Abstract
Monopolar spindle 1 (MPS1), a mitotic kinase that is overexpressed in several human cancers, contributes to the alignment of chromosomes to the metaphase plate as well as to the execution of the spindle assembly checkpoint (SAC). Here, we report the identification and functional characterization of three novel inhibitors of MPS1 of two independent structural classes, N-(4-{2-[(2-cyanophenyl)amino][1,2,4]triazolo[1,5-a]pyridin-6-yl}phenyl)-2-phenylacetamide (Mps-BAY1) (a triazolopyridine), N-cyclopropyl-4-{8-[(2-methylpropyl)amino]-6-(quinolin-5-yl)imidazo[1,2-a]pyrazin-3-yl}benzamide (Mps-BAY2a) and N-cyclopropyl-4-{8-(isobutylamino)imidazo[1,2-a]pyrazin-3-yl}benzamide (Mps-BAY2b) (two imidazopyrazines). By selectively inactivating MPS1, these small inhibitors can arrest the proliferation of cancer cells, causing their polyploidization and/or their demise. Cancer cells treated with Mps-BAY1 or Mps-BAY2a manifested multiple signs of mitotic perturbation including inefficient chromosomal congression during metaphase, unscheduled SAC inactivation and severe anaphase defects. Videomicroscopic cell fate profiling of histone 2B-green fluorescent protein-expressing cells revealed the capacity of MPS1 inhibitors to subvert the correct timing of mitosis as they induce a premature anaphase entry in the context of misaligned metaphase plates. Hence, in the presence of MPS1 inhibitors, cells either divided in a bipolar (but often asymmetric) manner or entered one or more rounds of abortive mitoses, generating gross aneuploidy and polyploidy, respectively. In both cases, cells ultimately succumbed to the mitotic catastrophe-induced activation of the mitochondrial pathway of apoptosis. Of note, low doses of MPS1 inhibitors and paclitaxel (a microtubular poison) synergized at increasing the frequency of chromosome misalignments and missegregations in the context of SAC inactivation. This resulted in massive polyploidization followed by the activation of mitotic catastrophe. A synergistic interaction between paclitaxel and MPS1 inhibitors could also be demonstrated in vivo, as the combination of these agents efficiently reduced the growth of tumor xenografts and exerted superior antineoplastic effects compared with either compound employed alone. Altogether, these results suggest that MPS1 inhibitors may exert robust anticancer activity, either as standalone therapeutic interventions or combined with microtubule-targeting chemicals.
Keywords: chromosomal instability, colon cancer, mitochondrial membrane permeabilization, mitotic spindle, SP600125
Mitosis is a sophisticated process that ensures the faithful inheritance of the genetic material by the cellular progeny while preventing aneuploidy and chromosomal instability (CIN), two established hallmarks of cancer.1, 2, 3 A set of protein kinases that are collectively known as mitotic kinases, including Aurora kinases (AURKs), mitotic cyclin-dependent kinases (CDKs), Polo-like kinases and monopolar spindle 1 (MPS1), regulates and coordinates multiple aspects of chromosome segregation during mitosis.4
MPS1 (also known as TTK) is a dual-specificity kinase (meaning that it phosphorylates both serine/threonine and tyrosine residues) with an established role in the proper alignment and orientation of chromosomes on the metaphase plate.5 MPS1 is a core component of the spindle assembly checkpoint (SAC), a control mechanism that monitors the attachment between kinetochores (KTs) and microtubules (MTs) during prometaphase, halting the metaphase-to-anaphase transition until all chromosomes are fully attached and bioriented at the metaphase plate.6 Although the precise contribution of MPS1 to SAC functions remains elusive,7 accumulating evidence suggests that MPS1 facilitates SAC activation by promoting the recruitment of SAC components to unoccupied KTs.8, 9, 10, 11, 12, 13, 14, 15 MPS1 has also been implicated in the so-called ‘error correction mechanism',16 a poorly characterized process that resolves the erroneous KT–MT attachments that frequently form in the early steps of mitosis.17, 18 According to current models, AURKB, one component of the chromosomal passenger complex (reviewed by Ruchaud et al.19), catalyzes this process by destabilizing incorrect attachments, hence generating free KTs that activate the SAC.20, 21 Contradictory reports place MPS1 either upstream or downstream of AURKB in this pathway.10, 16 Consistent with these observations, both the inhibition/depletion and the overexpression of MPS1 abrogate SAC functions, leading to aneuploidy/polyploidy and eventually cell death.9, 16, 22, 23, 24
MPS1 appears to play additional, more controversial, roles in mitosis or interphase. For instance, MPS1 not only may influence mitotic exit and cytokinesis25 but also seems to promote the assembly of a cytosolic anaphase-promoting complex/cyclosome inhibitory complex during interphase to define the overall timing of the M phase.11 In addition, MPS1 may be required for centrosome duplication.26 Finally, MPS1 has been implicated in surveillance mechanisms such as the G2/M checkpoint (by interacting with checkpoint kinase 2 and Bloom syndrome)27, 28 and the tetraploidy checkpoint (by phosphorylating the oncosuppressor protein p53),29 which prevent the initiation of mitosis in cells with damaged or unreplicated DNA30, 31 and the proliferation/survival of illicitly generated polyploids,32, 33, 34 respectively.
Although the MPS1-coding gene (TTK) is rarely mutated during oncogenesis (reviewed by Liu and Winey5), high MPS1 expression levels have been detected in several human neoplasms, including thyroid, breast and lung cancers.35, 36, 37, 38 In breast carcinoma patients, the overexpression of MPS1 correlates with increased histological grade and tumor aggressiveness and may contribute to the overriding of the so-called ‘intolerance to aneuploidy',37 a mechanism that normally prevents the proliferation/survival of aneuploid cells.39 On the basis of these findings, MPS1 is considered as one of the most promising drug targets for cancer therapy.40 During the last years, first-generation MPS1 inhibitors have been identified and characterized in preclinical assays (reviewed by Liu and Winey5).
Here, we report the characterization of the antineoplastic activity of three novel selective and potent MPS1 kinase inhibitors, N-(4-{2-[(2-cyanophenyl)amino][1,2,4]triazolo[1,5-a]pyridin-6-yl}phenyl)-2-phenylacetamide (Mps-BAY1), N-cyclopropyl-4-{8-[(2-methylpropyl)amino]-6-(quinolin-5-yl)imidazo[1,2-a]pyrazin-3-yl}benzamide (Mps-BAY2a) and N-cyclopropyl-4-{8-(isobutylamino)imidazo[1,2-a]pyrazin-3-yl}benzamide (Mps-BAY2b), in models of human colorectal and cervical carcinoma, in vitro and in vivo. These inhibitors abolish SAC activation, thus causing chromosome missegregation, aneuploidization/polyploidization and, ultimately, mitotic catastrophe, an oncosuppressive mechanism for the elimination of mitosis-insufficient cells.41 Cell death as induced by MPS1 inhibitors was under the control of Bcl-2-like proteins, implying that it was executed through the intrinsic pathway of apoptosis. Altogether, our results pave the way to additional preclinical and clinical studies on these MPS1 inhibitors, which may be used as monotherapeutic agents or in combination with MT-targeting compounds.
