Abstract
Apaf1 is an evolutionarily conserved component of the apoptosome. In mammals, the apoptosome assembles when cytochrome c is released from mitochondria, binding Apaf1 in an ATP-dependent manner and activating caspase 9 to execute apoptosis. Here we identify and characterize a novel mouse mutant, yautja, and find it results from a leucine-to-proline substitution in the winged-helix domain of Apaf1. We show that this allele of Apaf1 is unique, as the yautja mutant Apaf1 protein is stable, yet does not possess apoptotic function in cell culture or in vivo assays. Mutant embryos die perinatally with defects in craniofacial and nervous system development, as well as reduced levels of apoptosis. We further investigated the defects in craniofacial development in the yautja mutation and found altered Sonic hedgehog (Shh) signaling between the prechordal plate and the frontonasal ectoderm, leading to increased mesenchymal proliferation in the face and delayed or absent ossification of the skull base. Taken together, our data highlight the time-sensitive link between Shh signaling and the regulation of apoptosis function in craniofacial development to sculpt the face. We propose that decreased apoptosis in the developing nervous system allows Shh-producing cells to persist and direct a lateral outgrowth of the upper jaw, resulting in the craniofacial defects we see. Finally, the novel yautja Apaf1 allele offers the first in vivo understanding of a stable Apaf1 protein that lacks a function, which should make a useful tool with which to explore the regulation of programmed cell death in mammals.
Keywords: Apaf1, apoptosis, craniofacial development, Shh signaling, programmed cell death
Normal embryonic development hinges on the precise coordination of cell proliferation, differentiation, migration and death. The regulation of cell death is deeply conserved throughout the evolution. Indeed, work in Caenorhabditis elegans defined core components of the cell execution pathway, including CED-9, which inhibits cell death,1 and CED-3 and CED-4, which direct programmed cell death.2 Similar components have since been found to mediate apoptosis in other organisms, with caspase 3 and caspase 9 functioning in place of CED-3 in most metazoans,3, 4 and Apaf1 functioning as CED-45 to execute apoptosis.
There are two major pathways of apoptosis: extrinsic apoptosis, classically mediated by tumor necrosis factor (TNF)-family death receptors via a signaling complex that includes caspase 8, and intrinsic apoptosis mediated via mitochondrial release of cytochrome c, which depends on the balance of pro- and anti-apoptotic Bcl2 family members.6 Although both pathways lead to caspase-dependent regulated cell death, each has distinct roles in mammalian development. In the extrinsic pathway, caspase 8 is necessary to suppress the activation of RIP1 and RIP3 kinase-dependent cell death,7 but is otherwise dispensable for development. In contrast, the role of the intrinsic pathway is well defined in developing mammals. The earliest example of intrinsic apoptosis is during cavitation, the process whereby the preimplantation blastocyst forms.8 Better-known examples occur later in development, when apoptosis is needed to sculpt features of the embryo, such as the limbs, the inner ear, the nervous system and the heart.9
Following release from the intermembrane space of mitochondria into the cytoplasm during intrinsic apoptosis,10 cytochrome c binds and activates Apaf1,4 inducing a conformational change that facilitates the interaction of Apaf1 with ATP that drives its oligomerization into an apoptosome.11 Whereas the Apaf1 apoptosome in mammals exhibits sevenfold symmetry and forms a wheel-like particle,12 the CED-4 apoptosome in C. elegans is an octamer: a symmetric conical shape of four asymmetric dimers.13 Inside the cone is a space termed the ‘hutch' that can fit two CED-3 molecules.13 For both CED-3 and caspase 9, the processed form associates with the apoptosome and triggers the cleavage of a variety of targets associated with apoptotic cell death.14
Apaf1 is composed of six domains: an N-terminal caspase recruitment domain (CARD), a nucleotide-binding domain, a winged-helix domain (WHD), two helical domains and a domain at the C-terminus of the protein containing 12–13 WD-40 repeats. The CARD domain of Apaf1 interacts with the prodomain of caspase 9,4, 12 whereas the WD-40 repeats bind to cytochrome c.12, 15, 16 Once associated with cytochrome c, Apaf1 undergoes a conformational change11 such that ATP binds at the interface between the nucleotide-binding domain, the first helical domain and the WHD,17 allowing further intermolecular interactions that form the apoptosome, where the WHD is predicted to be an essential mediator of oligomerization.13
Mice with targeted deletions of Apaf1 exhibit embryonic lethality and morphological defects, highlighting the pivotal role of this protein in apoptosis during mammalian embryogenesis.18, 19 Loss of Apaf1 causes decreased apoptosis, best documented in the embryonic neuroectoderm, which results in failed neural tube closure and cleft palate, among other morphological defects.18, 19, 20, 21, 22 Although the cleft palate is attributed to decreased apoptosis between the two palatal shelves resulting in failed fusion,20 details of the craniofacial defects due to Apaf1 deficiency remain to be determined.
The development of vertebrate facial features results from coordinated signaling by multiple pathways on cells from multiple embryonic origins. The complexity of the process is highlighted by the incidence of disordered craniofacial development: about one-fifth of all birth defects involve craniofacial abnormalities.23 All three germ layers contribute to forming the vertebrate face. Ectoderm gives rise to the neural crest, which migrates from the dorsal midline of the embryo into the facial prominences and generates most of the cartilage, bone and connective tissue of the head and face. In addition, the morphology of the neural tube, composed of neuroectoderm, regulates the positioning of both the overlying cranial mesoderm that is responsible for the musculature and vasculature, as well as the positioning of adjacent endodermal tissue that develops into structures such as the pharyngeal pouches.24
Seven prominences contribute to the face: the frontonasal (FNP), paired maxillary, paired mandibular and paired nasal prominences. These prominences consist of an outer ectodermal cell layer and an internal neural crest-derived mesodermal core. Signaling interactions between adjacent prominences and from underlying mesenchyme and neuroectoderm direct the morphogenetic events that pattern the developing face.24 For example, upper jaw morphogenesis results from reciprocal signaling between the forebrain, the neural crest and the surface ectoderm. Precise cellular regions of Sonic hedgehog (Shh) expression in the ventral forebrain and fibroblast growth factor 8 (Fgf8) expression in the cephalic ectoderm are termed ‘signaling centers' as they convey the signal to the adjacent tissue.25, 26 Directions from these signaling centers drive bone morphogenetic protein (Bmp), retinoid and wingless (Wnt) expression in the mesenchyme and induce outgrowth of the facial prominences.27, 28, 29, 30, 31, 32 Shh is important not only for establishing craniofacial signaling centers, but also for controlling proliferation and directing cell migration,33 which make this signaling molecule a key player in craniofacial development.
