Abstract
In addition to protein-coding genes, the human genome makes a large amount of noncoding RNAs, including microRNAs and long noncoding RNAs (lncRNAs). Both microRNAs and lncRNAs have been shown to have a critical role in the regulation of cellular processes such as cell growth and apoptosis, as well as cancer progression and metastasis. Although it is well known that microRNAs can target a large number of protein-coding genes, little is known whether microRNAs can also target lncRNAs. In the present study, we determine whether miR-21 can regulate lncRNA expression. Using the lncRNA RT-PCR (reverse transcription-polymerase chain reaction) array carrying 83 human disease-related lncRNAs, we show that miR-21 is capable of suppressing the lncRNA growth arrest-specific 5 (GAS5). This negative correlation between miR-21 and GAS5 is also seen in breast tumor specimens. Of interest, GAS5 can also repress miR-21 expression. Whereas ectopic expression of GAS5 suppresses, GAS5–siRNA increases miR-21 expression. Importantly, there is a putative miR-21-binding site in exon 4 of GAS5; deletion of the miR-21-binding site abolishes this activity. Experiments with in vitro cell culture and xenograft mouse model suggest that GAS5 functions as a tumor suppressor. We further show that the biotin-labeled GAS5–RNA probe is able to pull down the key component (AGO2) of the RNA-induced silencing complex (RISC) and we subsequently identify miR-21 in this GAS5–RISC complex, implying that miR-21 and GAS5 may regulate each other in a way similar to the microRNA-mediated silencing of target mRNAs. Together, these results suggest that miR-21 targets not only tumor-suppressive protein-coding genes but also lncRNA GAS5.
Keywords: GAS5, post-transcriptional regulation, lncRNA, tumor suppressor, oncogene, miR-21
MicroRNAs are naturally occurring small noncoding RNAs capable of regulating target mRNAs at the post-transcriptional level.1, 2, 3 Overwhelming evidence suggests that in cancer, microRNAs are often dysregulated and they may serve as oncogenes or tumor suppressors.4 Among them, miR-21 is probably one of the most studied microRNAs and has been shown to function as an oncogene in various types of cancers. For example, miR-21 is upregulated in a variety of tumors including breast, prostate, colon, liver and lung cancer types,5, 6 squamous cell carcinoma7, 8 and cholangiocarcinoma.9 As an oncogene, miR-21 targets a number of genes involved in cell growth, proliferation and apoptosis as demonstrated in cell culture and animal models. Knockdown of miR-21 in cultured glioblastoma cells triggers the activation of caspases and leads to increased apoptotic cell death.10 Our previous studies showed that miR-21 not only promotes tumor growth11 but also enhances cell invasion and tumor metastasis.12 Furthermore, cell-specific modulations of miR-21 provides direct evidence that miR-21 functions as an oncogene in development of lymphoma.13
The fundamental consequence of miR-21 action may have to do with its ability to regulate numerous genes. The first experimentally validated miR-21 target was PTEN14 in human hepatocellular cancer cell lines. Given the role of PTEN in suppression of tumor growth, identification of PTEN as an miR-21 target may highlight the significance of miR-21 as an oncogene because as a repressor for PI3K, suppression of PTEN would promote Akt-mediated cell growth and proliferation. We have identified tropomyocin 1 (TPM1) as a direct target for miR-21 by proteomic approach combined with luciferase reporter and western blot.15 Both PTEN and TPM1 are known tumor suppressors. There are also other tumor suppressors such as PDCD4, maspin, RECK and TIMP3 that were shown to be targets for miR-21.12, 16, 17, 18 Of considerable interest, miR-21, similar to many other microRNAs, is able to simultaneously silence multiple genes involved in cell growth and apoptosis. To date, at least 42 experimentally validated targets have been reported based on miRecord (http://mirecords.biolead.org/) and it appears that the number of miR-21 targets is still growing.
However, all of these miR-21 targets are limited to protein-coding genes. It is well known that protein-coding genes account for only ∼2% of the human genome, whereas the vast majority of transcripts are noncoding RNAs. Among them are long noncoding RNAs (lncRNAs), with molecular weight of >200 bases in length. For example, both noncode database (http://www.noncode.org/NONCODERv3) and LNCipedia (http://www.lncipedia.org/) list over 30 000 human lncRNAs. Evidently, this number is even larger than the number of protein-coding genes and is expected to grow. Therefore, a question is whether microRNAs can also target lncRNAs, given their important role in cancer.19, 20, 21
The growth arrest-specific 5 (GAS5) is a noncoding gene that hosts a number of small nucleolar RNAs (snoRNAs). It was originally isolated from mouse NIH 3T3 cells using subtraction hybridization.22 GAS5 is induced by stress such as serum starvation and cell–cell contact inhibition. Human GAS5 has also been identified (NR_002578 and AF141346); at the primary sequence level, there is not much homology between human and mouse counterparts. However, it appears that their functions are conserved. Although GAS5 has been suggested to have a tumor-suppressive role,23 the underlying mechanism of GAS5-mediated gene expression having an impact on tumorigenesis is still elusive.
In the present study, we show that the human GAS5 is a direct target of miR-21 through interaction with the putative miR-21-binding site at exon 4 of GAG5. Moreover, GAS5 sensitizes tumor cells to UV or anticancer drug doxorubicin (doxo); GAS5 also suppresses cell invasion and tumor growth. Furthermore, we detect a negative correlation between GAS5 and miR-21 in clinical breast cancer specimens. Finally, GAS5 negatively regulates miR-21 possibly through the RNA-induced silencing complex (RISC), suggesting that there is a reciprocal repression feedback loop between miR-21 and GAS5.
Results
Identification of miR-21-regulated lncRNAs by the lncRNA RT-PCR array
As master gene regulators, microRNAs often have multiple targets of protein-coding genes. Among them, miR-21 has been implicated in the regulation of variety of cellular processes, in particular cancer development and metastasis. A number of tumor-suppressor genes have been identified as miR-21 targets; however, all of them are protein-coding genes. Therefore, in this study we asked whether miR-21 can target lncRNAs. Although there is an overwhelming number of lncRNAs that can be transcribed from the human and mouse genomes,24, 25 so far only a relatively small number of lncRNAs have been characterized and shown to be associated with human diseases (www.lncRNAdb.com), in particular human cancer. To focus on this group of lncRNAs, we chose a commercially available qRT-PCR (reverse transcription-polymerase chain reaction)-based array (Human Disease Related LncRNA Profiler, no. RA920D-1 from SBI) for lncRNA profiling.