Results
Identification of three MPS1 inhibitors with potent antineoplastic effects
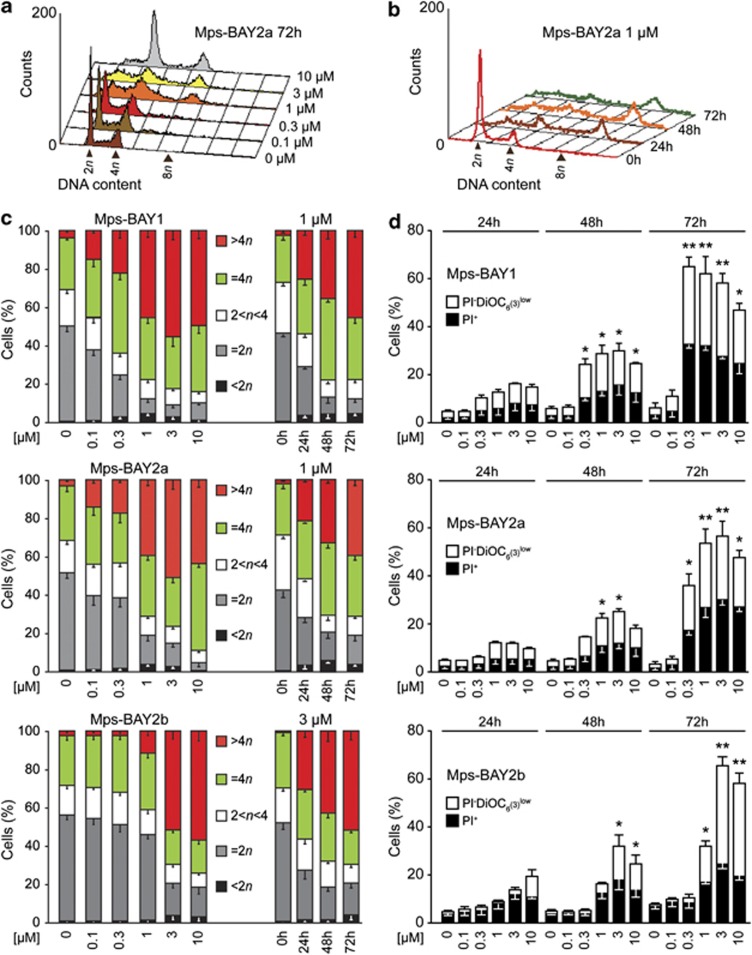
An in vitro kinase assay designed to measure the inhibition of MPS1 enzymatic activity led to the identification of three top-scoring compounds: Mps-BAY1, a triazolopyridine, and Mps-BAY2a and Mps-BAY2b, two imidazopyrazines (Supplementary Figure 1). Both these classes of compounds contain H-bond donor/acceptor nitrogen atoms, which are common among molecules that bind to the ATP pocket -and associated hinge region- of protein kinases. Mps-BAY1 Mps-BAY2a and Mps-BAY2b inhibited human MPS1 with an IC50 ranging between 1 and 10 nM (Supplementary Table 1). When used at a high concentration (10 μM), Mps-BAY1, Mps-BAY2a and Mps-BAY2b exhibited a restricted inhibitory effect on a panel of 220 human kinases compared with the broad-spectrum kinase inhibitors reversine and anthra[1,9-cd]pyrazole-6(2H)-one (SP600125) (Supplementary Table 2).10, 15 Of note, Mps-BAY1, Mps-BAY2a and Mps-BAY2b failed to inhibit several kinases that are known for their role in mitosis. Mps-BAY1, Mps-BAY2a and Mps-BAY2b inhibited the activation of the SAC with an IC50 of 130 nM, 95 nM and 670 nM, respectively, as monitored in an assay in which the disappearance of histone 3 (H3) phosphorylation (a post-translational modification occurring in prophase/metaphase) was assessed in HeLa cells responding to 300 nM nocodazole (data not shown). Thus, Mps-BAY1, Mps-BAY2a and Mps-BAY2b are efficiently taken up by cultured cells and can reach their molecular target. In line with this notion, all these MPS1 inhibitors reduced the proliferation of the vast majority of primary and transformed human and rat cells tested, and exerted even higher antiproliferative effects on mouse cells (Supplementary Table 3). Mps-BAY2a caused heterogeneous antiproliferative responses within a collection of human colon carcinoma cell lines, with sensitivities (IC50) ranging from 160 nM to >10 μM (Supplementary Table 4). Noteworthy, neither CIN nor microsatellite instability (MIN) was clearly associated with the resistance/sensitivity of human colorectal cancer cell lines to Mps-BAY2a (Supplementary Table 4). Mps-BAY1, Mps-BAY2a and Mps-BAY2b had a major impact on the cell cycle progression and survival of human colorectal carcinoma HCT 116 (Figure 1 and Supplementary Figure 2) and human cervical carcinoma HeLa cells (Supplementary Figures 3 and 4), both of which are particularly sensitive to these compounds (Supplementary Table 3). Thus, Mps-BAY1, Mps-BAY2a and Mps-BAY2b induced a dose- and time-dependent perturbation of the cell cycle, manifesting with an increase in the frequency of cells exhibiting a hyperploid DNA content (>4n) (Figures 1a–c and Supplementary Figures 2 and 3), as well as with a progressive accumulation of dying cells (i.e., cells that had lost their mitochondrial transmembrane potential, Δψm) and cell corpses (with ruptured plasma membranes) (Figure 1d and Supplementary Figure 4).
Figure 1.
Impact of MPS1 inhibitors on cell cycle progression and survival of human colorectal cancer cells. (a–d) Human colorectal carcinoma HCT 116 cells were left untreated or treated with Mps-BAY1, Mps-BAY2a or Mps-BAY2b as indicated, and then collected and either processed for the cytofluorometric assessment of cell cycle progression upon staining with the DNA-intercalating agent PI (a–c) or subjected to the cytofluorometric quantification of cell death-associated parameters upon costaining with the Δψm-sensitive dye DiOC6(3) and PI (d). Representative cell cycle distributions for Mps-BAY2a are shown in panels a and b, whereas quantitative data on DNA content (means±S.E.M.; n=6 for Mps-BAY1 and Mps-BAY2a; n=3 for Mps-BAY2b) are shown in panel c. (d) White and black columns illustrate the percentage (means±S.E.M.; n=3) of dying (PI− DiOC6(3)low) and dead (PI+) cells, respectively. *P<0.05, **P<0.01 (Student's t-test), compared with untreated cells (at the same time point). See also Supplementary Figures 2 and 3
These findings identify Mps-BAY1, Mps-BAY2a and Mps-BAY2b as new MPS1 inhibitors with potent antiproliferative and cytotoxic effects.
Characteristics of cell cycle perturbations as induced by Mps-BAY1 and Mps-BAY2a