In mammals, the medial part of the upper jaw and midface derive from the fusion of the nasal prominences and the FNP. Defects in the fusion of these prominences can result in cleft lip and cleft palate. The remainder of the upper jaw derives from the maxillary prominences, whereas the entire lower jaw develops from the mandibular prominences. Although overdevelopment (hyperplasia), underdevelopment (hypoplasia, agenesis and micrognathia) or mispositioning (retrognathia and prognathism) of the mandible is seen in humans, clefting of the lower jaw is rare.34
Regulation of apoptosis is important in the control of FNP signaling center size and activity. Increased apoptosis results in altered signaling centers and craniofacial defects, including hypoplasia of the facial bones in a murine model of Treacher Collins syndrome35 and a cleft nasal region in a model of frontonasal dysplasia (the Alx3/Alx4 double-mutant mouse line).36 Decreased apoptosis during development also results in craniofacial defects such as cleft palate in one caspase 3-deficient mouse line.37 Taken together, these defects underscore the fact that proper growth and interaction of the facial prominences is required for normal morphology and function.
In this report, we describe the developmental consequences of a novel point mutation, yautja, within the crucial WHD of Apaf1. Surprisingly, we found that yautja mutant Apaf1 protein was produced at normal levels during embryonic development. Consistent with the role of the WHD in the assembly of Apaf1 oligomers during apoptosis, we demonstrate the yautja mutant protein is a functional null. We analyzed the deleterious effects on craniofacial development where we found decreased apoptosis in facial mesoderm and a cleft midfacial region in yautja embryos and show that there is an expanded region of Shh-expressing cells in the ventral forebrain that correlates with increased cell proliferation in the FNP. These molecular changes likely explain the craniofacial widening and clefting we saw. Our data link the critical role of apoptosis with Shh signaling in the proper formation of the vertebrate face and also provide a critical in vivo model through which the field will be better able to define the mechanism underlying apoptosis.
Results
The yautja mutation causes multiple developmental defects
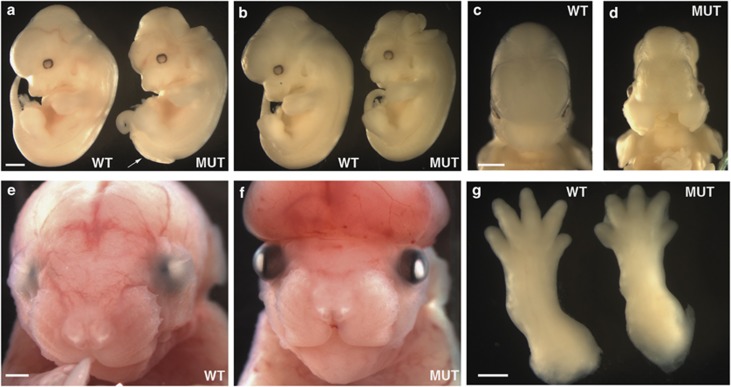
We identified the yautja mutation in a recessive N-ethyl-N-nitrosourea screen for embryos with abnormal morphology at E12.5. Yautja mutant embryos exhibited exencephaly, delayed removal of interdigital webs, failure of the eyelids to fuse, flattened forebrain vesicles and craniofacial abnormalities that included cleft maxilla and widened FNP (Figure 1). There was variation in the constellation of phenotypes yautja mutants displayed, as well as in the severity and penetrance for each of the phenotypes (Table 1). We examined 487 progeny from yautja heterozygous intercrosses and found 137 (28%) displayed one or more features of the mutant phenotype (Table 2). To assess viability, we examined embryos at various time points throughout gestation. We were able to obtain live mutant embryos at E17.5; however, on the day of birth (P0), the mutants were nonviable, indicating that yautja is late embryonic lethal (Table 2).
Figure 1.
Yautja embryos display multiple morphological defects. Wild-type (WT) and yautja mutant (MUT) embryos showing defects in: cranial neural tube closure (a,b,f), caudal neural tube closure (a, white arrow), craniofacial development (d and f), eyelid closure (f) and interdigital web removal (g). Embryos are E12.5 (a–d), E17.5 (e and f) and E15.5 (g). Scale bars=1 mm
Table 1. Incidence of phenotypic features in E12.5 yautja embryos.
| Phenotypic feature of mutant | Overall incidence of phenotypea | Incidence as sole mutant feature | Incidence of phenotype in WT or HET embryos |
|---|---|---|---|
| Cranial neural tube defect | 32/57 | 6/57 | 1/174 |
| Caudal neural tube defect | 43/57 | 3/57 | 0/174 |
| Compressed forebrain | 21/57 | 5/57 | 0/174 |
| Craniofacial abnormalityb | 12/57 | 0/57 | 0/174 |
Most mutants showed various combinations of the four phenotypic features listed
Craniofacial abnormality includes cleft lip and cleft face
Table 2. Viability of progeny from yautja mapping crosses.
|
Genotype |
||||
|---|---|---|---|---|
| Stage | No. of litters | No. of embryos | WT or HET | MUT (%) |
| E10.5 | 13 | 90 | 63 | 27 (30) |
| E11.5 | 4 | 26 | 16 | 10 (38) |
| E12.5 | 34 | 231 | 174 | 57 (25) |
| E13.5 | 6 | 37 | 26 | 11 (30) |
| E14.5 | 3 | 21 | 14 | 7 (33) |
| E15.5 | 7 | 46 | 31 | 15 (33) |
| E17.5 | 5 | 26 | 19 | 7 (27) |
| P0 | 2 | 10 | 7 | 3 (30)a |
| Total | 74 | 487 | 350 | 137 (28) |
The genotypes of mice at the embryonic (E) or postnatal (P) day indicated were determined by PCR analysis. Wild-type, heterozygous and yautja animals are indicated as WT, HET and MUT, respectively
None of the mutants recovered at P0 were alive
Yautja is a mutation in apoptotic protease-activating factor Apaf1
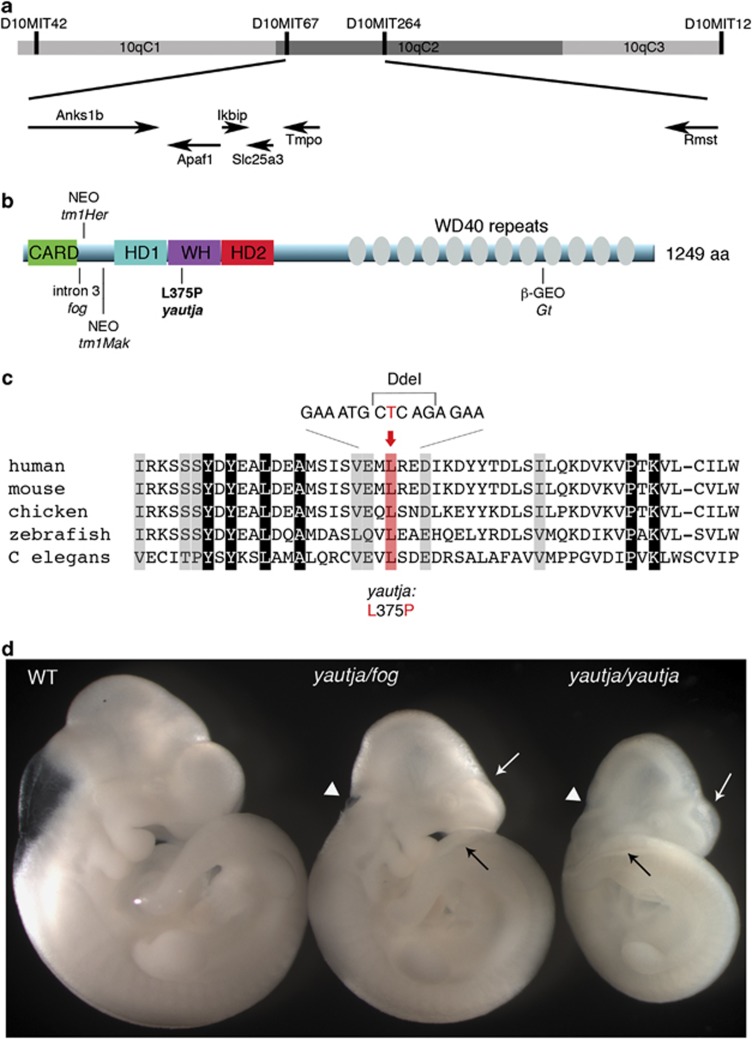
The yautja mutation, initially on a C57BL/6J background, was backcrossed to the C3H/HeJ strain. Using meiotic recombination and strain-specific polymorphisms, we mapped the yautja mutation to a 1.5-Mb region on chromosome 10. This region, bounded by microsatellite markers D10MIT67 and D10MIT264, included six genes: Anks1b, Apaf1, Ikbip, Rmst, Slc25a3 and Tmpo (Figure 2a). We prioritized sequencing the Apaf1 cDNA, as existing mutant alleles of Apaf1 share phenotypic similarities with yautja mutants, including exencephaly, retention of interdigital webbing and craniofacial anomalies.18, 19, 20 We identified a T-to-C transition in exon 8 of Apaf1 corresponding to the WHD of the Apaf1 protein (Figures 2b and c). This mutation alters an evolutionarily conserved leucine to a proline at amino acid 375 of Apaf1 (Figure 2c).