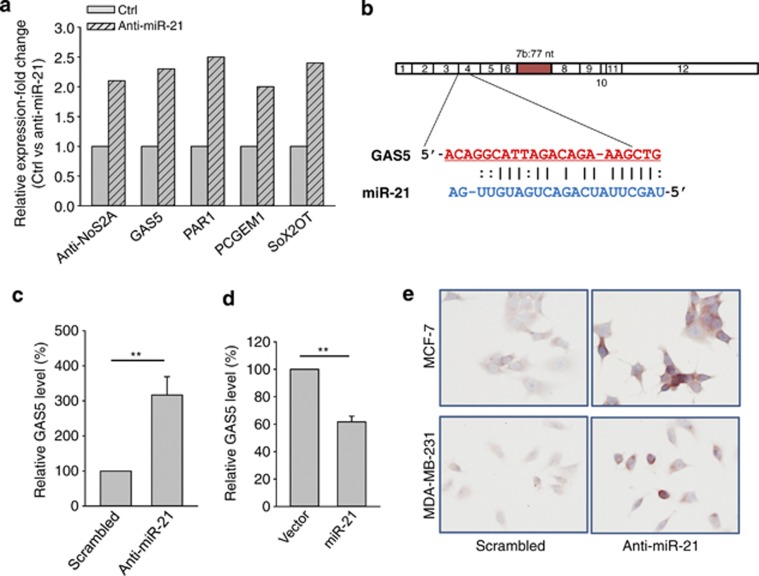
As miR-21 is often overexpressed in tumor specimens as well as in cancer cell lines, we knocked down miR-21 in breast cancer MCF-7 cells by anti-miR-21 and then determined the effect of anti-miR-21 on lncRNA expression. As shown in Supplementary Figure S1, suppression of miR-21 by anti-miR-21 caused over 80% reduction in miR-21 expression in breast cancer MCF-7 cells. Thus, total RNA extracted from these transfected cells was used for lncRNA profiling. From initial profiling, we found that five lncRNAs were induced by over a two-fold (Figure 1a). We were particularly interested in GAS5 because analysis with the RNA22 program (http://cbcsrv.watson.ibm.com/rna22.html) identified a complementary region with miR-21 (Figure 1b). To further confirm this finding, we designed a new set of primers. GAS5 transcripts are subjected to complex processing and several human isoforms were reported, and all carry exon 12.26 Thus, the new primer set (GAS5-RT-5.1 and GAS5-RT-3.1) was derived from exon 12 to cover all known isoforms. RT-PCR confirmed the profiling results. In addition, we also designed another set of primers (GAS5-RT-5.2 and GAS5-RT-3.2) for the measurement of GAS5 levels by qRT-PCR. As these two sets of primers provided similar results in multiple samples, only one set was used in the following experiments. We found that whereas anti-miR-21 increased GAS5 (Figure 1c), ectopic expression of miR-21 suppressed GAS5 (Figure 1d). To further demonstrate this upregulation of GAS5 by anti-miR-21, we performed in situ hybridization (ISH). As shown in Figure 1e, it is evident that the GAS5 signal was much stronger in the anti-miR-21 cells than in scrambled control cells in both breast cancer MCF-7 and MDA-MB-231 cells. These results suggest that, similar to protein-coding genes, lncRNA GAS5 can also be targeted by miR-21.
Figure 1.
Identification of GAS5 as an miR-21 target. (a) Identification of putative miR-21-regulated lncRNAs. MCF-7 cells were transfected with either scrambled oligo (Ctrl) or anti-miR-21. The cells were harvested 24 h after transfection and total RNA was isolated, followed by lncRNA profiling as described in the text. GAS5 was among five lncRNAs with over a two-fold increase by anit-miR-21. (b) Alignment of potential miR-21 base pairing with GAS5 as identified by RNA22 program (http://cbcsrv.watson.ibm.com/rna22.html). GAS5 (top) consists of 12 exons, where the putative binding site is in exon 4. The isoform b of GAS5 has 77 nucleotides at exon 7. (c) Detection of induction of GAS5 in the anti-miR-21 cells using newly designed primers (GAS5-RT-5.1 and GAS5-RT-3.1). Experimental procedure was same as in (a). (d) Ectopic expression of miR-21 suppresses GAS5. MCF-7 cells were transfected with vector alone or miR-21, and total RNA was isolated 24 h after transfection for RT-PCR. (e) Induction of GAS5 by anti-miR-21 as detected by in situ hybridization. MCF-7 or MDA-MB-231 cells were transfected with scrambled oligo or anti-miR-21. GAS5 signal was detected by in situ hybridization as detailed in the Materials and Methods section. Error bars represent S.E.M., n=3. **P<0.01
Reciprocal repression of miR-21 and GAS5
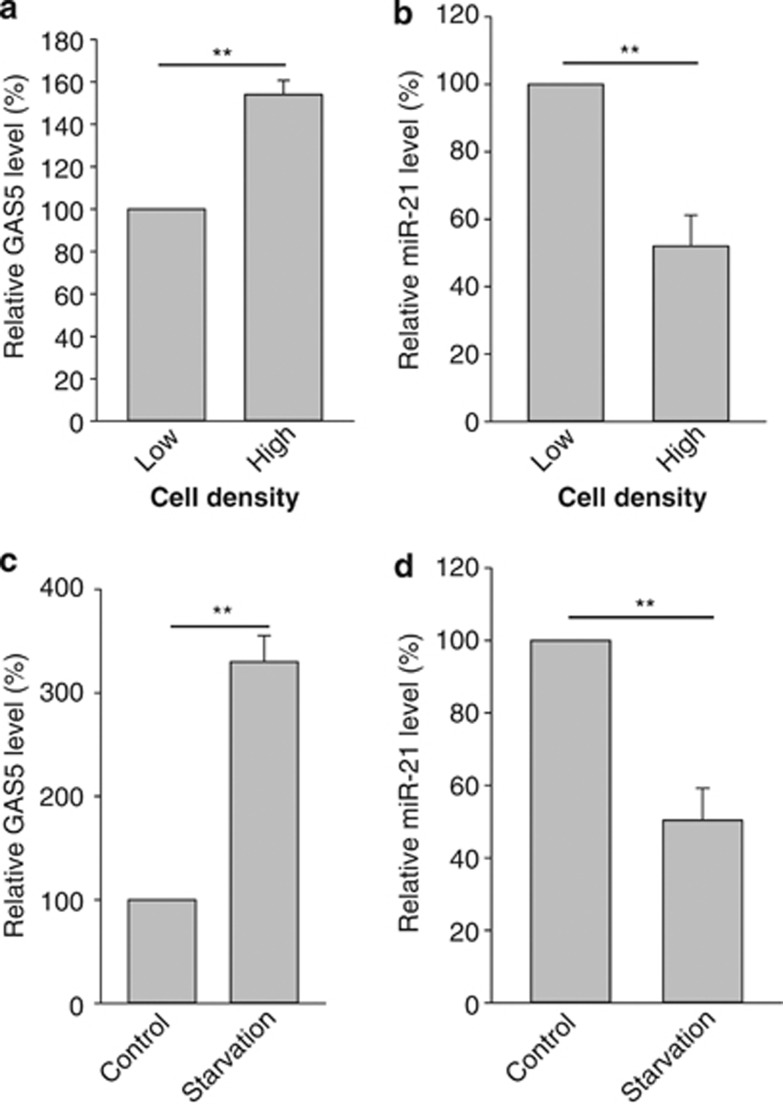
As a stress-inducible gene, GAS5 is expressed in response to cellular stress such as cell–cell contact inhibition and serum starvation. Hence, we characterized this negative correlation between GAS5 and miR-21 under these two stresses. As showed in Figure 2a, the GAS5 level at a high cell density (full confluence) was about 50% higher than that at a low cell density (∼40% cell confluence). In contrast, the miR-21 level at the high cell density was about 50% of the low cell density (Figure 2b). Similarly, serum starvation induced GAS5 by almost three-fold (Figure 2c), whereas miR-21 was downregulated by over 50% under the same condition (Figure 2d).