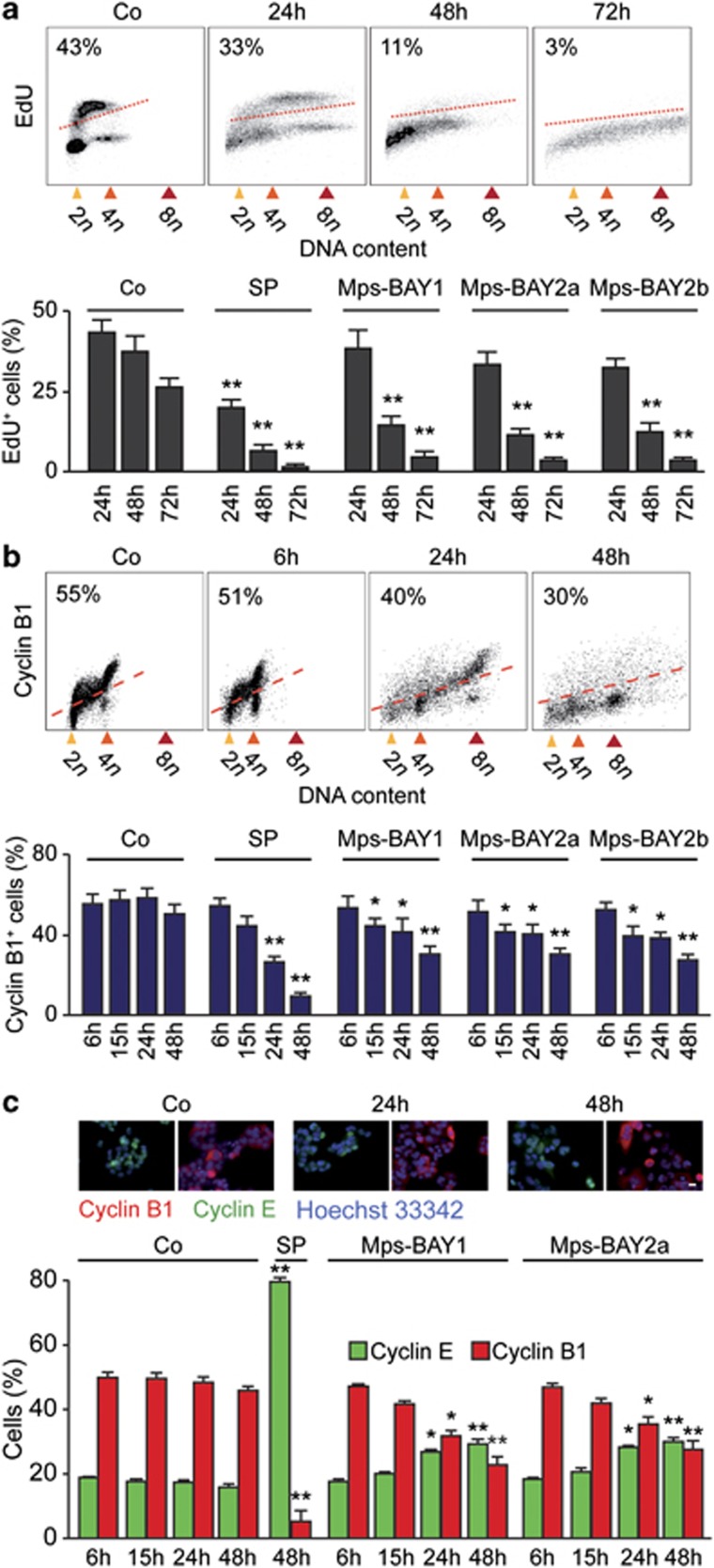
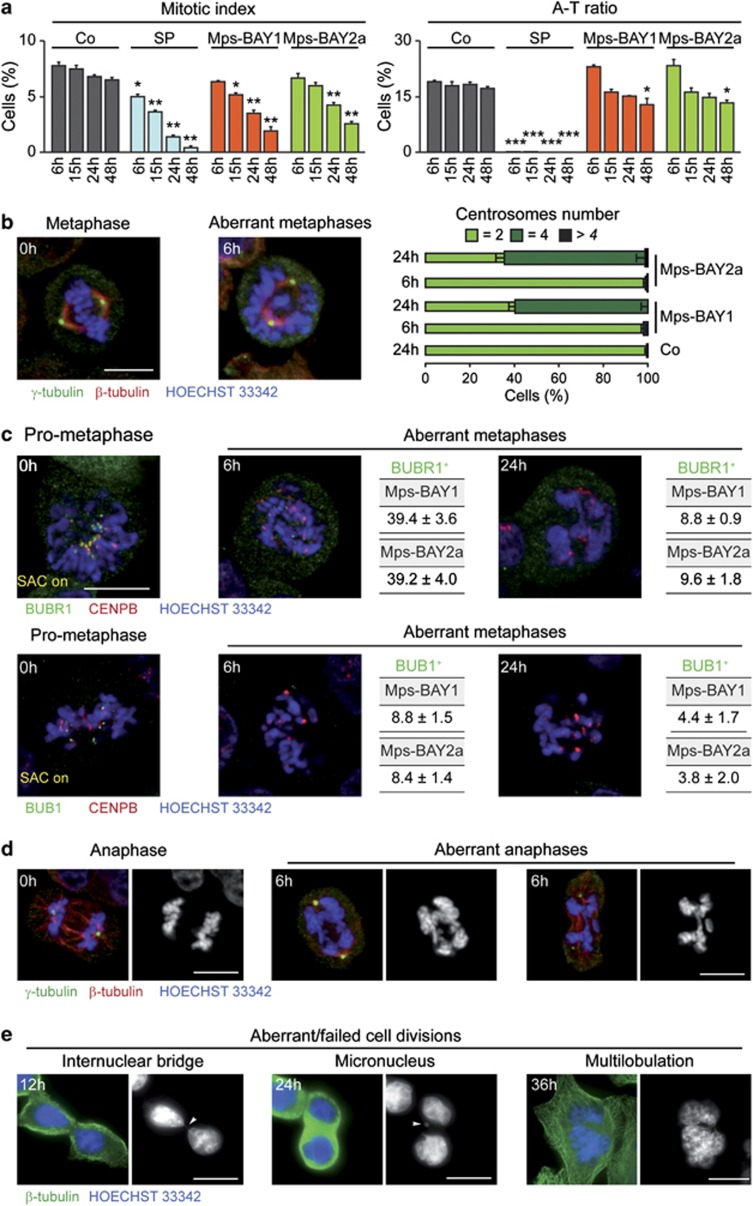
We then investigated the precise impact of MPS1 inhibitors on cell cycle progression. Upon the administration of Mps-BAY1, Mps-BAY2a or Mps-BAY2b, the fraction of HCT 116 cells that incorporated the DNA precursor 5-ethynyl-2′-deoxyuridine (EdU, which is only taken up in the S phase of the cell cycle) decreased over time, although such an inhibition was more pronounced with SP600125 (Figure 2a). Of note, a significant fraction of the cells still replicated their DNA even after 48 h of exposure to MPS1 inhibitors. We then performed an in-depth cytofluorometric and (fluorescence) microscopic analysis of the levels of cyclin E and B1, two markers that accumulate in the G1 and G2 phase of the cell cycle, respectively. In response to Mps-BAY1, Mps-BAY2a and Mps-BAY2b (standard dose: 1 μM, 1 μM and 3 μM, respectively), the frequency of cyclin B1+ HCT 116 cells diminished, although these effects were less consistent than those mediated by SP600125 (Figures 2b and c). The two novel classes of MPS1 inhibitors also induced a minor accumulation of cyclin E+ cells (∼30% in Mps-BAY2-treated cells compared with ∼16% in control conditions at 48 h), suggesting that a fraction of cells maintains the ability to proliferate in this setting (Figures 2b and c). Mps-BAY1 and Mps-BAY2a reduced – but not abrogated – the incidence of mitoses, yet failed to affect the anaphase–telophase ratio (Figure 3a). Moreover, HCT 116 cells that were cultured in the presence of Mps-BAY1 or Mps-BAY2a exhibited a major disorganization of mitoses, manifesting with non-aligned metaphase plates, incomplete centrosome separation, asymmetric chromosome distributions and a progressive increase in the number of centrosomes (Figure 3b). Of note, Mps-BAY1 and Mps-BAY2a inhibited the recruitment of the two SAC components BUB1B1 (best known as BUBR1) and BUB1 to CENPB+ centromeres (Figure 3c), although with distinct kinetics (with complete inhibition at 6 h and 24 h for BUB1 and BUBR1, respectively). These findings indicate that Mps-BAY1 and Mps-BAY2a abolish SAC activation. Accordingly, all anaphases observed among Mps-BAY1- or Mps-BAY2a-treated cells displayed multiple abnormalities that are associated with aberrant karyokinesis (as demonstrated by an increased frequency of cells bearing internuclear bridges or microchromosomes/micronuclei) or cytokinesis (as demonstrated by the accumulation of cells exhibiting multiple and/or multilobular nuclei) (Figures 3d and e).
Figure 2.
Detailed analysis of cell cycle-associated parameters in colorectal carcinoma cells treated with MPS1 inhibitors. (a–c) Human colorectal carcinoma HCT 116 cells were maintained in control (Co) conditions or treated with 1 μM Mps-BAY1 (a–c), 1 μM Mps-BAY2a (a–c) or 3 μM Mps-BAY2b (a and b) for the indicated time. Thereafter, cells were either labeled with the thymidine analog EdU and analyzed by cytofluorometry (a), or processed for the detection of cyclin B1 by flow cytometry (b) or cyclin E and cyclin B1 by immunofluorescence microscopy (c). Representative dot plots and quantitative assessments (means±S.E.M.; n=3) of EdU incorporation and cyclin B1+ cells are provided in panels a and b, respectively. PI was employed to monitor DNA content; numbers refer to the percentage of EdU-incorporating (a) or cyclin B1+ cells (b). Representative immunofluorescence microphotographs and quantitative data on the percentage of cells staining positively for cyclin E (cyclin E+) and cyclin B1 (cyclin B1+) (means±S.E.M.; n=4) are reported in panel c. Nuclei were counterstained with Hoechst 33342. Scale bar=10 μm. In all panels, SP600125 (SP, 30 μM) was employed as a positive control condition. *P<0.05, **P<0.01 (Student's t-test), compared with untreated cells (at the same time point)
Figure 3.