Figure 2.
Positional cloning of yautja identified a mutation in Apaf1. (a) Yautja was mapped to a 1.5-Mb interval bounded by markers D10MIT67 and D10MIT264 on chromosome 10. The arrows indicate the six genes in the interval and the direction of their transcription. (b) Schematic of the Apaf1 protein with domains indicated: caspase recruitment domain (CARD), helical domain I (HD1), winged-helix domain (WH), helical domain II (HD2) and multiple WD40-repeat domains. The existing Apaf1 mutant alleles are shown where they affect the protein: Apaf1fog inefficiently splices intron 3,22 Apaf1tm1Her has a neomycin cassette inserted at amino-acid 111,19 Apaf1tm1Mak inserted a neomycin cassette at exon 5,18 Apaf1yautja (L375P) is described in this paper and the Apaf1Gt has a gene trap insertion downstream of amino-acid 1018 (official symbol: Apaf1Gt(IRESBetageo)XIX18Pgr).20 (c) The yautja mutation changes a highly conserved leucine residue of Apaf1. Sequence comparison of a portion of the Apaf1 WHD showing evolutionary conservation; conserved residues are highlighted in gray. The red shading and text indicate the conserved leucine that is changed to proline by the yautja point mutation. Accession numbers used to produce the alignment were: human NP_863651, mouse NP_033814, chicken XP_416167, zebrafish NP_571683, Drosophila NP_7256347 and C. elegans NP_001021203. (d) The yautja mutation did not complement a functional null allele of Apaf1: Apaf1yautja/fog embryos share phenotypic features with Apaf1yautja and Apaf1fog: white arrow=flattened forebrain ventricles, black arrow=open caudal neural tube, white arrowhead=open rostral neural tube. Lateral view of E10.5 embryos
To determine whether the Apaf1L375P mutation underlies the yautja phenotype, we performed a complementation cross with an existing Apaf1 allele. The Apaf1fog allele is functionally deficient in Apaf1 protein due to aberrant transcript processing.21, 22 Embryos heteroallelic for the yautja and fog alleles failed to complement and recapitulated features of both phenotypes: we saw defects of the forebrain, lumbosacral and facial regions, including forebrain encephalocele, exencephaly, open fourth ventricle, compressed forebrain ventricles, spina bifida with or without a kinked tail and wide or split face (Figure 2d). We saw these features either in isolation or in complex, consistent with the individual mutant phenotypes for yautja and fog. In addition, the variability in severity and penetrance of the phenotypic features was reminiscent of the yautja mutant and reported in the fog allele.21 None of these defects were observed in embryos heterozygous for either allele. Thus, we conclude the yautja mutation is a novel loss-of-function Apaf1 allele.
The yautja mutation does not destabilize Apaf1 protein, but eliminates Apaf1 function
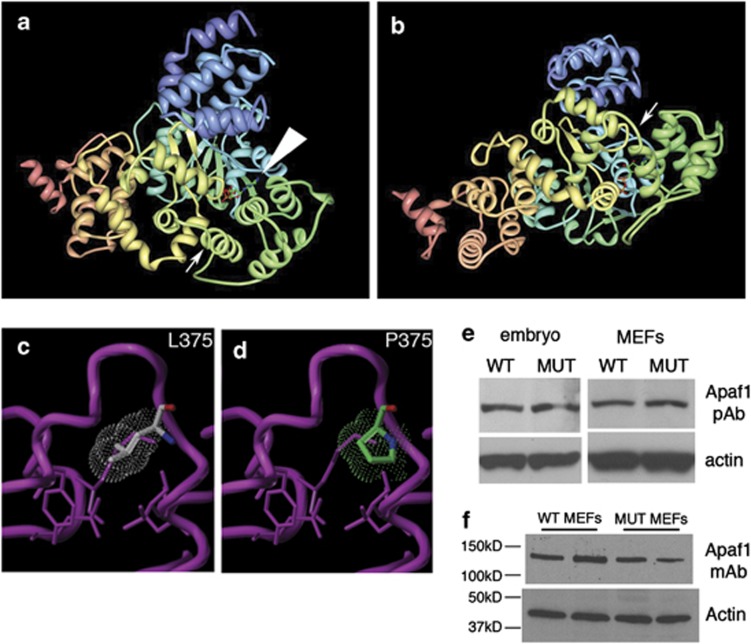
To investigate the mechanism of yautja loss of function, we analyzed Apaf1 protein expression in whole embryo and mouse embryonic fibroblast (MEF) lysates. The proline introduced by the Apaf1yautja mutation would be predicted to alter the protein backbone structure, most likely introducing rigidity at L375P, immediately adjacent to the nucleotide-binding pocket (Figures 3a–d). By immunoblot using two distinct Apaf1 antibodies, we detected no change in protein size and only a moderate decrease in intensity with one of the antibodies, indicating that Apaf1 protein stability is generally intact in Apaf1yautja mutants (Figures 3e and f).38, 39 This finding makes the Apaf1yautja allele unique compared with existing targeted Apaf1 mutant alleles, which all lack protein expression.18, 19, 20
Figure 3.
