scholar]
- Veitia RA (2002) Exploring the etiology of haploinsufficiency. Bioessays 24:175–184 [DOI] [PubMed] [Google Scholar]
- Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M (1994) Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA 91:9218–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Hayasaka S, Matsumoto M, Budu, Esa T, Hayasaka Y, Endo M (2001) Heterozygous 17-bp deletion in the forkhead transcription factor gene, FOXL2, in a Japanese family with blepharophimosis-ptosis-epicanthus inversus syndrome. J Hum Genet 46:733–736 [DOI] [PubMed] [Google Scholar]
- Zlotogora J, Sagi M, Cohen T (1983) The blepharophimosis, ptosis, and epicanthus inversus syndrome: delineation of two types. Am J Hum Genet 35:1020–1027 [PMC free article] [PubMed] [Google Scholar]