Mitotic perturbations induced by MPS1 inhibitors. (a–d) Human colorectal carcinoma HCT 116 cells were maintained in control (Co) conditions or treated with 30 μM SP600125 (SP), 1 μM Mps-BAY1 or 1 μM Mps-BAY2a for the indicated time, and then processed for the fluorescence microscopy-assisted quantification of mitotic figures, upon staining with the DNA-binding fluorochrome Hoechst 33342 (a), or mitotic spindle organization (b, d and e) and SAC activation (c), upon immunostaining with antibodies specific for components of MTs (i.e., β-tubulin), centrosomes (i.e., γ-tubulin), SAC (i.e., BUB1, BUBR1) and KTs (i.e., CENPB). (a) The mitotic index and the anaphase–telophase (A–T) ratio – representing the frequency of mitoses among the entire cell population and the proportion of anaphases plus telophases among all mitotic figures, respectively – are illustrated (means±S.E.M.; n=5). *P<0.05, **P<0.01, ***P<0.001 (Student's t-test), compared with untreated cells (at the same time point). Representative immunofluorescence microphotographs of cells in normal/abnormal metaphase (b and c), prometaphase (c) or normal/abnormal anaphase (d) as well as of cells that were generated by abnormal cell divisions (e) are shown. (b) Bars illustrate the percentage of cells bearing 2, 4 or >4 centrosomes (means±S.E.M.; n=3). The percentage of abnormal metaphases manifesting with BUBR1 and BUB1 at KTs (BUBR1+ and BUB+, respectively) – which is indicative of SAC activation – is shown in panel c (means±S.E.M.; n=3). Note that all anaphases and metaphases observed in the presence of MpsBAY1 and MpsBAY2a were abnormal. (e) Arrowheads indicate an internuclear bridge and a micronucleus. Scale bars=10 μm
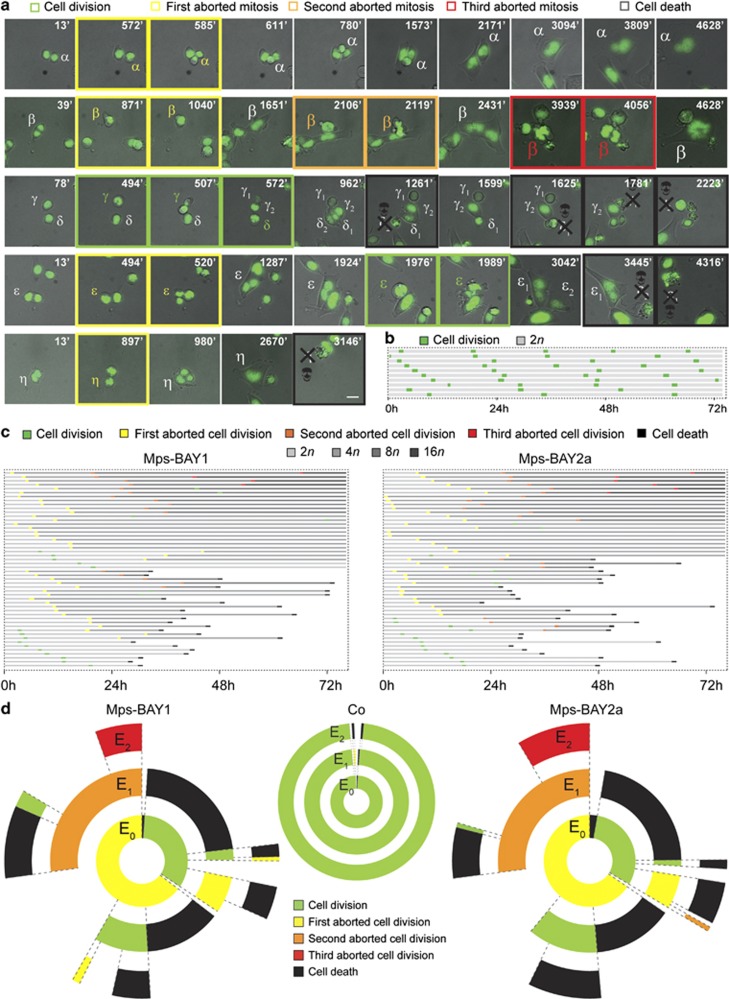
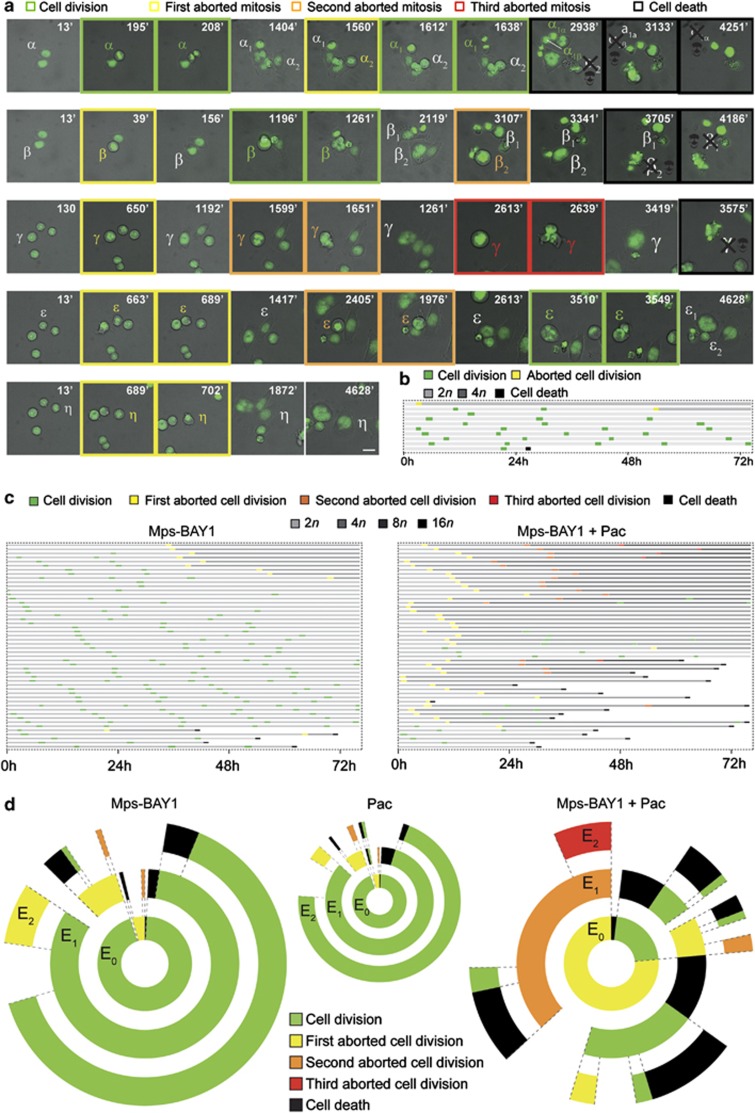
To quantify these changes, HCT 116 cells stably expressing a histone 2B-green fluorescent protein (H2B-GFP) fusion protein, which allows for the visualization of chromatin, were subjected to live fluorescence videomicroscopy. This analysis revealed major alterations in cell cycle progression and mitosis execution among cells exposed to Mps-BAY1 or Mps-BAY2a (Figure 4 and Supplementary Movies 1–5). Indeed, upon premature anaphase onset and in the absence of a proper metaphase plate, cells exposed to MPS1 inhibitors attempted to divide in the presence of misaligned chromosomes, generating either one single polyploid cell (when the cytokinesis furrow regressed) or two daughter cells (when abscission was successful) (Figure 4a and Supplementary Movies 1–5). In this latter case, however, cell division was manifestly asymmetric in ∼35% of the cases. Irrespective of their apparent symmetry or asymmetry, the vast majority (>95%) of apparently successful cell divisions were followed by the death of one or both daughter cells. This observation points to an incorrect segregation of chromosomes between daughter cells, leading to the generation of an unviable, aneuploid progeny. Often, polyploid HCT 116 cells generated in the presence of Mps-BAY1 or Mps-BAY2a as a result of cytokinesis failure progressively hyperploidized through consecutive rounds of abortive mitoses (Figure 4a and Supplementary Movie 2). Alternatively, such polyploid cells remained inert, divided asymmetrically or underwent apoptosis (Figure 4a and Supplementary Movies 1, 4 and 5). In this latter case, cell death occurred in interphase, 13–60 h after the latest of (1–2 rounds of) aberrant mitosis. In several instances, daughter cells originating from an initially normal, close-to-successful cell division remained connected by an internuclear DNA-containing bridge and re-fused later, forming one single cell (Figure 4a). Systematic cell fate profiling performed on 50 cells revealed that death affected more than 50% of cell populations exposed to Mps-BAY1 and Mps-BAY2a, with a relatively homogeneous latency from the last aborted cell division of 25.4±2.5 h (mean±S.E.M., n=25) and 26.1±2.2 h (mean±S.E.M., n=28), respectively (Figures 4b and c). Transgenerational cell cycle profiling,42 an experimental setting in which the destiny of 150 cells is followed over three consecutive generations, confirmed that both these drugs arrest cell cycle progression and efficiently kill colorectal carcinoma cells (Figure 4d). Hence, MPS1 inhibitors trigger a type of mitotic catastrophe whereby cells die in the interphase that follows an attempted/asymmetric division.41 Importantly, cell death instances observed upon the administration of Mps-BAY1 and Mps-BAY2a were preceded by chromatin condensation (pyknosis) and accompanied by nuclear fragmentation (karyorrhexis), two morphological hallmarks of apoptosis.43, 44
Figure 4.