The yautja mutation is predicted to disrupt the structure of Apaf1, but protein size and expression appear intact in mutants. (a) Image from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB; http://www.pdb.org) of PDB ID 1Z6T,17 with small arrow indicating the location of the yautja L375P mutation. Large arrowhead indicates the bound ATP molecular structure. (b) Same image as in a, but rotated 90° away from viewer, to show the yautja mutation location in the loop between α-helices in the same orientation as depicted in the close-up models (c and d). Close-up of the winged-helix domain in wild-type (WT; L375) (c) and mutant (MUT; P375) (d) versions of Apaf1 showing steric hindrance (dot cloud) on the protein backbone (purple pipe/ribbon) generated by proline substitution (Sybyl models provided by Kim Gernert at Emory University School of Medicine Biomolecular Computing Resource). (e and f) Apaf1 protein is stably expressed in Apaf1yautja mutant embryos and MEFs. Western blot of whole-cell lysates from WT and Apaf1yautja MUT embryos or embryonic fibroblasts (MEFs) using either polyclonal (e) or monoclonal (f) antibodies against Apaf1
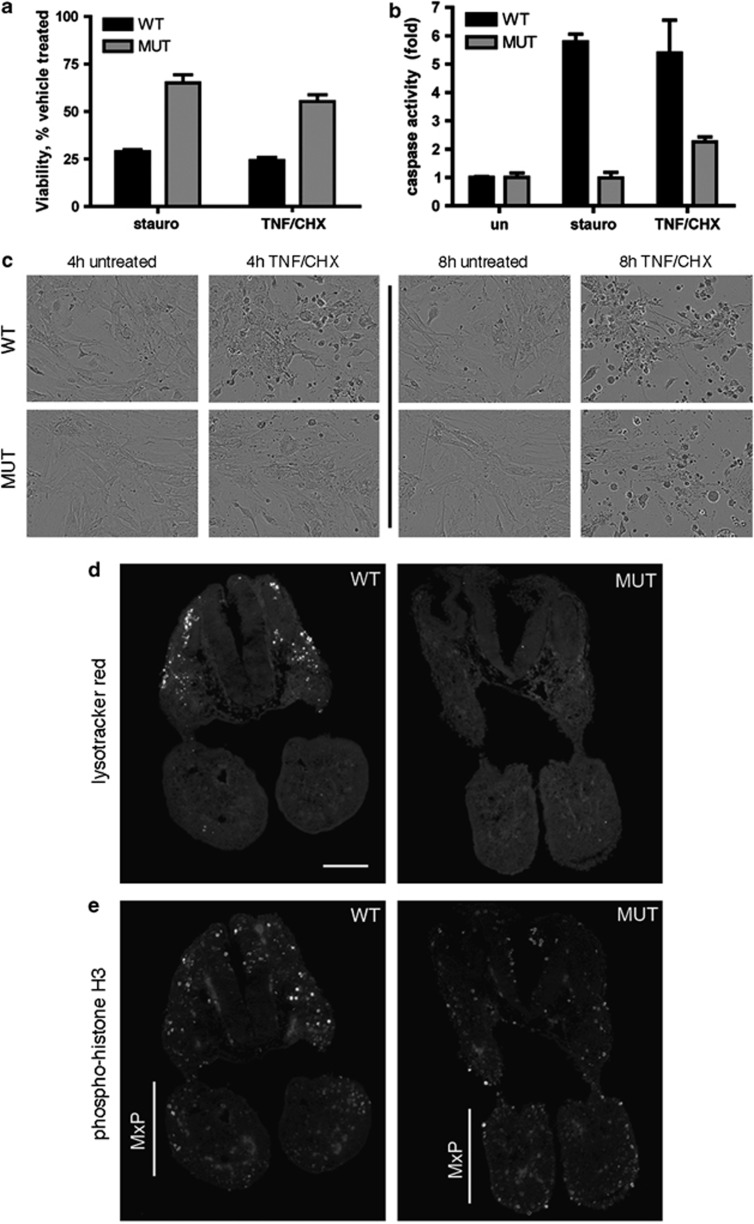
We next examined the susceptibility of Apaf1yautja MEFs to apoptotic stimuli. We treated MEFs with either staurosporine or a combination of TNF and cycloheximide (CHX) to induce apoptosis, then assayed cell viability. After 4 h of treatment, 25–30% of wild-type MEFs were viable, whereas 55–65% of Apaf1yautja were alive, indicating that apoptosis is reduced or delayed by the Apaf1yautja allele (Figure 4a). Direct viewing of the MEFs after 8 h of TNF and CHX treatment showed dying Apaf1yautja and wild-type cells in similar proportions, supporting the conclusion that apoptosis is reduced and delayed in the mutant cells (Figure 4c).
Figure 4.
Decreased apoptosis in Apaf1yautja mutant embryos. (a) Apaf1yautja MEFs are less susceptible to death by apoptotic stimulation than wild-type cells. Viability of WT or MUT MEFs following 4-h stimulation with staurosporine (2 μg/ml) or with TNF-α (25 ng/ml) and cycloheximide (CHX; 5 μg/ml; n=4). (b) Apaf1yautja MEFs are unable to activate effector caspases. Levels of DEVDase-specific caspase activity relative to untreated WT MEFs determined 4 h after stimulation (n=3). Error bars indicate standard deviation (S.D.) of the mean. (c) Representative images of MEFs in culture 4 and 8 h after treatment with TNF and CHX. (d) LysoTracker Red staining shows decreased cell death, and (e) anti-phospho-histone H3 immunofluorescence shows increased proliferation in the maxillary prominences (MxP) of E9.5 Apaf1yautja embryos. Horizontal sections at the level of the maxillary component of the first branchial arch. Dorsal is to the top. Scale bar=100 μm
To determine whether the reduction in apoptosis occurred before or after caspase activation, we assayed cleavage of a caspase 3/7 substrate using lysates from treated MEFs. Normally, wild-type MEFs display a five- to sixfold increase in caspase activity upon treatment with staurosporine or a combination of TNF and CHX. However, we saw a 2.5-fold increase in caspase activity when the Apaf1yautja MEFs were treated with TNF and CHX, and no increase in caspase activation upon staurosporine treatment (Figure 4b). This reduced level of caspase activity in the Apaf1yautja MEFs indicates the mutation affects a step upstream of caspase activation.
In the developing embryo, the vital dye LysoTracker Red stains apoptotic cells.40 We examined horizontal brain sections of wild-type embryos at E9.5 and E12.5 and saw stained cells indicating cell death. In contrast, analogous sections from Apaf1yautja mutant embryos exhibited significantly fewer stained cells (Figure 4d and data not shown), pointing to a defect in apoptosis in vivo, consistent with the decrease in cell death we observed in MEFs in vitro. This is also concordant with the established role of Apaf1 in apoptosis and with the Apaf1yautja mutant phenotype behaving as a recessive loss-of-function allele. Together, our in vitro and in vivo data suggest that the yautja mutation destroys Apaf1 function as a critical component of the intrinsic apoptotic pathway.
