The p53 tumor suppressor is well known for its ability to trigger cell death by apoptosis or induce cellular senescence. p53 is also involved in a wide range of other biological processes, such as differentiation, metabolism, fecundity, and aging.1, 2 p53 responds to DNA damage, induced by, for instance, gamma irradiation or chemotherapeutic drugs, and triggers cell cycle arrest, DNA repair, apoptosis, or senescence.1, 2 A key function of p53 is to respond to oncogenic stress. Activation of p53 by oncogenic signalling may occur via ARF, an inhibitor of the p53 antagonist Mdm2, or by induction of a DNA damage response that involves activation of ATM, ATR, Chk1, and/or Chk2 kinases. The p53 response to oncogenic stress allows efficient elimination of incipient tumor cells and is fundamental for suppression of tumor development.3 However, p53 is mutated in a large fraction of human tumors, leading to evasion of p53-dependent cell death.4, 5 Restoration of wild-type p53 expression causes rapid elimination of tumors in vivo.6, 7, 8
p53 exerts its biological activities mainly through transcriptional transactivation of downstream target genes.1 Upregulation of the cell cycle inhibitor p21 is responsible for p53-dependent cell cycle arrest and senescence. Likewise, p53 transactivates genes that play important roles in mitochondrial apoptosis, including the Bcl-2 family proteins Bax, Puma, and Noxa.2 p53 can also translocate to the mitochondria and trigger apoptosis in a transcription-independent manner.9 Moreover, p53 transactivates genes involved in cell metabolism, for example, TIGAR and GLS2,2 and recent evidence indicates that regulation of these genes is essential for p53-dependent tumor suppression.10
Reactive oxygen species (ROS) are generated during the normal metabolic activities of the cell, and can also be induced by cytotoxic drugs. One consequence of ROS induction is DNA damage. ROS-induced DNA damage activates the protein PARP-1 or poly(ADP-ribose) polymerase-1.11 PARP-1 responds to DNA damage and orchestrates DNA repair via recruitment and modification of various proteins, including histones, topoisomerases, and DNA helicases.11, 12 PARP-mediated incorporation of NAD into poly(ADP-ribose) leads to depletion of cellular NAD and ATP, which results in energetic collapse and cell death by necrosis.13
In an article in this issue, Montero et al.14 have examined how ROS induces cell death. They demonstrate that PARP activation in response to ROS is dependent on p53. Mouse embryonic fibroblasts (MEFs) that lack p53 showed a markedly reduced cell death upon treatment with the ROS hydrogen peroxide, whereas MEFs lacking both Bak and Bax were efficiently killed. Similarly, p53 loss in human breast or colorectal cancer cells conferred increased resistance to ROS and PARP-mediated cell death. p53 is apparently critical for hydrogen peroxide-induced cell death, whereas Bak and Bax are not, implying that classical mitochondrial apoptosis is not involved. Rather, this ROS-induced cell death is due to activation of PARP and rapid depletion of NAD and ATP, causing necrotic cell death. Montero et al. thus show that p53 not only induces apoptosis and senescence but also has a key role in the regulation of necrosis upon exposure to ROS, and they show that this effect is mediated by PARP-1.
The work of Montero et al. allows several important conclusions and poses a number of provocative questions. First, the demonstration that p53 can promote non-apoptotic cell death through necrosis widens the panorama of p53-dependent biological responses and has implications for our understanding of how p53 suppresses tumor development. In particular, these findings raise questions regarding the role of the p53-PARP crosstalk in the response of tumor cells to chemotherapy and radiotherapy.
Second, PARP activity is evidently dependent on p53. Montero et al.14 do not provide any data regarding the mechanism at the molecular level, but speculate that SIRT1 is involved, as they observed increased SIRT1 expression in p53 knockdown and knockout cells, and as SIRT1 modulates PARP activity by deacetylation.15 This needs further investigation. Does p53 influence the activity of PARP in DNA repair? Do p53 family members p63 and p73 also talk to PARP, and if so, what is the biological significance?