Figure 2.
Effect of cell stress on the expression of GAS5 and miR-21. (a and b) Cell contact inhibition increases GAS5, but suppresses miR-21. MCF-7 cells were grown in culture media at a low (∼40% confluence) or high cell density (full confluence) when the cells were harvested for RNA isolation. (c and d) Upregulation of GAS5 and downregulation of miR-21 by serum starvation. MCF-7 cells were treated with 0% (serum starvation) or 10% FBS for 24 h before isolation of total RNA and qRT-PCR. Error bars represent S.E.M., n=3. **P<0.01
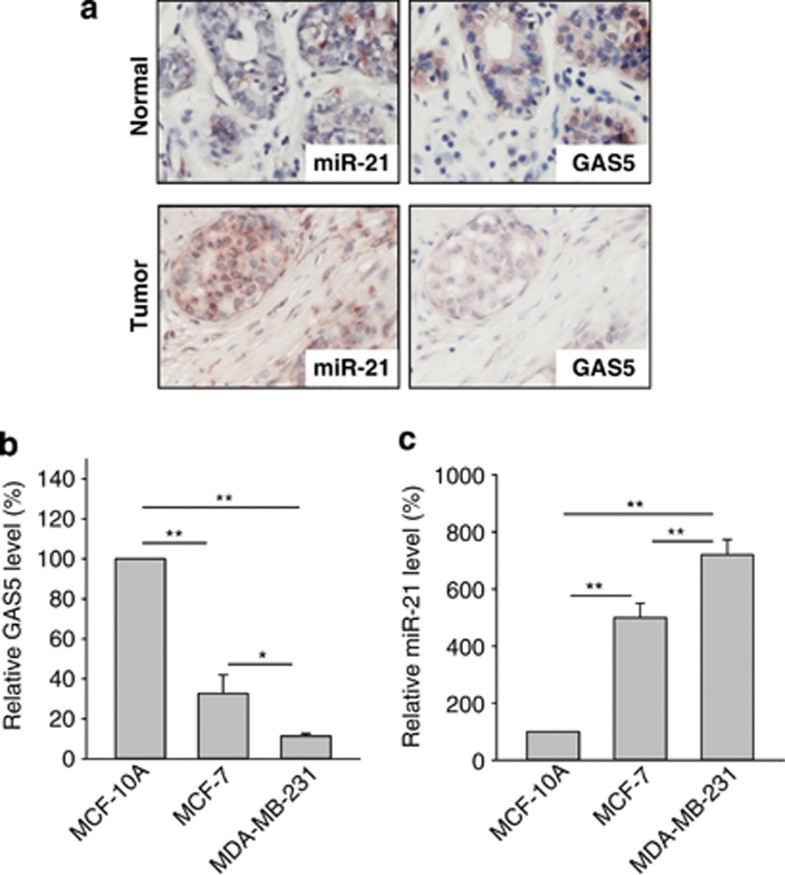
This negative correlation between GAS5 and miR-21 was also detected in clinical specimens. For example, ISH showed that whereas GAS5 was downregulated in breast tumors, miR-21 was upregulated in the same breast tumors (Figure 3a). Consistent with cell-culture staining, GAS5 was localized predominantly in the cytoplasm. Statistical analysis of breast cancer tissue microarray (TMA) revealed a significant negative correlation between GAS5 and miR-21 (Supplementary Figure S2). Moreover, we found that GAS5 was differentially expressed in breast cell lines and the GAS5 level in the non-malignant breast MCF-10A cells was higher than in breast cancer MCF-7 or MDA-MB-231 cells as detected using qRT-PCR; the lowest level of GAS5 was seen in aggressive MDA-MB-231 cells (Figure 3b). In contrast, miR-21 level was in opposite trend in these cell lines (Figure 3c).
Figure 3.
A negative correlation between GAS5 and miR-21 in clinical specimens and cell lines. (a) GAS5 is downregulated, whereas miR-21 is upregulated in breast tumor. Consecutively cut sections of normal breast tissue and tumor tissue were subjected to in situ hybridization to detect GAS5 (left panel) and miR-21 (right panel). (b and c) Expression of GAS5 and miR-21 in MCF-10A, MCF-7 and MDA-MB-231 cells. Levels of GAS5 and miR-21 were detected using qRT-PCR as described in the Materials and Methods section. Error bars represent S.E.M., n=3. **P<0.01; *P<0.05
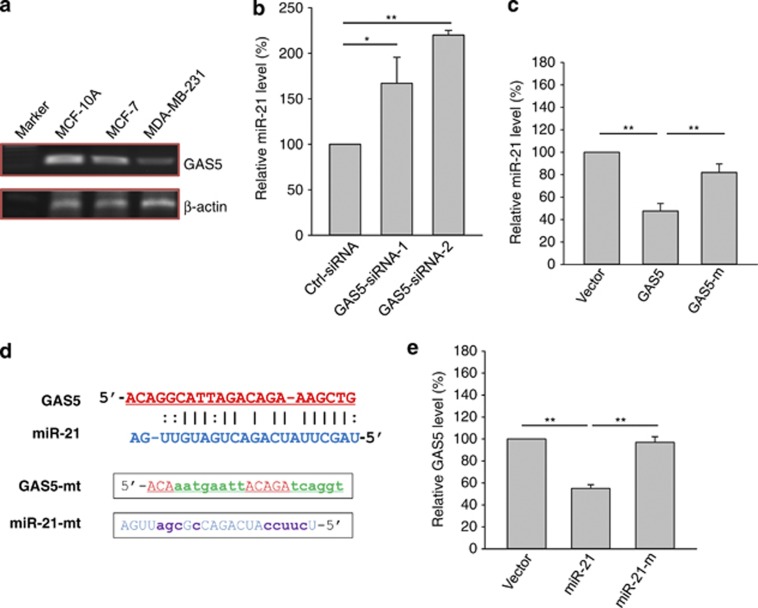
A previous report showed that human GAS5 is expressed as two alternative isoforms, GAS5a and GAS5b in addition to five GAS5 EST sequences.26 The only difference between GAS5a and GAS5b was at exon 7. Whereas GAS5b (NR_002578) has 77 nt, GAS5a has only 45 nt, missing 32 nt at the 3′ end. To determine which isoform is predominantly expressed in cancer cells, we designed a set of primers covering exon 7 (GAS5-RT-5.2 and GAS5-RT-3.1A). RT-PCR revealed that GAS5b was a major isoform in three cell lines (Figure 4a), which was confirmed by DNA sequencing. Therefore, we cloned the entire sequence of this isoform b (GAS5b) into pCDH-MSCV-MCS-EF1-GFP-T2A-Pu, but it was still simply called GAS5 in this study. Ectopic expression of this construct was confirmed using qRT-PCR.