Apaf1 yautja mutants have defects in cranium morphology
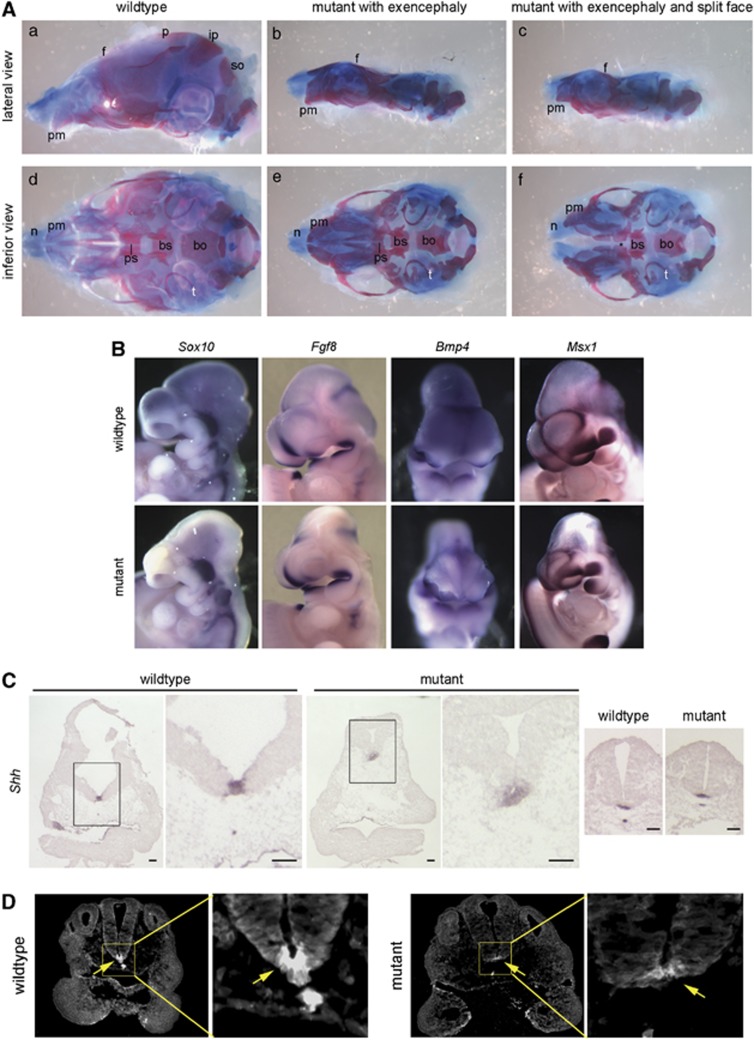
Although previous reports of Apaf1 alleles noted craniofacial defects, these defects were not associated with any particular pathway or developmental mechanism.18, 19, 20 Approximately 20% of our Apaf1yautja embryos exhibit craniofacial defects of variable severity when examined at E12.5 (Table 1). To investigate the role of the mutation on cranial skeletal development, we examined the skulls of stillborn pups immediately after birth (P0). In Apaf1yautja mutants with exencephaly, the frontal bones malformed around protruding brain tissue (Figures 5Ab and c). In addition, the parietal, interparietal and supraoccipital bones that compose the roof of the skull were absent (Figures 5Ab and c). In a mutant with the split-face phenotype, the nasal bones and cartilages were separate at the midline (Figure 5Af). Viewed from the inferior perspective (Figure 5Af), the presphenoid bone was absent in this specimen. This skeletal defect was variable: of 10 E17.5-P0 mutant skulls, 4 lacked a presphenoid, 1 had an abnormally shaped presphenoid and 5 presphenoids appeared the same as wild type (data not shown). Whereas all 10 mutants were exencephalic, only 1 yautja pup had a split face, suggesting that the absence or abnormal morphology of the presphenoid bone does not correlate with the facial clefting phenotype.
Figure 5.
Craniofacial defects: getting at a mechanism. (A) Preparations of P0 skulls from wild-type and yautja, stained with alcian blue and alizarin red. (b and c) In mutants with exencephaly, the frontal bone (f) is malformed and the parietal (p), interparietal (ip) and supraoccipital (so) bones are absent. (d–f) The inferior perspective allows visualization of skull base elements, including the tympanic rings (t), basisphenoid (bs), basioccipital (bo) and presphenoid (ps) bones. (f) A mutant with a split mid-face has widely spaced premaxillary bones (pm) and nasal cartilages (n), and also lacks the presphenoid bone (asterisk). (B) Expression of genes important during craniofacial development does not differ between wild-type and yautja. Whole-mount in-situ hybridization of E10.5 embryos with probes for Sox10, Fgf8, Bmp4 and Msx1. (C) Shh expression is expanded in the prechordal plate of yautja embryos. In-situ hybridization of E10.5 coronal sections through the head; the box in the first panel of each pair is enlarged in the second panel to show detail. Normal Shh expression is seen in the caudal neural tube floor plate and notochord of yautja mutants (last two panels). Scale bars=1 mm. (D) Shh protein is seen in a broader region of the prechordal plate of E9.5 yautja embryos. Anti-Shh immunofluorescence of horizontal sections at the level of the maxillary component of the first branchial arch; dorsal is to the top. The box in the first panel of each pair is enlarged in the second panel to show detail of the floor plate and Shh expression (arrows)
The deformed frontal bones and missing skull vault components in the exencephalic mutants may be due to the interference of the expanded neural tissue; however, the presphenoid ossification defect may be more complex. The mammalian skull is formed from the mesoderm and the neural crest lineages. The presphenoid derives from neural crest.41 As this structure was abnormal or missing in 50% of Apaf1yautja mutant samples, we assayed the existence and localization of neural crest cells by examining Sox10 expression. There were no differences between the wild-type and the Apaf1yautja embryos (Figure 5B). This finding, coupled with the presence of other neural crest-derived skull elements (the basisphenoid and nasal bones),41, 42 suggests that the neural crest is not disrupted in an extreme manner.
To investigate the mechanism(s) underlying the craniofacial defects observed, we examined the expression of additional genes known to be important for craniofacial morphogenesis. The frontonasal ectodermal zone is one important signaling center that regulates the morphogenesis of the upper jaw.25, 26 This signaling center, defined by the boundary between Fgf8 and Shh expression in the FNP surface ectoderm, can induce expression of Bmp2, Bmp4 and Bmp7 in the underlying mesenchyme. Bmps are known to regulate the growth of facial primordia,26 with Bmp2- and Bmp4-regulating species-specific upper jaw patterning in birds.43 Using whole-mount in situ hybridization, we determined that the expression patterns of Fgf8, Bmp2 and Bmp4 were similar in wild-type and yautja embryos (Figure 5B and data not shown).