Cell fate profiling of colorectal carcinoma cells treated with MPS1 inhibitors. (a–d) Human colorectal carcinoma HCT 116 cells expressing a H2B-GFP chimera were left untreated (Co) or treated with 1 μM Mps-BAY1 or Mps-BAY2a and then monitored by live videomicroscopy for approximately 72 h. Representative snapshots are shown in panel a (scale bar=10 μm), whereas single cell fate profiles of untreated cells (n=10), single cell fate profiles of Mps-BAY1- or Mps-BAY2a-treated cells (n=50) and transgenerational cell cycle profiles of untreated as well as Mps-BAY1- or Mps-BAY2a-treated cells (n=150) are depicted in panels b, c and d, respectively. (a) Greek letters refer to individual cells that underwent a successful cell division (framed in green), generating two daughter cells (numbers depicted in subscript), or – alternatively – an abortive cell division (framed in yellow, orange and red for one, two and three consecutive rounds of abortive divisions, respectively), leading to one single cell or apoptosis (the latter framed in black and labeled with a skull). (a) Increased font size of Greek letters depicts an increase in cell size (and hence in ploidy). (b and c) The figure key relative to cell fate and ploidy is indicated. (d) Green, yellow, orange and red illustrate the fraction of cells that underwent successful replication (symmetric and asymmetric divisions all confounded) and one, two and three consecutive rounds of abortive divisions, respectively. Black sections depict the fraction of dying cells. Concentric circles depict three consecutive generational events (E), starting from E0 (inner circle). As cells presenting internuclear bridges upon an aberrant mitosis always fused during the next interphase or mitosis (e.g., α in panel a, see text for further details), we arbitrarily included them in the category of ‘aborted cell division', in both panels c and d. In these panels, cell divisions were considered to be successful only when daughter cells were clearly separated. Of note, successful cell divisions often generated an anysokaryotic and anysocytotic progeny (e.g., γ and ɛ in panel a, see text for further details). Full-length movies are provided as Supplementary Movies 1–5
Mechanisms of apoptosis induction by Mps-BAY1 and Mps-BAY2a
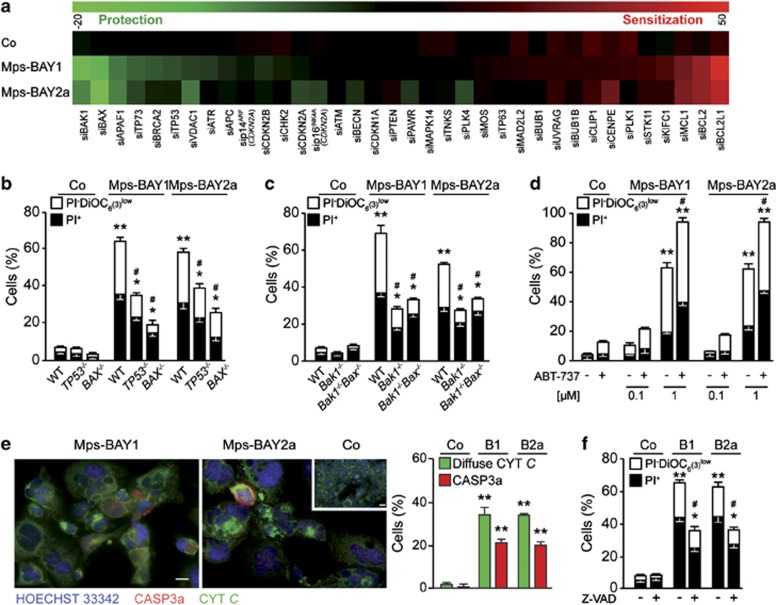
To gain insights into the molecular mechanisms whereby MPS1 inhibitors induce apoptosis upon the activation of mitotic catastrophe, we transfected HCT 116 cells with 36 distinct small interfering RNAs (siRNAs) that target cell cycle- or cell death-relevant proteins. Within this collection, siRNAs that deplete antiapoptotic proteins of the Bcl-2 family (i.e., BCL2; BCL2L1, best known as BCL-XL; and MCL1) were found to be particularly efficient at sensitizing HCT 116 cells to Mps-BAY1- or Mps-BAY2a-induced cell death (Figure 5a). Conversely, siRNAs targeting two multidomain proapoptotic proteins of the Bcl-2 family (i.e., BAX and BAK1) prevented the loss of viability provoked by Mps-BAY1 or Mps-BAY2a (Figure 5a). Along similar lines, HCT 116 cells were protected from the cytotoxic effect of MPS1 inhibitors by the depletion of APAF-1, the essential coactivator of caspase-9 that operates downstream of mitochondria in the intrinsic pathway of apoptosis.43 Accordingly, the knockout of BAX, BAK1 or both greatly reduced cell killing by Mps-BAY1 and Mps-BAY2a (Figures 5b and c), whereas the neutralization of BCL2 and BCL-XL with the chemical BH3-mimetic ABT-737 (employed at the sublethal concentration of 1 μM) sensitized HCT 116 cells to death as triggered by MPS1 inhibition (Figure 5d and Supplementary Figure 5). In this setting, the knockdown of p53 as well as the knockout of TP53 also mediated partial cytoprotective effects (Figures 5a and b). In line with an involvement of mitochondrial apoptosis,45 HCT 116 cells treated with MPS1 inhibitors manifested the release of cytochrome c (CYT C) into the cytosol followed by the activation of caspase-3 (Figure 5e). Inhibition of caspases with the broad-spectrum caspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (Z-VAD-fmk) significantly reduced the cytotoxic activity of MPS1 inhibitors, although this effect was partial, suggesting that caspase-independent mechanisms contribute to the execution of cell death in this setting (Figure 5f). Altogether, these results indicate that abortive mitoses induced by MPS1 inhibition result in the activation of the intrinsic pathway of apoptosis.
Figure 5.
Mechanisms of cell death triggered by MPS1 inhibitors. (a) Human colorectal carcinoma HCT 116 cells were transfected with a control siRNA (siUNR) or with previously validated, non-toxic siRNAs targeting a panel of cell cycle- or cell death-modulatory proteins for 24 h, and then either maintained in control (Co) conditions or treated with 1 μM Mps-BAY1 or Mps-BAY2a for additional 48 h. Thereafter, apoptosis-associated parameters were assessed by cytofluorometry upon costaining with the Δψm-sensitive dye DiOC6(3) and PI. Green and red boxes depict siRNA-mediated cytoprotection (Δ<0) and chemosensitization (Δ>0), respectively. For a given siRNA, siXXX, Δ represents the difference between the percentage of siXXX-transfected and siUNR-transfected cells succumbing (PI−DiOC6(3)low+PI+) in response to the indicated experimental condition. (b–d) HCT 116 cells (b) and immortalized mouse embryonic fibroblasts (c) with the indicated genotype as well as wild-type (WT) HCT 116 cells that had been pre-exposed or not to 0.1 or 1 μM ABT-737 (d) were kept in Co conditions or incubated with 1 μM Mps-BAY1 or Mps-BAY2a for 72 h. Thereafter, cell were processed for the cytofluorometric assessment of apoptosis-associated parameters as described in the legend to panel a. (e) HCT 116 cells maintained in Co conditions or treated with 1 μM Mps-BAY1 (B1) or Mps-BAY2a (B2a) for 72 h were costained with Hoechst 33342 and antibodies specific for CYT C and activated caspase-3 (CASP3a), followed by the quantification of cells exhibiting diffuse CYT C staining or caspase-3 activation by fluorescence microscopy. Representative fluorescence microphotographs and quantitative results (means±S.E.M., n=4) are shown. Scale bars=10 μm. (f) Upon preincubation with the pan-caspase inhibitor Z-VAD-fmk (Z-VAD, for 24 h), HCT 116 cells were exposed or not to 1 μM Mps-BAY1 (B1) or Mps-BAY2a (B2a), followed by the cytofluorometric assessment of cell death-related parameters as described in the legend to panel a. (b, c, d and f) White and black columns illustrate the percentage of dying (PI− DiOC6(3)low) and dead (PI+) cells, respectively (means±S.E.M., n=3). *P<0.05, **P<0.01 (Student's t-test), compared with untreated cells of the same genotype (b, c, d, e and f); #P<0.05 (Student's t-test), compared with equally treated WT cells (b and c) or compared with cells exposed to MPS1 inhibitors only (d and f). See also Supplementary Figure 5
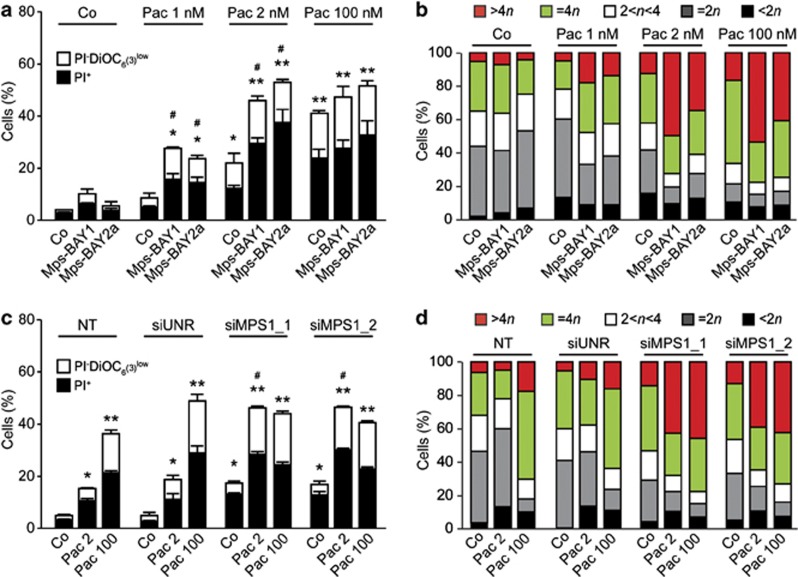
Synergistic effects of low-dose paclitaxel and MPS1 inhibitors
As MPS1 inhibitors turned out to block SAC activation, we wondered whether combining them with the MT-targeting agent paclitaxel (which activates SAC) might promote synergistic effects. To address this question, sublethal concentrations of Mps-BAY1 or Mps-BAY2a (100 nM) and paclitaxel (1 or 2 nM) were coadministered to HCT 116 cells and, indeed, were found to mediate synergistic effects. Such a synergistic interaction was observed with respect to cell death induction (Figure 6a) as well as polyploidization (Figure 6b). The knockdown of MPS1 using two distinct, previously validated46 siRNAs had a similar sensitizing effect on cell death (Figure 6c) and polyploidization (Figure 6d) triggered by low-dose paclitaxel. The combination of MPS1 inhibitors and paclitaxel frequently induced tetraploidization or hyperploidization after one or several consecutive rounds of abortive mitoses, respectively, eventually generating non-proliferating cells or cells that died 28.0±3.2 h (mean±S.E.M., n=21) after the last mitosis (Figures 7a–c and Supplementary Movies 6–10). In some instances, HCT 116 cells responding to MPS1 inhibitors plus paclitaxel attempted to divide in a bipolar manner, generating two daughter cells destined to die. Sublethal doses of paclitaxel or Mps-BAY1 had a relatively mild effect on the cell cycle progression and viability of HCT 116 cells, that is, >75% of the cells proliferated normally, although displaying a significant increase in the rate of chromosome missegregation. Conversely, the combination of these two agents altered the cell cycle progression or affected the survival of the vast majority of cells, as determined by single cell fate and transgenerational cell cycle profiling (Figures 7b–d).42 Similar results were obtained when paclitaxel was combined with a low dose of imidazopyrazines (data not shown).
Figure 6.