Figure 4.
Reciprocal repression of GAS5 and miR-21. (a) Identification of GAS5 b isoform as a predominant form in MCF-10A, MCF-7 and MDA-MB-231 cells. Primers used for RT-PCR are GAS5-RT-5.2 and GAS5-RT-3.1A. (b) Upregulation of miR-21 by GAS5–siRNA. MCF-7 cells were transfected with control siRNA or GAS5–siRNA, and total RNA was isolated 24 h after transfection. (c) Downregulation of miR-21 by ectopic expression of GAS5. MCF-7 cells were transfected with vector control or GAS5 or mutant GAS5, and total RNA was isolated 24 h after transfection. (d) The mutant GAS5 and mutant miR-21 at the putative binding site. (e) The binding site in miR-21 is important for miR-21 to suppress GAS5. MCF-7 cells were transfected with vector control or miR-21 or mutant miR-21, and the rest procedure was same as in (c). Error bars represent S.E.M., n=3. **P<0.01; *P<0.05
Our recent studies showed that whereas miR-211 can negatively regulate lncRNA loc285194, at the same time, loc285194 can also repress miR-211 expression.27 Therefore, we asked whether this is also the case for miR-21 and GAS5. GAS5–siRNAs significantly reduced the endogenous GAS5 (Supplementary Figure S3); at the same time, GAS5–siRNAs increased the miR-21 level (Figure 4b). In contrast, ectopic expression of GAS5 suppressed miR-21 (Figure 4c). To determine whether this suppression is through the potential interaction at the putative miR-21-binding site, we generated a GAS5 mutant (Figure 4d). This mutant GAS5 clone revealed no significant suppression of miR-21 compared with the wild-type GAS5 (Figure 4c). To further determine whether this reduction in GAS5 is specific to miR-21, we generated a mutant with changes of eight nucleotides in miR-21 (Figure 4d). As expected, whereas wild-type miR-21 was able to suppress GAS5 by over 50%, the mutant miR-21 lost this activity (Figure 4e), demonstrating the importance of this binding site for the reciprocal repression of GAS5 and miR-21.
GAS5 functions as a potential tumor suppressor
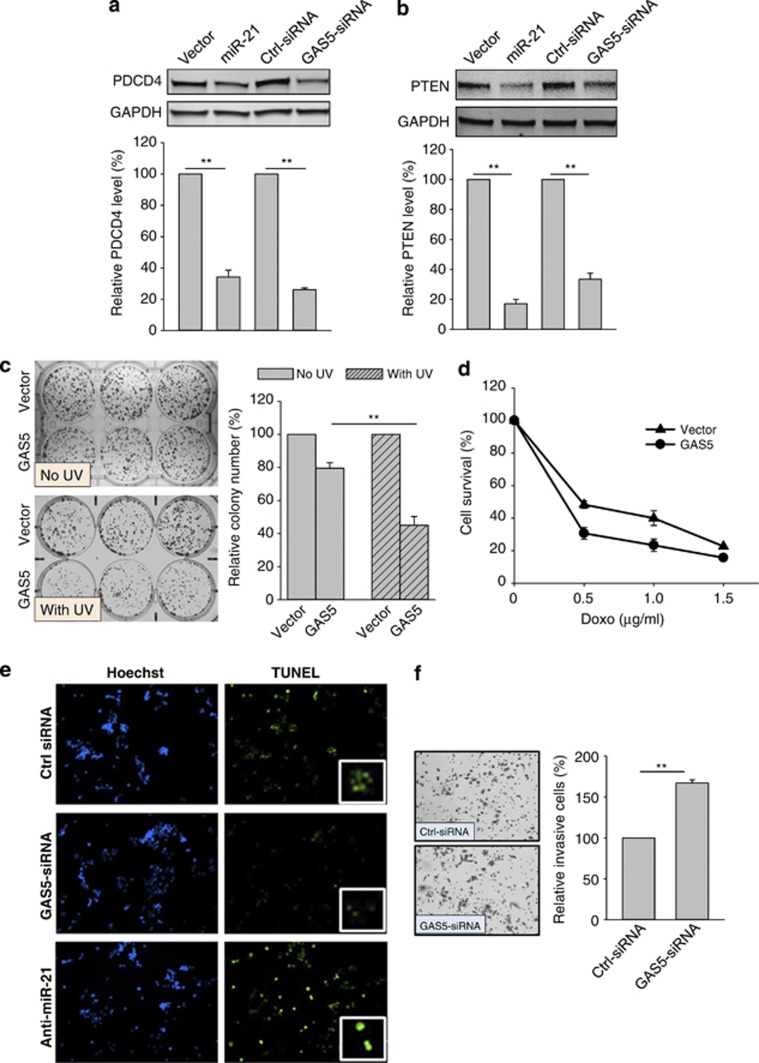
Having demonstrated the reciprocal negative regulation between miR-21 and GAS5, we then examined the functional effect of GAS5. We first determined whether GAS5 has any effect on miR-21 targets and found that GAS5–siRNA suppressed both PTEN and PDCD4 – two well-known miR-21 targets14, 17 (Figures 5a and b). As a control, ectopic expression of miR-21 also suppressed both PTEN and PDCD4. These results suggest that GAS5-mediated suppression of miR-21 has functional consequences. To further determine the effect of GAS5 on cell growth, we performed both MTT and colony-formation assays after the transfection of GAS5. Although GAS5 did not significantly affect cell growth under normal culture conditions (not shown), GAS5 was able to enhance the UV-induced cell growth inhibition (Figure 5c), in agreement with the previous report.23 Moreover, GAS5 also increased doxo-induced cell growth inhibition (Figure 5d), which might be through GAS5-regulated apoptosis. For example, GAS5–siRNA suppressed, whereas anti-miR-21 enhanced doxo-induced apoptosis (Figure 5e). Of interest, GAS5 significantly suppressed cell invasion in MDA-MB-231 cells (Figure 5f). Matrigel chamber assays revealed that GAS5–siRNA was able to increase cell invasion by over 60% in MDA-MB-231 cells. Together, these results suggest that GAS5 serves as an important miR-21 target to impact the stress-induced cell growth inhibition, apoptosis and tumor cell invasion.
Figure 5.