Msx1 is strongly expressed in the developing craniofacial region and is involved in coordinating patterning events in the orofacial region.44 Recently, Wnt1 was also identified as an important factor in the regional specification of the vertebrate face.32 Both of these genes were expressed at normal levels and locations in yautja embryos at all stages examined (Figure 5B and data not shown). Taken together, the expression of genes we would expect to drive the underlying outgrowth and patterning of the upper jaw is not altered in Apaf1yautja mutants.
Craniofacial phenotype in yautja mutants correlates with expanded Shh signaling from the prechordal plate
Shh signaling within the forebrain is required for the induction of Shh in the frontonasal ectodermal zone.45 To monitor the Shh signaling, we examined the expression of a Shh target gene, Ptch1 (also the Shh receptor). Using either a Shh reporter line, PtcD-lacZ (Ptch1tm2Mps), or in situ hybridization with a Shh probe, we found that the Shh response was expanded throughout the prechordal plate of Apaf1yautja mutant embryos, whereas expression in wild-type embryos was restricted to the ventral-most cells (Figure 5C and data not shown). Because of the abnormal craniofacial morphology of the E10.5 mutant, we examined the Shh expression before the clear FNP divergence. At E9.5, we found a broadened region of Shh expression in the ventral neural tube of the mutants (Figure 5D). This broadened region of Shh activity could affect the neighboring frontonasal mesenchyme, directing increased proliferation or outgrowth. Consistent with this hypothesis, we found an increased number of phospho-histone H3-positive proliferating cells in the medial FNP of the Apaf1yautja mutant embryos compared with their wild-type littermates (Figures 4d and e). Alterations in Shh activity in either the forebrain or facial mesenchyme are known to affect the spatial organization and mitotic activity of midfacial growth zones, resulting in a continuous phenotypic spectrum from cyclopia to hypertelorism.33, 46 Therefore, our data are consistent with the loss of Apaf1 cell-death function causing an increase in Shh signaling that results in the craniofacial development defects we see in the Apaf1yautja mutants (Figure 6).
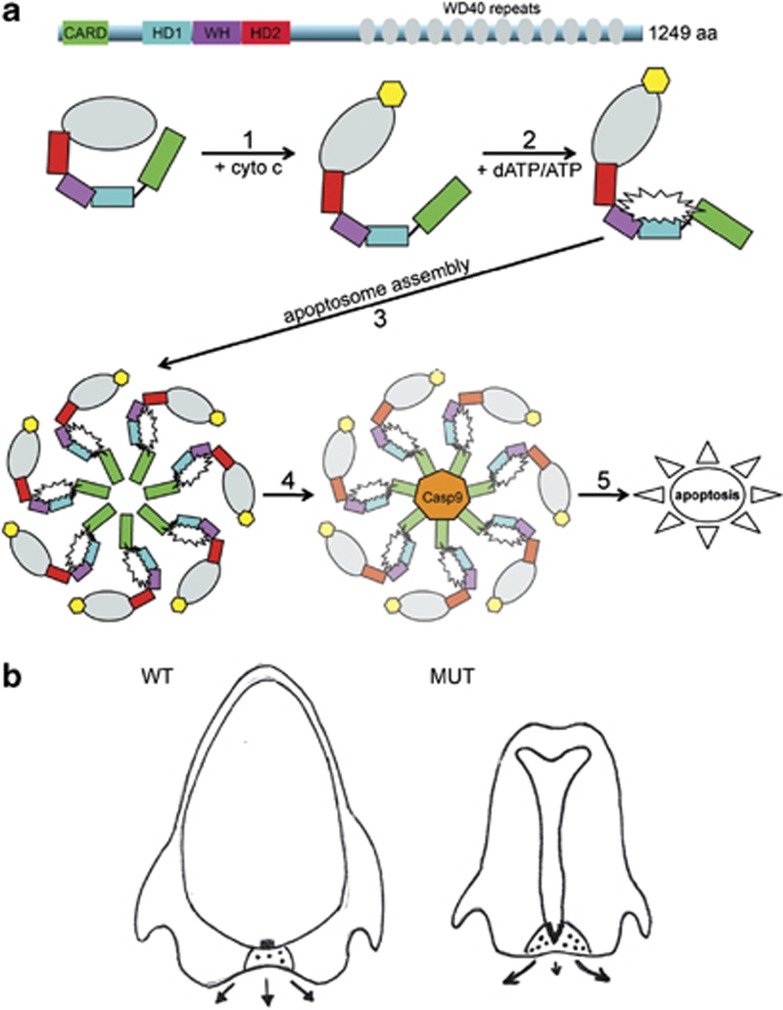
Figure 6.
Model of Apaf1yautja action. (a) In the cytoplasm, Apaf1 exists as an autoinhibited monomer, wherein the WD40 repeats (gray) interact with the N-terminal domains. Step 1: cytochrome c (yellow), released from the mitochondria, binds to the WD40 repeats and a conformational change occurs in Apaf1. Step 2: the binding of dATP/ATP (white starburst) to the interface between the nucleotide-binding domain (black line), HD1 (blue) and the WH domain (purple), allows further conformational changes that promote apoptosome assembly. Step 3: seven molecules of Apaf1 associate with each other to form the apoptosome, with protein–protein interactions being critical for stability and function. Step 4: procaspase 9 (orange) associates with the apoptosome and becomes active, causing the downstream activation of effector caspase 3. Step 5: cleavage of targets associated with programmed cell death and apoptosis of the cell. The L375P mutation in Apaf1yautja, in the WH domain, could alter the ability of steps 2, 3 and/or 4 to occur, or decrease their efficiency. (b) Expanded Shh (black boxes) in the prechordal plate of Apaf1yautja embryos (MUT) acts on the mesenchyme of the neighboring FNP (black dots). This causes increased proliferation and directed growth away from the midline (arrows, with length of arrow indicating amount of outgrowth)
Discussion
We have identified a novel loss-of-function allele of Apaf1, Apaf1yautja, which disrupts nervous system and craniofacial development. As expected from previous studies of Apaf1 deletion and reduced expression mutants, we saw decreased cell death in Apaf1yautja embryos. Moreover, our allele phenocopies previously characterized Apaf1 deletion alleles that exhibit defects in neural tube closure, delayed interdigital web removal, embryonic lethality and craniofacial malformations.18, 19, 20, 21, 22 Our data suggest that Shh-producing cells persist and direct a lateral outgrowth of the FNP that results in the craniofacial defects we observe. Because Apaf1yautja encodes a stable, non-functional protein, this allele provides a valuable tool for examining apoptosome formation and activation.