Synergistic lethality of low-dose MPS inhibitors and microtubular poisons. (a–d) Human colon carcinoma HCT 116 cells were kept in control (Co) conditions or exposed to 0.1 μM Mps-BAY1 or Mps-BAY2a, alone or combined with the indicated dose of paclitaxel (Pac), for 48 h (a and b). Alternatively, HCT 116 cells were left untransfected (NT) or transfected with a control siRNA (siUNR) or with the indicated, previously validated siRNAs for 24 h, and then maintained in Co conditions or treated with the indicated concentration of Pac for 48 h (c and d). Thereafter, cells were processed for the cytofluorometric quantification of cell death-associated parameters upon costaining with the Δψm-sensitive dye DiOC6(3) and PI (a and c), or for the cytofluorometric assessment of cell cycle progression upon staining with PI only (b and d). (a and c) White and black columns illustrate the percentage of dying (PI− DiOC6(3)low) and dead (PI+) cells, respectively (means±S.E.M., n =3). *P<0.05, **P<0.01 (Student's t-test), compared with untreated (siUNR transfected) cells; #P<0.05 (Student's t-test), compared with cells exposed to the same concentration of paclitaxel only (a) or compared with equally treated siUNR-transfected cells (c). (b and d) Data from one representative experiment (out of three yielding similar results) are shown
Figure 7.
Cell fate profiling of colorectal carcinoma cells treated with low-dose MPS1 inhibitors and microtubular poisons. (a–d) Human colorectal carcinoma HCT 116 cells expressing a H2B-GFP chimera were left untreated or exposed to 100 nM Mps-BAY1, 2 nM paclitaxel (Pac) or both, and then followed by live videomicroscopy for more than 72 h. Representative snapshots are shown in panel a (scale bar=10 μm), whereas single cell fate profiles for Pac-treated cells (n=10), single cell fate profiles for Mps-BAY1- and Mps-BAY1+Pac-treated cells (n=50), and transgenerational cell cycle profiles for Pac-, Mps-BAY1- and Mps-BAY1+Pac-treated cells (n=150) are shown in panels b, c and d, respectively. (a) Individual cells are depicted with Greek letters and labeled as detailed in the legend to Figure 4. (b, c and d) Single cell fate profiles and transgeneration cell cycle profiles are depicted as detailed in the legend to Figure 4. Full-length movies are provided as Supplementary Movies 6–10
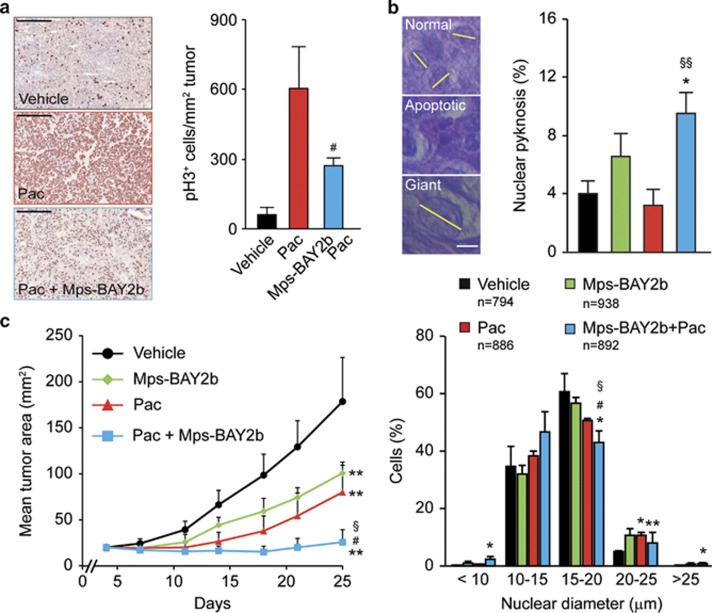
Finally, we evaluated the therapeutic potential of Mps-BAY2b plus paclitaxel, in vivo, on HeLa-Matu cervical carcinomas growing in immunodeficient mice. We used Mps-BAY2b because it displayed a higher in vivo stability than Mps-BAY1 and Mps-BAY2a (Supplementary Table 5). Twenty-four hours after the administration of paclitaxel, HeLa-Matu cell-derived xenografts displayed higher levels of phosphorylated H3 than untreated tumors, as determined by immunohistochemistry. A short (1 h) exposure of tumor-bearing, paclitaxel-treated mice to Mps-BAY2b resulted in the decrease of H3 phosphorylation (Figure 8a). This finding indicates that Mps-BAY2b is efficiently distributed in vivo, reaches xenotransplanted tumors and penetrates cancer cells to inhibit MPS1. In this xenograft model, the combination of Mps-BAY2b and paclitaxel induced higher levels of apoptosis and a higher incidence of giant mononuclear cells (nuclear diameter >25 μm) than either agent employed as a standalone intervention (Figure 8b). Moreover, the coadministration of paclitaxel and Mps-BAY2b exerted superior antineoplastic effects compared with the administration of vehicle and either paclitaxel or Mps-BAY2b alone (Figure 8c). Altogether, these data underscore the possibility to advantageously combine MPS1 inhibitors with MT-targeting agents.
Figure 8.
Therapeutic effect of MPS1 inhibitors and paclitaxel in vivo. (a and b) Human cervical carcinoma HeLa-Matu cells were subcutaneously inoculated in athymic nu/nu mice. When tumor area reached 40–80 mm2, mice were treated with vehicle or 30 mg/kg paclitaxel (Pac) i.p., followed (after 24 h) by the administration of vehicle or the indicated dose of Mps-BAY2b p.o. (a). Alternatively, tumor-bearing mice were treated with vehicle, 8 mg/kg Pac i.v. once, 30 mg/kg Mps-BAY2b p.o. twice daily for 2 days or 8 mg/kg Pac i.v. once+30 mg/kg Mps-BAY2b p.o. twice daily for 2 days (b). (a) Tumors were recovered 1 h after the administration of Mps-BAY2b and processed for the immunohistochemical detection of phosphorylated histone 3 (pH3). Scale bar=500 μm. (b) Tumors were recovered 72 h after the first treatment and stained with hematoxylin and eosin for the light microscopy-assisted determination of nuclear diameter and nuclear pyknosis. Scale bar=10 μm. Representative microphotographs and quantitative data (means±S.E.M., n=3) are shown. (c) Athymic nu/nu mice carrying HeLa-Matu-derived xenografts were treated with vehicle, 10 mg/kg Pac i.v. once weekly, 30 mg/kg Mps-BAY2b p.o. twice daily or 10 mg/kg Pac i.v. once weekly+30 mg/kg Mps-BAY2b p.o. twice daily, and tumor area was routinely monitored by means of a common caliper. Data from one representative experiment are shown (means±S.D.). *P<0.05, **P<0.01 (Student's t-test), compared with vehicle-treated mice; #P<0.05 (Student's t-test), compared with Pac-treated mice; §P<0.05, §§P<0.01 (Student's t-test), compared with Mps-BAY2b-treated mice
Discussion
Here, we reported the identification and functional characterization of three novel and potent MPS1 inhibitors, the triazolopyridine Mps-BAY1 and the imidazopyrazines Mps-BAY2a and Mps-BAY2b. All these agents were capable of abrogating the functionality of the SAC, as demonstrated by the incapacity of cells exposed to MPS1 inhibitors to sustain a mitotic arrest upon exposure to MT poisons. Even in the absence of SAC activators, both classes of MPS1 inhibitors markedly increased the rate of chromosome misalignments resulting from erroneous MT–KT attachments and promoted a premature anaphase entry (i.e., before the formation of a correct equatorial metaphase plate). These results are in line with previous findings obtained with other MPS1-specific inhibitors,8, 9, 10, 12, 15, 47 upon MPS1 depletion16, 24 or following the conditional knockout of TTK,11 confirming the central implication of this mitotic kinase in SAC function and chromosome congression.
Upon exposure to MPS1 inhibitors, colorectal carcinoma cells displayed severely abnormal anaphases, and, as they progressed in mitosis, they underwent two alternative catastrophic fates: (i) they divided in a bipolar (often asymmetrical) manner, generating two aneuploid cells that most often died in the following interphase, or (ii) they failed to complete cytokinesis and hence became hyperploid. These hyperploid cells either failed to progress through the cell cycle or died before or upon a second round of aberrant divisions.
It should be noted that the impact of MPS1 inhibition on polyploidization is a matter of debate. Some authors sustain that MPS1 inhibition/depletion neither perturbs proper chromosome orientation nor affects the correct execution of cytokinesis.10, 15, 47 Conversely, it has been reported that the absence of MPS1 favors cytokinesis failure and polyploidy.8, 16, 48 This discrepancy may reflect differences in the experimental models or the strategy used for MPS1 inhibition.7, 11 Indeed, as the inactivation of >90% of MPS1 is required for the induction of mitotic abnormalities,7, 11 in some studies MPS1 inhibition may have been partial, yielding suboptimal effects on mitosis. Here, we observed that even low doses of Mps-BAY1 and Mps-BAY2a inhibit SAC functions and induce polyploidization as well as apoptosis in a fraction of cells. These results support the contention that Mps-BAY1 and Mps-BAY2a efficiently attain the subcellular compartment(s) containing the pool of MPS1 that is relevant for mitotic progression, hence inhibiting its activity to a point that polyploidization is induced.