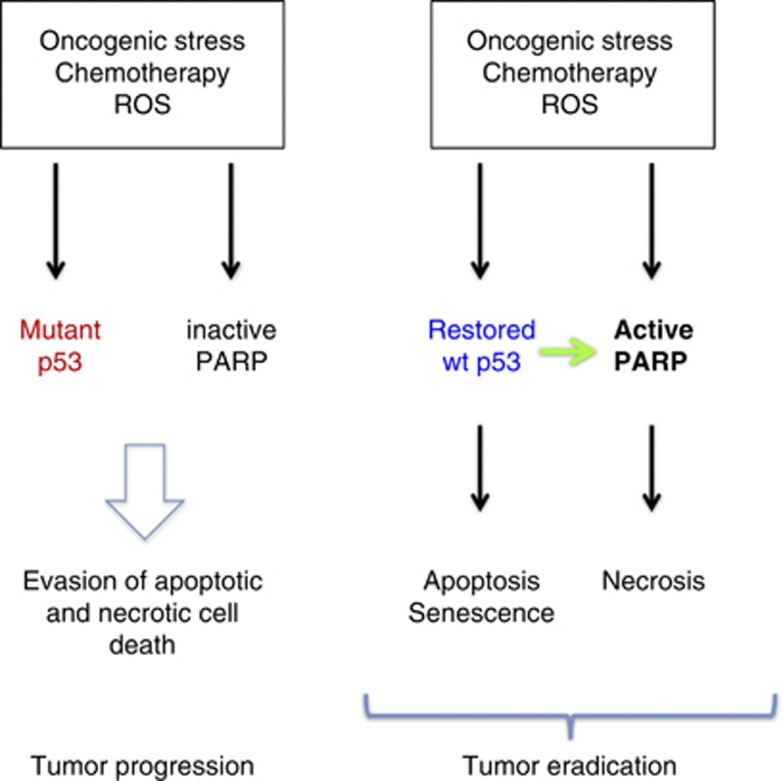
Another important question is the effect of common tumor-associated p53 mutations on PARP. This has not been tested directly by Montero et al. If missense mutant p53 proteins are deficient for PARP activation, which is a reasonable assumption, it is clear that loss of p53 function by mutation during tumor development will provide a double protection against cell death, knocking out both p53-dependent apoptosis and PARP-mediated necrosis (Figure 1).
Figure 1.
Loss of p53 function in tumors affects both apoptosis and necrosis. Left panel: Tumor cells carrying mutant p53 are deficient for both p53-dependent apoptosis and PARP-dependent necrosis in response to ROS-inducing stress, fueling tumor progression. Right panel: Restoration of wild-type p53 function should not only rescue p53-dependent apoptosis and senescence but also enhance PARP activity and rescue PARP-dependent necrosis, allowing efficient tumor eradication
Do the findings by Montero et al. open avenues for improved cancer therapy? Clearly, these results emphasize the notion that cell death upon chemotherapy can be complex, having both a necrosis and an apoptosis component. Both death pathways can be regulated by p53 and are presumably dysfunctional in cancer cells carrying inactivating p53 mutations. One prediction from the data of Montero et al.14 is that restoration of wild-type p53 function in cancer cells should not only rescue p53-dependent apoptosis but also enhance PARP-mediated necrosis, and thereby allow efficient elimination of the tumor (Figure 1).
In general, cancer therapy based on concomitant activation of several parallel death pathways makes sense, given the heterogeneity and plasticity of tumors and the existence of mixed tumor cell populations with different propensities to enter cell death by apoptosis or necrosis. The work of Montero et al. raises the possibility of utilizing necrosis as a death pathway in tumor cells in which the apoptotic pathway has been inactivated by alterations in key regulatory components, for instance, Bcl-2. For clinical oncologists, it will be crucial to learn how to modulate and exploit these processes in order to achieve maximal tumor cell death with minimal side effects. With such knowledge, we may be in a good position to design more efficient cancer therapy in the future.
KGW is cofounder, shareholder, and board member of Aprea AB, a company that develops p53-based cancer therapy.
References
- Vogelstein B, Lane D, Levine AJ. Nature. 2000. pp. 307–310. [DOI] [PubMed]
- Vousden KH, Prives C. Cell. 2009. pp. 413–431. [DOI] [PubMed]
- Halazonetis TD, Gorgoulis VG, Bartek J. Science. 2008. pp. 1352–1355. [DOI] [PubMed]
- Petitjean A, et al. Oncogene. 2007. pp. 2157–2165. [DOI] [PubMed]
- Soussi T, Wiman KG. Cancer Cell. 2007. pp. 303–312. [DOI] [PubMed]
- Martins CP, Brown-Swigart L, Evan GI. Cell. 2006. pp. 1323–1334. [DOI] [PubMed]
- Ventura A, et al. Nature. 2007. pp. 661–665. [DOI] [PubMed]
- Xue W, et al. Nature. 2007. pp. 656–660. [DOI] [PMC free article] [PubMed]
- Moll UM, et al. Curr Opin Cell Biol. 2005. pp. 631–636. [DOI] [PubMed]
- Li T, et al. Cell. 2012. pp. 1269–1283. [DOI] [PMC free article] [PubMed]
- Zong W-X, Thompson CB. Genes Dev. 2006. pp. 1–15. [DOI] [PubMed]
- Schreiber V, et al. Nat Rev Mol Biol. 2006. pp. 517–528. [DOI] [PubMed]
- Zong W-X, et al. Genes Dev. 2004. pp. 1272–1282. [DOI] [PMC free article] [PubMed]
- Montero J, et al. Cell Death Differ 2013. e-pub ahead of print 25 May 2013doi: 10.1038/cdd.2013.52 [DOI]
- Rajamohan SB, et al. Mol Cell Biol. 2009. pp. 4116–4129. [DOI] [PMC free article] [PubMed]