Although previous studies of Apaf1 mutants noted craniofacial abnormalities, the underlying mechanism(s) were not explored. We found the craniofacial defects in the Apaf1yautja mutants characterized here to result from fewer cells dying in the prechordal plate of the Apaf1yautja mutant embryos. Furthermore, when we evaluated the expression of a number of genes important in craniofacial development, including Sox10, Fgf8, Bmp2, Bmp4, Msx1, Wnt1 and Shh, we found that only Shh was affected. Apaf1yautja embryos exhibited a broader distribution of Shh transcripts and protein in the prechordal plate, consistent with recent observations showing craniofacial width is modified by alterations in Shh activity.47, 48 The current understanding is that the size of the Shh expression region is strictly controlled to coordinate craniofacial development.33, 46 We found increased cell proliferation in the FNP mesenchyme, as would be predicted by increased or persistent Shh signaling from the ventral forebrain signaling center. The broadened, diffuse Shh expression patterns we saw in yautja mutants are concordant with published results in which ectopically expanded Shh activity resulted in a widened midface.33 Taken together, our data suggest that decreased cell death resulting from loss of Apaf1 function leads to an expansion of Shh-expressing cells and cell proliferation that results in the wide FNP and split-face phenotype (Figure 6b).
We observed variability in the penetrance and severity of phenotypes among individual Apaf1yautja mutants. Not every mutant displayed a wide or cleft upper jaw, suggesting the existence of compensatory mechanisms or a specific threshold beyond which the system must be pushed before craniofacial defects manifest. Moreover, there are clear distinctions in phenotypes among the identified Apaf1 alleles: a gene-trap cassette inserted in the WD40 repeat domain (Apaf1Gt(IRESBetageo)XIX18Pgr);20 neomycin cassettes inserted in the nucleotide-binding domain (Apaf1tm1Her and Apaf1tm1Mak);18, 19 and the splicing defect just C-terminal to the CARD domain in Apaf1fog.21, 22 Although a subset of the phenotypic differences may be due to strain modifiers, it is also likely that the phenotypic distinctions reflect the complexity of the interactions among the processes that coordinate development, including the complex nature of apoptosis itself. Programmed cell death results from a series of pro- and anti-apoptotic signals at many steps. From the release of mitochondrial contents, through the formation of the apoptosome, to the activation of caspases and final destruction of the cell, there are many proteins and small molecules either pushing the cell toward death or attempting to halt the process.49 The developmental processes that lead to a morphologically ‘normal' face may be highly sensitive to minor environmental or stochastic events, reflected in the prevalence of craniofacial defects in human births and the lack of a single genetic cause.
Because the previously published Apaf1 mutants either lack or produce lower levels of Apaf1 protein, the fact that Apaf1yautja produces stable yet non-functional protein gives us a remarkable tool with which to better understand the formation and activation of the apoptosome. There are several potential explanations for the loss of Apaf1 function. First, in 3D-structural models, the L375P mutation in the WHD is located immediately adjacent to the nucleotide-binding domain (Figure 3a and b), raising the possibility that this change alters the ability of Apaf1 to bind or hydrolyze ATP. This would compromise the conformational changes and oligomerization necessary for apoptosome assembly. Second, the cyclic side chain of proline would be predicted to interfere with the protein backbone (Figures 3c and d), affecting the ability to interact with itself or another protein. Third, based on the CED4 structure of the apoptosome, the WHD is predicted to encircle the ‘hutch' space into which caspase 9 fits,13 suggesting the domain could be critical for caspase 9 placement or function. Finally, several proteins have been reported to bind Apaf1 in either inhibitory or potentiating manners; thus, it is plausible that the L375P mutation alters (either stabilizing or disrupting) such an interaction, resulting in a loss-of-function phenotype (Figure 6a). Although the exact mechanism remains to be worked out, the Apaf1yautja allele reveals that all the phenotypic consequences of Apaf1 are mediated via the essential WHD.
Materials and Methods
Mice and cells (MEFs)
Mice carrying the forebrain overgrowth mutation21 (Apaf1fog) were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). These mice are functionally deficient in Apaf1 protein because of aberrant transcript processing.22 The yautja mutation, generated on a C57BL/6J background and maintained on a C3H/HeJ background, was identified in a screen for recessive N-ethyl-N-nitrosourea mutations that caused morphological defects during midgestation (http://mouse.mskcc.org = 03αR1).50
Timed pregnant females were killed by cervical dislocation, and embryos were dissected in cold phosphate-buffered saline (PBS) containing 0.4% bovine serum albumin. The morning a copulatory plug was seen was considered E0.5. All procedures involving mice were performed in accordance with protocols approved by Emory's Institutional Animal Care and Use Committee.
Primary MEFs were isolated according to standard procedures from decapitated and eviscerated E13.5 embryos. Cells were maintained in DMEM with 10% FBS (Atlanta Biologicals, Lawrenceville, GA, USA), 2 mM L-glutamine, 100 U/ml penicillin and 100 U/ml streptomycin (Invitrogen, Grand Island, NY, USA).
Mapping and identification of the yautja mutation
The yautja mutation was mapped using C3H/HeJ (The Jackson Laboratory) as the outcross strain, as previously described.51 MIT SSLP markers were used to map yautja on mouse chromosome 10. In a mapping cross of 964 opportunities for recombination, yautja was mapped to a 1.45 Mb interval between D10Mit67 and D10Mit264, which contains six genes: Anks1b, Apaf1, Ikbip, Rmst, Slc25a3 and Tmpo. Apaf1 cDNA was amplified by RT-PCR (SuperScript, Invitrogen) using RNA isolated from E12.5 yautja embryos (TRIzol, Invitrogen) and Sanger sequenced (Macrogen USA, Rockville, MD, USA).
The yautja mutation, a T-to-C substitution in exon 8 of Apaf1, destroys a DdeI restriction site (Figure 2c). PCR using primers F6 (5′-GCATACTACCTCAGACAGC-3′) and R7 (5′-GGTCTGTGTAATAGTCTTTG-3′) followed by digestion with DdeI was used to genotype weanlings and embryos.
Western blot
Lysates were prepared from E12.5 embryos or MEFs, separated on Criterion gels (Bio-Rad, Hercules, CA, USA), transferred to Immobilon PVDF membranes (Millipore, Billerica, MA, USA) and subjected to immunoblot analysis. The following antibodies were used: anti-β-actin (clone AC-74; Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-Apaf1 (clone H324; Santa Cruz Biotechnology, Dallas, TX, USA; Figure 3e) or rat anti-Apaf1 (clone 18H2; Enzo Life Sciences Inc., Farmingdale, NY, USA; Figure 3f), anti-rat IgG-HRP (GE Healthcare, Piscataway, NJ, USA) and anti-rabbit IgG-HRP (Vector Laboratories, Burlingame, CA, USA).
Histology and antibody staining
Alcian blue and alizarin red staining were performed on newborn pups as previously described.52, 53
For immunofluorescence analysis, embryos were fixed for 1 h in 4% paraformaldehyde in PBS, washed in PBS and cryoprotected with 30% sucrose in phosphate buffer overnight at 4 °C. Embryos were embedded in Tissue-Tek OCT compound (Sakura Finetek USA Inc., Torrance, CA, USA), and 8-μm sections were cut on a Leica cryostat. Anti-phospho-histone H3 (Millipore) was used at 1 : 1000, followed by goat anti-rabbit secondary (AlexaFluor488, Invitrogen) at a 1 : 200 dilution. The anti-Shh hybridoma 5E1 (developed by Jessell and Brenner-Morton and obtained from the Developmental Studies Hybridoma Bank, which was developed under the auspices of the NICHD and is maintained by the University of Iowa, Department of Biology, Iowa City, IA, USA) was used at a 1 : 5 dilution, followed by anti-mouse secondary (AlexaFluor488, Invitrogen) at a 1 : 200 dilution.