Identification of tumor-suppressive role of GAS5. (a and b) Effect of miR-21 and GAS5–siRNA on expression of the tumor suppressors PDCD4 and PTEN. MCF-7 cells were transfected with vector or miR-21 and control siRNA or GAS5–siRNA. Cellular protein was isolated from the transfected cells 24 h later and used for western blot. Top panels are representative western blots; bottom panels are relative expression of these proteins from three separate experiments. (c) GAS5 sensitizes cells to UV treatment. MCF-7 cells were transfected with vector or GAS5 for 24 h. The cells were seeded in 12-well plates and then UV radiated with 40 J/m2. Fourteen days later, colonies were stained with crystal violate and counted. (d) GAS5 sensitizes cells to doxo. The same transfected cells (as in c) were treated with indicated concentrations of doxo for 3 days, and the relative cell survival was determined using MTT assay. (e) Detection of apoptosis using the TUNEL assay. MCF-7 cells were first transfected with control siRNA, GAS5–siRNA or anti-miR-21 and then treated with Doxo at 1 μg/ml for 24 h before the TUNEL assay, as described in the text. Inserts are enlarged apoptotic cells. Error bars represent S.E.M., n=3. *P<0.05; **P<0.01. (f) Suppression of cell invasion by GAS5. MDA-MB-231 cells were transfected with control siRNA or GAS5–siRNA. Matrigel chamber invasion assay was carried out 24 h after transfection
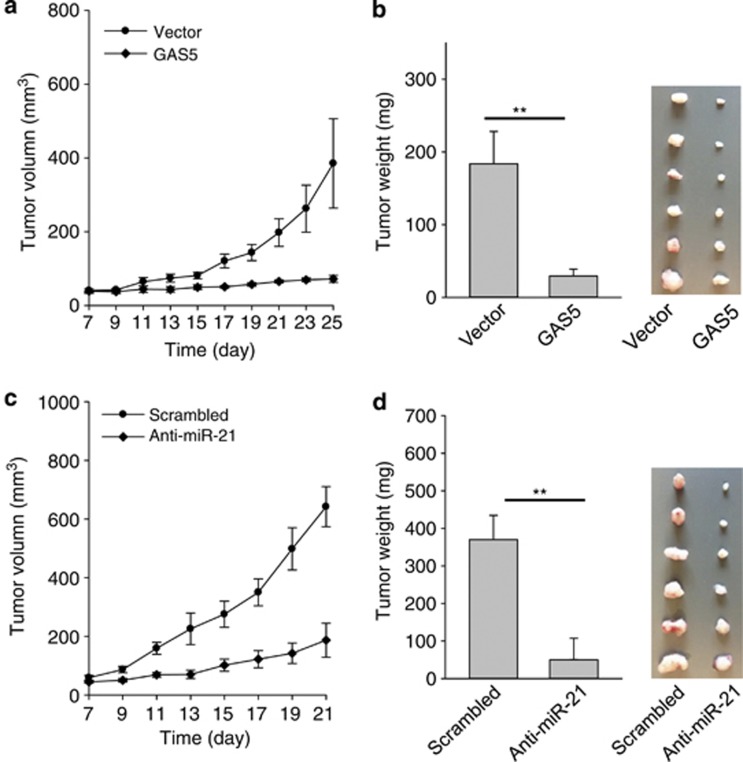
To further demonstrate the tumor-suppressive role of GAS5, we injected MCF-7 cells infected with GAS5 into female nude mice. As shown in Figures 6a and b, tumors derived from the GAS5 cells grew much more slowly than the vector control, and the tumor weight was also significantly lesser than the vector control. Similar to GAS5, anti-miR-21 also suppressed tumor growth and reduced tumor weight (Figures 6c and d). To determine whether there is a negative correlation between GAS5 and miR-21 in these tumors, we isolated total RNA from three tumors separately. Although ectopic expression of GAS5 only yielded a moderate increase in the GAS5 level, we were able to detect a significant reduction in miR-21 (Supplementary Figure S4).
Figure 6.
Suppression of tumor growth by GAS5 as well as anti-miR-21 in the xenograft mouse model. (a and b) Suppression of tumor growth by GAS5. MCF-7 cells were infected with vector or GAS5 and then injected into mammary fat pads as described in the text. Tumor growth was measured every other day after 7 days of injection (a) and tumors were then harvested on day 25 and weighed (b). Actual tumor size after the harvest was shown in the right panel. (c and d) Suppression of tumor growth by anti-miR-21. Experimental procedure and tumor measurement were same as for GAS5, which was shown in (c). Tumors were harvested on day 21 and weighed (d). Error bars represent S.E.M., n=6. **P<0.01
Association of both miR-21 and GAS5 with the RISC complex
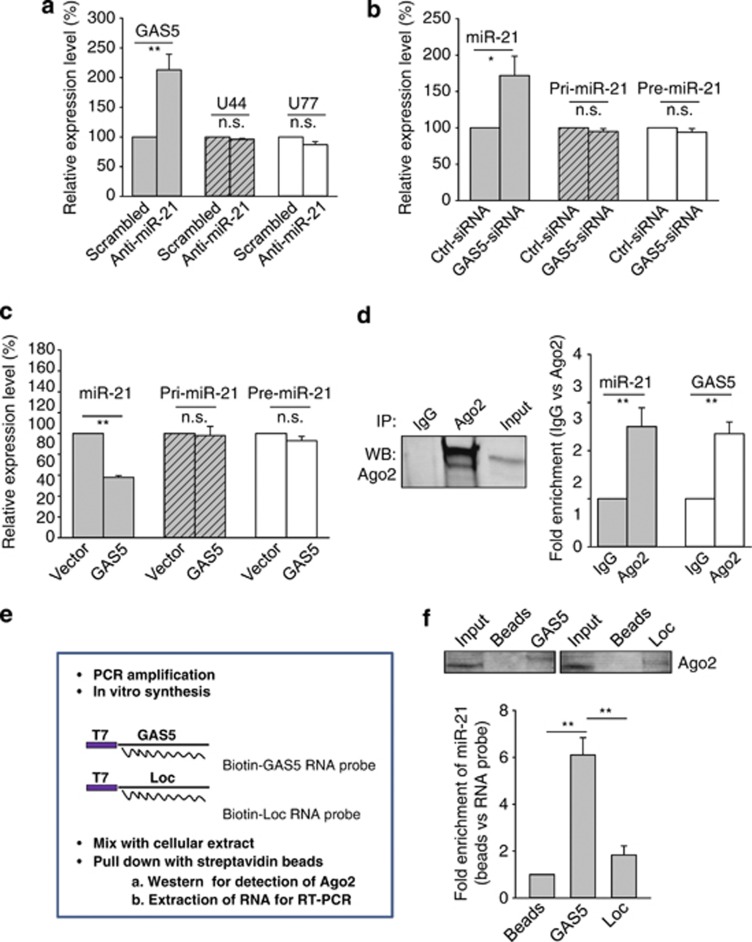
To better characterize this reciprocal negative regulation, we first examined whether miR-21 affects the level of U77 and U44, both of which are imbedded between exons 4 and 5, and exons 5 and 6 of the GAS5 gene26 (Supplementary Figure S5). As shown in Figure 7a, although we detected significant upregulation of GAS5 by anti-miR-21, no such upregulation of U44 or U77 was seen, suggesting that this negative regulation of GAS5 by miR-21 might be through a post-transcriptional mechanism, consistent with a previous report suggesting that GAS5 can be controlled at the post-transcriptional level.28 We then determined the effect of GAS5 on levels of pri-miR-21, pre-miR-21 and mature miR-21. Ectopic expression of GAS5 caused a significant reduction in mature miR-21 in GAS5-transfected cells (Figure 7b). However, GAS5 had no effect on pri-miR-21 and pre-miR-21 (Figure 7b). On the other hand, GAS5–siRNA caused the upregulation of mature miR-21 but not of pri-miR-21 or pre-miR-21 (Figure 7c), suggesting that this GAS5-mediated regulation of miR-21 is also likely through a post-transcriptional mechanism. Thus, we reasoned that microRNAs may regulate the expression of lncRNAs possibly through RNAi pathways, which would imply that both GAS5 and miR-21 might be in the RISC complex. Therefore, we performed RNA immunoprecipitation using antibody against AGO2 – a key component of the RISC complex. We first confirmed that the AGO2 antibody precipitated the AGO2 protein from our cellular extract (Figure 7d, left panel). As expected, we detected both miR-21 and GAS5 in the AGO2 pellet (Figure 7d, right panel). Both miR-21 and GAS5 had over a two-fold enrichment (AGO2 antibody versus IgG). However, the identification of the association of AGO2 with miR-21 and GAS5 cannot rule out the possibility that miR-21 and GAS5 are in a separate AGO2 complex. Therefore, we synthesized a GAS5 RNA probe, labeled it with biotin and then mixed it with cellular extract (Figure 7e). After precipitation with streptavidin beads, we detected AGO2 by western blot (Figure 7f, top panel), suggesting that GAS5 RNA interacts with AGO2. Importantly, we also detected a significant amount of miR-21 in this GAS5-pulled down pellet carrying AGO2 (Figure 7f, bottom panel), whereas there was only a slight increase in the miR-21 when loc285194 RNA was used as a probe. Thus, these results suggest that both GAS5 and miR-21 are likely in the same AGO2 complex.