The completion of cytokinesis requires an accurate segregation of chromosomes. Thus, the presence of chromatin trapped in the cytokinesis plane may cause the regression of the cleavage furrow. As this delays or even prevents abscission and promotes polyploidy,49, 50, 51, 52 misaligned chromosomes resulting from an accelerated mitosis and SAC abrogation may account for the abortion of cytokinesis induced by MPS1 inhibitors. Consistently, videomicroscopic studies revealed that the progenies of Mps-BAY1- or Mps-BAY2a-treated cells often remain interconnected through a thin chromatin bridge and then fuse to form one single cell with a double chromosomal content. These internuclear connections presumably are fragile and do not withstand the shear stress induced by trypsinization and flow cytometry, implying that the actual frequency of polyploid cells generated in this (and similar) setting(s) is prone to underestimation. A direct role of MPS1 in cytokinesis has recently been suggested.25 In particular, the depletion of the MPS1-interacting protein 1 appears to perturb the actin cytoskeleton, provoking the accumulation of multinucleated cells. Here, we provide additional lines of evidence supporting the capacity of MPS1 inhibitors to induce polyploidy, although the exact mechanism underlying this effect requires further investigation.
In agreement with previous studies,9, 12, 16, 53 we found that MPS1 inhibition potentially kills cancer cells. We observed a heterogeneous response to MPS1 inhibitors in a panel of human colon carcinoma cell lines. Of note, such a response did not clearly correlate to the pattern of genomic instability (CIN and MIN),54 the activity of several proteins relevant for oncogenesis (e.g., APC, BRAF, CDKN2A and p53) or the functionality of the SAC, as SAC-proficient cells were found to be either sensitive (e.g., HCT 116, RKO, LoVo and SW480 cells)55 or resistant (e.g., SW837, HT29 and DLD-1 cells)55 to Mps-BAY2a.
Mps-BAY1 and Mps-BAY2a activated mitotic catastrophe following a premature/aberrant mitotic exit. The molecular pathways regulating distinct types of mitotic catastrophe have not yet been precisely identified, and, in particular, the involvement of caspases remains a matter of debate.41 In our experimental setting, MPS1 inhibitors activated the intrinsic pathway of apoptosis, as indicated by Δψm dissipation as well as the release of CYT C into the cytosol. Moreover, the depletion or pharmacological inhibition of antiapoptotic members of the Bcl-2 protein family sensitized cancer cells to the cytotoxic effects of MPS1 inhibitors, whereas the knockdown of the proapoptotic proteins BAX and BAK1 limited such a cytotoxic response. This suggests that the overall equilibrium among pro- and antiapoptotic Bcl-2 proteins might constitute a potential biomarker to predict the cytotoxic potential of MPS1 inhibitors.
In line with previous results,53 sublethal doses of paclitaxel and MPS1 inhibitors synergized in the killing of colorectal cancer cells both in vitro and in vivo. Of note, such a synergy seems to involve an obligatory step of tetraploidization. Presumably, polyploid, giant mononuclear cells accumulated when paclitaxel and MPS1 inhibitors were combined in vitro and in vivo, suggesting that this histopathological feature may be used as a biomarker to assess the efficacy of such a combinatorial therapeutic regimen in cancer patients.
Altogether, our results reveal that MPS1 inhibitors, alone or in combination with MT inhibitors, are capable of inducing mitotic aberrations and apoptosis in rapidly proliferating cancer cells. Further investigation is needed to understand which particular class of malignancies might benefit from MPS1-targeting therapeutic regimens.
Materials and Methods
Chemicals
Mps-BAY1, Mps-BAY2a and Mps-BAY2b were provided by Bayer Pharma AG (Berlin, Germany) and stored at −20 °C as 10 mM solutions in dimethylsulfoxide. The pan-caspase inhibitor Z-VAD-fmk was obtained from Bachem Bioscience (Bubendorf, Switzerland); SP600125 was purchased from Sigma-Aldrich (St. Louis, MO, USA); and ABT-737 was obtained from Selleck Chemicals (Houston, TX, USA). All commercial chemicals were dissolved and stocked as recommended by the manufacturers. An appropriate volume of solvent was invariably employed to provide negative control conditions.
Cytofluorometric studies
For the simultaneous quantification of plasma membrane integrity and Δψm, cells were collected and stained with 1 μg/ml propidium iodide (PI; Sigma-Aldrich) and 40 nM 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3); Molecular Probes-Life Technologies, Eugene, OR, USA) for 30 min at 37 °C.56 For the assessment of cell cycle distribution, cells were collected, stained with 50 μg/ml PI and analyzed by cytofluorometry as previously described.46, 57 For EdU incorporation assays, cells were incubated with 10 μM EdU for 30 min at 37 °C, fixed, permeabilized and stained with the fluorescent dye azide (Click-iT reaction cocktail; Molecular Probes-Life Technologies) and PI, according to the manufacturer's instructions. For the simultaneous measurement of DNA content and cyclin B1 levels, fixed cells were costained with 10 μM 4′,6-diamidino-2-phenylindole (Molecular Probes-Life Technologies) and mouse antiserum specific for cyclin B1 (mouse monoclonal IgG1 no. 610219; BD Biosciences, San Diego, CA, USA) as previously reported.46, 57 Cytofluorometric acquisitions were performed on a FACSCalibur cytofluorometer (BD Biosciences) equipped with a 70-μm nozzle or a Gallios cytofluorometer (Beckman Coulter, Miami, FL, USA). Data were statistically evaluated using the CellQuest (BD Biosciences) or Kaluza (Beckman Coulter) software. Only events characterized by normal forward scatter and side scatter were included in statistical determinations.
Immunofluorescence and videomicroscopy
Immunofluorescence microscopy was performed according to conventional procedures.58 Images were captured using a Zeiss Axio Observer.Z1 microscope equipped with the ApoTome system (Carl Zeiss, Oberkochen, Germany). For videomicroscopy, HCT 116 cells stably expressing a H2B-GFP chimera were grown in black/clear 96-well imaging plates (BD Biosciences) under standard conditions and subjected to pulsed observations (every 13 min for up to 72 h) with a BD pathway 855 automated live-cell microscope (BD Biosciences). Images were analyzed with the open-source software ImageJ (freely available from the National Institutes of Health, Bethesda, MD, USA, at the address http://rsb.info.nih.gov/ij/). Cell fate profiles are illustrated as previously described.46, 57
In vivo studies
The housing and handling of animals was performed in strict compliance with the European and German Guidelines for Laboratory Animal Welfare. Animals received food and water ad libitum. For the quantification of circulating MPS1 inhibitors, Mps-Bay1, Mps-BAY2a and Mps-BAY2b were administered to female athymic nu/nu mice (Taconic, Hudson, NY, USA) p.o. in a solubilized form (n=2 mice per compound and time point). Serum samples were prepared 1, 7 and 24 h after administration and precipitated with ice-cold 1 : 5 (v:v) acetonitrile/water. Supernatants were analyzed for Mps-BAY1, Mps-BAY2a and Mps-BAY2b content via liquid chromatography–tandem mass spectroscopy (AB Sciex, Framingham, MA, USA). For tumor xenograft studies, 50-day-old female athymic nu/nu mice with an average body weight of 20–22 g were used after an acclimation period of 14 days. Human HeLa-Matu cervical carcinoma cells derived from exponentially growing cultures were resuspended in 1 : 1 (v:v) FBS-free growth medium/Matrigel (BD Biosciences) to a final concentration of 1.5 × 107 cells/ml. Thereafter, 1.5 × 106 cells were subcutaneously implanted into the inguinal region. Tumor area (monitored with a common caliper and approximated to the product of the longest diameter by its perpendicular) and body weight were determined twice a week. When tumors reached an area of approximately 21 mm2, animals were randomized into the following groups (eight mice per group): control, receiving 3 : 1 (v:v) polyethylene glycol/water (vehicle) once a week p.o.; Mps-BAY2b, receiving 30 mg/kg Mps-BAY2b in 3 : 1 polyethylene glycol/water once a week p.o.; paclitaxel, receiving 10 mg/kg paclitaxel in 1 : 1 : 18 (v:v:v) cremophor/ethanol/PBS once a week i.v.; and Mps-BAY2b plus paclitaxel, receiving 30 mg/kg Mps-BAY2b in 3 : 1 polyethylene glycol/water p.o. plus 10 mg/kg paclitaxel in 1 : 1 : 18 cremophor/ethanol/PBS once a week i.v. When tumor area exceeded 150 mm2, animals were euthanized according to the German Animal Welfare Guidelines. For immunohistochemical studies, when tumors reached a size of 40–50 mm2, animals were randomized into the following groups (three mice per group): control, receiving 3 : 1 polyethylene glycol/water (vehicle) once p.o.; Mps-BAY2b, receiving 30 mg/kg Mps-BAY2b in 3 : 1 polyethylene glycol/water once p.o.; and paclitaxel, receiving 30 mg/kg paclitaxel in 1 : 1 : 18 cremophor/ethanol/PBS once i.p. For hematoxylin and eosin staining, when tumors reached a size of 50–80 mm2, animals were randomized into the following groups (four mice per group): control, receiving 3 : 1 polyethylene glycol/water (vehicle) twice daily for 2 days p.o.; Mps-BAY2b, receiving 30 mg/kg Mps-BAY2b in 3 : 1 polyethylene glycol/water twice daily for 2 days p.o.; paclitaxel, receiving 8 mg/kg paclitaxel in 1 : 1 : 18 cremophor/ethanol/PBS once i.v.; and Mps-BAY2b plus paclitaxel, receiving 30 mg/kg Mps-BAY2b in 3 : 1 polyethylene glycol/water twice daily for 2 days p.o. plus 10 mg/kg paclitaxel in 1 : 1 : 18 cremophor/ethanol/PBS i.v. once. Seventy-two hours after the first treatment, tumors were recovered, fixed with 4% (w/v) PFA for 4 h and embedded into paraffin. Ten-micrometer-thick tissue sections were then stained with hematoxylin and eosin according to standard protocols and analyzed as previously described.59
Statistical procedures
Unless otherwise specified, all experiments were carried out in triplicate parallel instances and independently repeated at least twice. Data were analyzed with Microsoft Excel (Microsoft, Redmont, WA, USA), and statistical significance was assessed by means of two-tailed unpaired Student's t-tests. The threshold for statistical significance was set to P<0.05.