Figure 7.
Post-transcriptional regulation of GAS5 and miR-21. (a) Whereas anti-miR-21 upregulates GAS5, it has no effect on U44 and U77, which are imbedded in the GAS5 gene. MCF-7 cells were transfected with scrambled oligo or anti-miR-21 and the rest of the procedure was same as in Figure 1c. Primers for U44 and U77 were U44-RT-5.1 and U44-RT-3.1; U77-RT-5.1 and U77-RT-3.1, respectively. (b and c) Effect of ectopic expression of GAS5 or GAS5–siRNA on pri-miR-21, pre-miR-21 and mature miR-21. MCF-7 cells were transfected with vector alone or GAS5, or control siRNA or GAS5–siRNA. Total RNA was isolated from the cells 24 h later, followed by qRT-PCR. Primers for pri-miR-21 were Pri-miR-21-RT-5.1 and Pri-miR-21-RT-3.1; Pre-miR-21-RT-5.1 and Pre-miR-21-RT-3.1. (d) Association of GAS5 and miR-21 with AGO2. Cellular lysates from MCF-7 cells were used for RNA immunoprecipitation with AGO2 antibody. Detection of AGO2 using IP-western (left panel), and detection of GAS5 and miR-21 using qRT-PCR (right panel). (e and f) Identification of GAS5 and miR-21 in the same RISC complex by RNA precipitation. (e) A procedure for making biotin-labeled RNA probes and RNA precipitation. In vitro transcribed RNA probes were made using T7 RNA polymerase, followed by precipitation assays as described in the Materials and Methods section. (f) Detection of miR-21 in the GAS5 RNA-precipitated samples. Error bars represent S.E.M., n=3. *P<0.05; **P<0.01; n.s., not significant
Discussion
As an oncogenic microRNA, miR-21 has been well characterized for its role in cancer. For instance, miR-21 has been experimentally shown to target a large number of protein-coding genes that have a critical role in tumor growth and metastasis. We present evidence here that miR-21 targets not only protein-coding genes but also noncoding lncRNAs such as GAS5, which is a putative tumor suppressor. Thus, identification of GAS5 as an miR-21 target expands the repertoire of miR-21 targets. Furthermore, GAS5 is also able to repress miR-21, thus forming a reciprocal repression-regulatory loop. These results provide further supporting evidence for the endogenous competitive RNA (ceRNA)-regulatory network.29
GAS5 is a stress-inducible gene, and it is differentially expressed in normal and tumor tissues, as well as in various cell lines. We show a negative correlation between GAS5 and miR-21 in breast tumor specimens and cell lines. For instance, GAS5 is downregulated and miR-21 is upregulated in the breast tumors; in contrast, GAS5 is highly expressed but miR-21 is underexpressed in the normal breast tissues. Furthermore, the non-malignant MCF-10A cells express the highest level of GAS5 among the three cell lines tested, whereas the aggressive breast cancer MDA-MB-231 cells express the lowest level of GAS5 among them. Ectopic expression of GAS5 sensitizes tumor cells to UV and doxo treatment. Finally, the suppression of GAS5 with RNAi is able to increase the invasiveness of MDA-MB-231 cells. Together, these results support the notion that GAS5 is a tumor suppressor.
A previous report with the mouse GAS5 (BC004622 IMAGE:3585621) clone has suggested its tumor-suppressive role in breast cancer cells.23 However, it was not clear about the role of the human GAS5 in this aspect. Our data suggest that, similar to the mouse GAS5, the human GAS5 also has a tumor-suppressive role. Importantly, our study may provide a possible mechanism for GAS5 as a tumor suppressor, which may be attributed to its ability to suppress the oncogenic miR-21. For example, we show that similar to miR-21, GAS5–siRNA is able to suppress both the well-known tumor suppressors PTEN and PDCD4. There is overwhelming evidence that these two tumor suppressors are major miR-21 targets in various types of cancer, including breast cancer. Therefore, demonstration of the ability of GAS5 to regulate PTEN and PDCD4 can at least in part explain why GAS5 can sensitize tumor cells to UV and doxo, as well as suppress cell invasion.