Acknowledgments
We are indebted to Bert Vogelstein (Johns Hopkins University, Baltimore, MA, USA) for wild-type (WT) and TP53−/− HCT 116 cells. MJ is funded by the Ligue Nationale contre le Cancer, and LG is funded by the LabEx Immuno-Oncology. IV is funded by the Associazione Italiana per la Ricerca sul Cancro (AIRC). This work is supported by grants to GK from the Ligue Nationale contre le Cancer (Equipes Labellisée), Agence Nationale pour la Recherche (ANR), European Commission (ArtForce), European Research Council, Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa), Cancéropôle Ile-de-France, Fondation Bettencourt-Schueller, AXA Chair for Longevity Research, the LabEx Immuno-Oncology and the Paris Alliance of Cancer Research Institutes.
Glossary
- AURK
Aurora kinase
- CDK
cyclin-dependent kinase
- CIN
chromosomal instability
- CYT C
cytochrome c
- DiOC6(3)
3,3′-dihexyloxacarbocyanine iodide
- GFP
green fluorescent protein
- EdU
5-ethynyl-2′-deoxyuridine
- H2B
histone 2B
- H3
histone 3
- KT
kinetochore
- MIN
microsatellite instability
- Mps-BAY1
N-(4-{2-[(2-cyanophenyl)amino][1,2,4]triazolo[1,5-a]pyridin-6-yl}phenyl)-2-phenylacetamide
- Mps-BAY2a
N-cyclopropyl-4-{8-[(2-methylpropyl)amino]-6-(quinolin-5-yl)imidazo[1,2-a]pyrazin-3-yl}benzamide
- Mps-BAY2b
N-cyclopropyl-4-{8-(isobutylamino)imidazo[1,2-a]pyrazin-3-yl}benzamide
- MPS1
monopolar spindle 1
- MT
microtubule
- PI
propidium iodide
- SAC
spindle assembly checkpoint
- siRNA
small interfering RNA
- SP600125
anthra[1,9-cd]pyrazole-6(2H)-one
- Z-VAD-fmk
N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone
MB, UB, MK, PL, SP, VS, GS, AMW, DM and KZ are employees of Bayer Pharma AG. GK received a grant from Bayer Pharma AG. MJ, IV and GK are listed as inventors on a patent application describing the combination effects of taxanes and MPS1 inhibitors.
Footnotes
Supplementary Information accompanies this paper on the Cell Death and Differentiation website (http://www.nature.com/cdd).
Edited by G Melino
Supplementary Material
References
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability'. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Malumbres M. Physiological relevance of cell cycle kinases. Physiol Rev. 2011;91:973–1007. doi: 10.1152/physrev.00025.2010. [DOI] [PubMed] [Google Scholar]
- Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem. 2012;81:561–585. doi: 10.1146/annurev-biochem-061611-090435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Lan W, Cleveland DW. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J Cell Biol. 2010;190:21–24. doi: 10.1083/jcb.201006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N, Jelluma N, Filippakopoulos P, Soundararajan M, Manak MS, Kwon M, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliedrecht T, Zhang C, Shokat KM, Kops GJ. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PLoS One. 2010;5:e10251. doi: 10.1371/journal.pone.0010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R, Caldarelli M, Mennecozzi M, Giorgini ML, Sola F, Cappella P, et al. Targeting the mitotic checkpoint for cancer therapy with NMS-P715, an inhibitor of MPS1 kinase. Cancer Res. 2010;70:10255–10264. doi: 10.1158/0008-5472.CAN-10-2101. [DOI] [PubMed] [Google Scholar]
- Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR, III, Lampson MA, Fukagawa T, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Liang H, Lim HH, Venkitaraman A, Surana U. Cdk1 promotes kinetochore bi-orientation and regulates Cdc20 expression during recovery from spindle checkpoint arrest. EMBO J. 2012;31:403–416. doi: 10.1038/emboj.2011.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Staples O, Taylor S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol. 2008;181:893–901. doi: 10.1083/jcb.200712028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison CP, Stumpff J, Wordeman L, Winey M. Mip1 associates with both the Mps1 kinase and actin, and is required for cell cortex stability and anaphase spindle positioning. Cell Cycle. 2011;10:783–793. doi: 10.4161/cc.10.5.14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike AN, Fisk HA. Centriole assembly and the role of Mps1: defensible or dispensable. Cell Div. 2011;6:9. doi: 10.1186/1747-1028-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YH, Huang YF, Lin TY, Shieh SY. The cell cycle checkpoint kinase CHK2 mediates DNA damage-induced stabilization of TTK/hMps1. Oncogene. 2009;28:1366–1378. doi: 10.1038/onc.2008.477. [DOI] [PubMed] [Google Scholar]
- Leng M, Chan DW, Luo H, Zhu C, Qin J, Wang Y. MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability. Proc Natl Acad Sci USA. 2006;103:11485–11490. doi: 10.1073/pnas.0601828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Chang MD, Shieh SY. TTK/hMps1 mediates the p53-dependent postmitotic checkpoint by phosphorylating p53 at Thr18. Mol Cell Biol. 2009;29:2935–2944. doi: 10.1128/MCB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Senovilla L, Criollo A, Jemaa M, Castedo M, et al. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2011;18:1403–1413. doi: 10.1038/cdd.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, et al. Apoptosis regulation in tetraploid cancer cells. EMBO J. 2006;25:2584–2595. doi: 10.1038/sj.emboj.7601127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore G, Nappi TC, Salerno P, Jiang Y, Garbi C, Ugolini C, et al. A cell proliferation and chromosomal instability signature in anaplastic thyroid carcinoma. Cancer Res. 2007;67:10148–10158. doi: 10.1158/0008-5472.CAN-07-1887. [DOI] [PubMed] [Google Scholar]
- Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- Daniel J, Coulter J, Woo JH, Wilsbach K, Gabrielson E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc Natl Acad Sci USA. 2011;108:5384–5389. doi: 10.1073/pnas.1007645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, Malumbres M. Killing cells by targeting mitosis. Cell Death Differ. 2012;19:369–377. doi: 10.1038/cdd.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–392. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- Jemaa M, Galluzzi L, Kepp O, Castedo M, Rello-Varona S, Vitale I, et al. Transgenerational cell fate profiling: a method for the graphical presentation of complex cell cycle alterations. Cell Cycle. 2013;12:183–190. doi: 10.4161/cc.23046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Jemaa M, Vitale I, Kepp O, Berardinelli F, Galluzzi L, Senovilla L, et al. Selective killing of p53-deficient cancer cells by SP600125. EMBO Mol Med. 2012;4:500–514. doi: 10.1002/emmm.201200228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif KD, Rogers A, Cassiano J, Roth BL, Cimbora DM, McKinnon R, et al. Characterization of the cellular and antitumor effects of MPI-0479605, a small-molecule inhibitor of the mitotic kinase Mps1. Mol Cancer Ther. 2011;10:2267–2275. doi: 10.1158/1535-7163.MCT-11-0453. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Wurzenberger C, Schmitz MH, Held M, Guizetti J, Maar S, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- van der Waal MS, Hengeveld RC, van der Horst A, Lens SM. Cell division control by the chromosomal passenger complex. Exp Cell Res. 2012;318:1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Maddox AS. Cytokinesis, ploidy and aneuploidy. J Pathol. 2012;226:338–351. doi: 10.1002/path.3013. [DOI] [PubMed] [Google Scholar]
- Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemaa M, Galluzzi L, Kepp O, Boileve A, Lissa D, Senovilla L, et al. Preferential killing of p53-deficient cancer cells by reversine. Cell Cycle. 2012;11:2149–2158. doi: 10.4161/cc.20621. [DOI] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science. 2012;337:1678–1684. doi: 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.