Our study further provides a possible regulation of miR-21. It is well known that microRNAs are regulated by several mechanisms. At the transcriptional level, several important transcription factors such as c-Myc and p53 have been shown to have a key role in the regulation of microRNA expression. Early studies indicate that the miR-17∼92 cluster has been shown to be induced by c-Myc.30, 31 The p53 status can also affect the expression of many microRNAs in colon cancer cells, leading to differential expression of many potential microRNA target genes.32 Moreover, p53 is a key regulator for expression of the miR-34 family.33 At the transcriptional level, miR-21 has been shown to be regulated by several transcription factors including Stat3 and AP-1. For instance, miR-21 can be induced by IL-6 in a STAT3-dependent manner wherein the miR-21 promoter carries two putative STAT3-binding sites.34 Subsequently, a study identified a double-negative feedback mechanism regarding miR-21 promoter involving NFI and AP-1.35 In addition, miR-21 is upregulated via the MAPK (ERK1/2) pathway upon stimulation of HER2/neu signaling in breast cancer cells, and the overexpression of other ERK1/2 activators such as RASV12 is sufficient to induce miR-21 upregulation in HER2/neu-negative breast cancer cells.36 Whereas transcriptional activators can lead to an increased level of miR-21, transcriptional repressors can suppress miR-21 expression. For example, the neuronal repressor REST (RE1-silencing transcription factor) transcriptionally represses miR-21 and this repression of miR-21 appears to be critical to maintain self-renewal and pluripotency in mouse ES cells.37
In addition to transcription regulation, epigenetic factors, such as acetylation and methylation,38, 39 or factors involved in microRNA biogenesis or processing40, 41, 42, 43 can also have a regulatory role in microRNA expression. The most characterized factor is probably lin-28 that can bind to the loop region of let-7 to block its processing.40, 41, 42, 43 Similarly, the expression of miR-21 is also subjected to post-transcriptional regulation. For example, TGFβ and BMP signalings have been implicated in the processing of pri-miR-21 into pre-miR-21 by Drosha. Specifically, TGFβ- and BMP-specific SMAD signal transducers are recruited to pri-miR-21 in a complex with the RNA helicase p68, suggesting the importance of ligand-mediated post-transcriptional regulation of miR-21 biogenesis.44 Furthermore, a recent study suggests that PTEN negatively regulates the expression of miR-21 at the post-transcriptional level.45 Of interest, the RNA-regulatory protein RNH1 directly interacts with the Drosha complex and PTEN blocks this interaction through which PTEN suppresses miR-21 processing. Our present study suggests another possible post-transcriptional regulation mechanism for miR-21. In addition to targeting both protein-coding and noncoding genes, miR-21 itself is also subjected to regulation by lncRNAs, such as GAS5. Therefore, this reciprocal repression of miR-21 and GAS5 may highlight the significance of RNA–RNA interaction involving microRNAs and lncRNAs in tumorigenesis, adding another piece of puzzle to the microRNA regulatory network.
We have recently shown that lncRNA loc285194 suppresses tumor cell growth in part through the suppression of miR-211. Ectopic expression of loc285194 reduces the miR-211 level and the miR-211-binding sites are critical for this loc285194-mediated repression.27 This is similar to the reciprocal repression between GAS5 and miR-21. Evidently, GAS5 may function as the endogenous sponge. It has been reported that ncRNA UC34A,46 HULC47 and linc-MD148 can interact with corresponding microRNAs, in which microRNA response elements (MREs) may serve as letters of a new language.29 Furthermore, recent studies suggest that the tumor suppressor PTEN is a key component of the CeRNA regulatory system.49, 50, 51 Given that GAS5 can also regulate PTEN through miR-21, this would serve as another layer of regulation. Finally, our study also suggests that this reciprocal repression of miR-21 and GAS5 is likely through the pathway involving the RISC complex. Accordingly, a better understanding of the microRNA–lncRNA interaction and their regulation will provide new insight into mechanisms underlying various aspects of tumorigenesis including tumor growth, invasion and metastasis, and tumor-drug resistance.
Materials and Methods
Reagents
Primary antibodies Ago2 and PTEN were purchased from Cell Signaling (Danvers, MA, USA); PDCD4 from Epitomics (Burlingame, CA, USA); GAPDH from Protein Tech (Chicago, IL, USA) and β-actin from Sigma (St. Louis, MO, USA). Secondary antibodies conjugated with IRDye 800CW or IRDye 680 were purchased from LI-COR Biosciences (Lincoln, NE, USA). PCR primers were purchased from IDT (Coralville, IA, USA) (Supplementary Table S1). GAS5–siRNAs and control siRNA were purchased from ThermoFisher Scientific (Waltham, MA, USA). Biotin-labeled GAS5-LNA probe and control oligos for ISH, and anti-miR-21 LNA were purchased from Exiqon (2950 Vedbaek, Denmark). Breast cancer TMAs were purchased from US Biomax (Rockville, MD, USA).
Cell culture
Breast cancer MCF-7 and MDA-MB-231 cells (both from ATCC) were grown in RPMI 1640 (Lonza, ThermoFisher Scientific) supplemented with 10% FBS (Sigma-Aldrich), 2 mM glutamine, 100 units of penicillin/ml and 100 μg of streptomycin/ml (Lonza). MCF-10A cells were grown in the serum-free MEGM medium (Lonza). Cells were incubated at 37 °C and supplemented with 5% CO2 in the humidified chamber.
LncRNA profiling
We used Human Disease Related LncRNA Profiler (CAT no. RA920D, System Biosciences, Mountain View, CA, USA) consisting of 83 lncRNAs, which were selected from the lncRNA database (www.lncRNAdb.org)52 or RNA database (http://www.ncrna.org/frnadb/). Total RNA was isolated from MCF-7 cells transfected with either scrambled siRNA or anti-miR-21, which was described previously.12 Reverse transcription was carried out by using RevertAid Reverse Transcriptase (Fermentas, ThermoFisher Scientific) and random primer mix (New England BioLabs). The values for scrambled control after normalization by the internal controls served as a basal level of expression of GAS5; delta–delta Ct values (scrambled control versus anti-miR-21) were used to determine their relative expression as fold changes, as previously described.11
Transfection
Cells were transfected with siRNAs using RNAfectin reagent (Applied Biological Materials) or plasmid DNA using DNAfectin (Applied Biological Materials) following the manufacturer's protocol.
Plasmid construction
PCR reactions for cloning purpose used Phusion enzyme (New England BioLabs, Ipswich, MA, USA). The entire GAS5 sequence was amplified with RT-PCR using primers GAS5-R1-5.1 and GAS5-Not1-3.1, and then cloned into the expression vector pCDH-MSCV-EF1-GFP-T2A-Pu (System Biosciences) using Cold Fusion kit (SBI). The same strategy was used to generate other constructs in this study, otherwise stated. Construction of miR-21 has been described previously.12 For the cloning of GAS5 with mutations at the putative miR-21-binding site, two-step PCR procedure was used as described previously.53 Briefly, we used two sets of primers (GAS5-R1-5.1 and GAS5-miR21-BS-M-3.1; GAS5-miR21-BS-M-5.1 and GAS5-Not1-3.1) along with the wild-type GAS5 as a template to generate two overlapped PCR products. We then used these two products as templates along with primers GAS5-R1-5.1 and GAS5-Not1-3.1 to generate a single but mutated product and subsequently cloned it into pCDH-MSCV-EF1-GFP-T2A-Pu. To construct an miR-21 carrying a mutant binding site to GAS5, we used the same two-step PCR approach along with primers miR-21-pCDH-R1-5.1 and miR-21-GAS5-mt-3.1; miR-21-GAS5-mt-5.1 and miR-21-pCDH-Not1-3.1. The final PCR product was cloned into pCDH-MCV-EF1-copGFP (SBI). All PCR products were verified by DNA sequencing.
qRT-PCR
LncRNA profiling by qRT-PCR was performed according to the manufacturer's protocol. To specifically detect the expression levels of GAS5, pri-miR-21 and pre-miR-21, we used the SYBR Green method, with primers listed in Supplementary Table S1. β-actin or U6 was used as an internal control. To detect the expression of mature miR-21, we use the poly A polymerase-based SYBR Green method (SBI). U6 RNA was used as an internal control. In both cases, delta–delta Ct values were used to determine their relative expression as fold changes, as previously described.11
Western blot
Cells were harvested, and protein was extracted from transfected cells and quantified as previously described53 using pre-casted gels (ThermoFisher Scientific).
RNA precipitation
To determine whether GAS5 is associated with the RISC complex, we performed an RNA precipitation assay using synthesized GAS5 as a probe and then detected AGO2 from the pellet using western blot and miR-21 using qRT-PCR. In brief, the DNA fragment covering the entire GAS5 sequence was PCR-amplified using T7-containing primers (T7-GAS5-5.1 and T7-GAS5-Not1-3.1) and then cloned into pCR8 (Invitrogen, Carlsbad, CA, USA). In addition, another lncRNA, loc285194, was also cloned as a negative control and used in precipitation experiments for comparison. The resultant plasmid DNA was linearized with restriction enzyme NotI, which was introduced from the reverse PCR primer, and then used to synthesize RNA by T7 polymerase. The rest of the procedure was same as previously described.54 The pellets were used either for the extraction of RNA for RT-PCR or for western blot according to the standard procedures.
RNA immunoprecipitation
RNA immunoprecipitaion used the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) and the AGO2 antibody according to the manufacturer's protocol. After the antibody was recovered by protein A+G beads, qRT-PCR was performed to detect GAS5 and miR-21 in the precipitates.
In situ hybridization
ISH was used to detect GAS5 in clinical specimens and cancer cell lines after doxo treatment based on a previously described method55 with some modifications. A biotin-labeled antisense LNA probe was derived from nt 241–267 of GAS5 (Supplementary Table S1). For paraffin-imbedded tissue, after deparaffinization and rehydration, the samples were treated with peroxidase-quenching solution; proteinase K was added to digest tissues before prehybridization and hybridization, which were carried out at 55 °C for 30 min and 4 h, respectively. Then streptavidin–HRP was used to react with the bound biotin-labeled probe. The signal was further amplified using TSA amplification kit (Perkin Elmer). Finally, the signal was revealed with Ultra Vision One polymer and AEC chromogen (ThermoFisher Scientific). To detect GAS5 induction by anti-miR-21, we first transfected MCF-7 cells with scrambled siRNA or GAS5–siRNA and then seeded them on coverslips overnight before fixing the cells with 2% formaldehyde for 15 min at room temperature.
TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay
MCF-7 cells were transfected with control siRNA, GAS5–siRNA or anti-miR-21 and then treated with doxo at 1 μg/ml for 24 h. The rest of the procedure has been previously described.56
MTT assay
MTT assay was performed to determine the effect of GAS5 in cell growth in response to dox as described previously.57 The measurement was carried out 3 days after doxo treatment.
Clonogenic assay
Long-term survival of cells transfected with GAS5-expressing constructs or vector alone was assessed by the ability of the cells to form colonies in culture dishes for 14 days. Colonies were fixed and stained with 0.05% crystal violate before counting.
Invasion assay
Invasion assays were carried out using matrigel chambers (BD Biosciences, San Jose, CA, USA) as per the manufacturer's protocol. In brief, transfected MDA-MB-231 cells were harvested, resuspended in serum-free medium and then transferred to the hydrated matrigel chambers (∼25 000 cells per well). The chambers were then incubated for 24 h in culture medium with 10% FBS in the bottom chambers before examination. The cells on the upper surface were scraped and washed away, whereas the invaded cells on the lower surface were fixed and stained with 0.05% crystal violet for 2 h. Finally, invaded cells were counted under a microscope and the relative number was calculated.
Animal work
Female nude (nu/nu) mice (4–5 weeks old) were purchased from Charles River (Wilmington, MA, USA). All animal studies were conducted in accordance with the NIH animal use guidelines and a protocol was approved by the UMMC Animal Care Committee. MCF-7 cells were transfected with scrambled control (100 nM) and anti-miR-21 (100 nM), respectively, or infected with vector control and GAS5, respectively. The cells at exponential stage were harvested and were then mixed with 50% matrigel (BD Biosciences). Tumor-cell injection was performed as described previously.11 Freshly frozen tumors were used for the extraction of total RNA and RT-PCR.
Statistical analysis
Statistical analysis of data was performed using the Student's t-test. Tissue microarray data were analyzed using the McNemar's Test method.
Acknowledgments
This work was in part supported by the NIH grant R01 CA154989 (YM).
Glossary
- Doxo
doxorubicin
- GAS5
growth arrest-specific 5
- ISH
in situ hybridization
- lncRNA
long noncoding RNA
- PCR
polymerase chain reaction
- RT
reverse transcription
- snoRNAs
small nucleolar RNAs
- UTR
untranslated region
- qRT-PCR
quantitative RT-PCR
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA. RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- Selaru FM, Olaru AV, Kan T, David S, Cheng Y, Mori Y, et al. MicroRNA-21 is overexpressed in human cholangiocarcinoma and regulates programmed cell death 4 and tissue inhibitor of metalloproteinase 3. Hepatology. 2009;49:1595–1601. doi: 10.1002/hep.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Ajioka I, Maeda T, Nakajima K. Large-scale correlation of DNA accession numbers to the cDNAs in the FANTOM full-length mouse cDNA clone set. Keio J Med. 2006;55:107–110. doi: 10.2302/kjm.55.107. [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Cicala C, Charlesworth A, Ciccarelli C, Rossi GB, Philipson L, et al. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol Cel Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12 (7 Pt 1:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- He X, He L, Hannon GJ. The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–11101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Huang TH, Wu F, Loeb GB, Hsu R, Heidersbach A, Brincat A, et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem. 2009;284:18515–18524. doi: 10.1074/jbc.M109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006;66:1277–1281. doi: 10.1158/0008-5472.CAN-05-3632. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Park SJ, Choi EY, Kim S, Kwak HJ, Yoo BC, et al. PTEN modulates miR-21 processing via RNA-regulatory protein RNH1. PLoS One. 2011;6:e28308. doi: 10.1371/journal.pone.0028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS.lncRNAdb: a reference database for long noncoding RNAs Nucleic Acids Res 201139:Database issueD146–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2012;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Mo YY. Detection of microRNAs in cultured cells and paraffin-embedded tissue specimens by in situ hybridization. Methods Mol Biol. 2011;676:73–83. doi: 10.1007/978-1-60761-863-8_6. [DOI] [PubMed] [Google Scholar]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Chiocca S, Beck WT, Mo YY. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol Cancer Ther. 2007;6:1823–1830. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.