LysoTracker Red (Invitrogen) staining was done as previously described40 with slight modifications. Freshly dissected embryos were incubated in LysoTracker Red diluted to a final concentration of 5 μM in PBS for 30 min at 37 °C. Following three 15-min washes with PBS, the embryos were fixed for 1 h in 4% paraformaldehyde and embedded in OCT as described above.
In situ hybridization (whole mount and sections)
In situ hybridization on whole-mount embryos54 or frozen sections55 was done as previously described.56
Viability assay
MEFs were seeded at 10 000 per well in 96-well culture dishes and 18 h later were treated with either 2 μg/ml staurosporine (Sigma-Aldrich) for 18 h or 25 ng/ml recombinant mouse TNFα (PeproTech, Rocky Hill, NJ, USA) and 5 μg/ml CHX for 16 h. DMSO concentration was constant in all samples. Cell viability was determined indirectly by measuring the intracellular levels of ATP as a marker for cell survival using the CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, Madison, WI, USA) according to the manufacturer's directions. Data were graphed relative to control cultures (n=4).
Caspase activity assay
Effector caspase DEVDase activity was measured using the Caspase-Glo 3/7 Assay Kit (Promega) according to the manufacturer's instructions. Briefly, MEFs in 96-well plates were allowed to reach room temperature before adding 50 μl of Caspase-Glo 3/7 Reagent to each well, and then mixed for 10 min. Four hours following stimulation with either staurosporine or TNF/CHX, 50 μl of reagent was added to each well and incubated for 1 h. Luminescence was measured on a Synergy HT Multi-Detection Microplate Reader (BioTek, Winooski, VT, USA). Caspase activity was graphed relative to WT MEF cultures (n=3). Error bars indicate the standard deviation of the mean.
Acknowledgments
We are grateful to Scott Weatherbee who helped identify the Apaf1yautja line and Kathryn Anderson, in whose lab at Sloan Kettering Institute the screen was performed. We also thank Laura Mariani, Nicole Umberger and Pratyusha Mandal for technical assistance. The work was funded by NS056380 to TC and NIEHS and NIMH postdoctoral training grants (T32ES012870 and T32MH087977) to ABL. We thank Cheryl Timms Strauss for comments on the manuscript. The Apaf1yautja allele has been submitted to the Mouse Genome Informatics (MGI) database.
Glossary
- Bmp
bone morphogenetic protein
- CARD
caspase recruitment domain
- CHX
cycloheximide
- E
embryonic day
- Fgf8
fibroblast growth factor 8
- FNP
frontonasal prominence
- MEF
mouse embryonic fibroblasts
- P
postnatal day
- PBS
phosphate-buffered saline
- Shh
sonic hedgehog
- TNF
tumor necrosis factor
- WHD
winged-helix domain
- Wnt
wingless
The authors declare no conflict of interest.
Footnotes
Edited by JC Marine
References
- Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Xue D, Shaham S, Horvitz HR. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnermri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 2012. 19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83:279–287. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- Penaloza C, Lin L, Lockshin RA, Zakeri Z. Cell death in development: shaping the embryo. Histochem Cell Biol. 2006;126:149–158. doi: 10.1007/s00418-006-0214-1. [DOI] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ding L, Spencer DM, Nunez G. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J Biol Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, et al. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/s0092-8674(00)81733-x. [DOI] [PubMed] [Google Scholar]
- Honarpour N, Du C, Richardson JA, Hammer RE, Wang X, Herz J. Adult Apaf-1-deficient mice exhibit male infertility. Dev Biol. 2000;218:248–258. doi: 10.1006/dbio.1999.9585. [DOI] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Harris BS, Franz T, Ullrich S, Cook S, Bronson RT, Davisson MT. Forebrain overgrowth (fog): a new mutation in the mouse affecting neural tube development. Teratology. 1997;55:231–240. doi: 10.1002/(SICI)1096-9926(199704)55:4<231::AID-TERA3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Honarpour N, Gilbert SL, Lahn BT, Wang X, Herz J. Apaf-1 deficiency and neural tube closure defects are found in fog mice. Proc Natl Acad Sci USA. 2001;98:9683–9687. doi: 10.1073/pnas.171283198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004-2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Young NM, Hu D, Hallgrimsson B. Mechanisms that underlie co-variation of the brain and face. Genesis. 2011;49:177–189. doi: 10.1002/dvg.20710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. Unique organization of the frontonasal ectodermal zone in birds and mammals. Dev Biol. 2009;325:200–210. doi: 10.1016/j.ydbio.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashique AM, Fu K, Richman JM. Endogenous bone morphogenetic proteins regulate outgrowth and epithelial survival during avian lip fusion. Development. 2002;129:4647–4660. doi: 10.1242/dev.129.19.4647. [DOI] [PubMed] [Google Scholar]
- Foppiano S, Hu D, Marcucio RS. Signaling by bone morphogenetic proteins directs formation of an ectodermal signaling center that regulates craniofacial development. Dev Biol. 2007;312:103–114. doi: 10.1016/j.ydbio.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Fu KK, Hui JN, Richman JM. Noggin and retinoic acid transform the identity of avian facial prominences. Nature. 2001;414:909–912. doi: 10.1038/414909a. [DOI] [PubMed] [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M, Helms JA. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development. 2001;128:2755–2767. doi: 10.1242/dev.128.14.2755. [DOI] [PubMed] [Google Scholar]
- Song Y, Hui JN, Fu KK, Richman JM. Control of retinoic acid synthesis and FGF expression in the nasal pit is required to pattern the craniofacial skeleton. Dev Biol. 2004;276:313–329. doi: 10.1016/j.ydbio.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, et al. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- Young NM, Chong HJ, Hu D, Hallgrimsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010;137:3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostrom CA, Vermeij-Keers C, Gilbert PM, van der Meulen JC. Median cleft of the lower lip and mandible: case reports, a new embryologic hypothesis, and subdivision. Plast Reconstr Surg. 1996;97:313–320. doi: 10.1097/00006534-199602000-00006. [DOI] [PubMed] [Google Scholar]
- Dixon J, Brakebusch C, Fassler R, Dixon MJ. Increased levels of apoptosis in the prefusion neural folds underlie the craniofacial disorder, Treacher Collins syndrome. Hum Mol Genet. 2000;9:1473–1480. doi: 10.1093/hmg/9.10.1473. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–3986. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Moran JL, Turbe-Doan A, Driver E, Kelley M, Beier DR. Focusing forward genetics: a tri-partite ENU screen for neurodevelopmental mutations in the mouse. Genetics. 2011;188:615–624. doi: 10.1534/genetics.111.126862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden VS, Kaufmann T, O'Reilly LA, Adams JM, Strasser A. Apaf-1 and caspase-9 are required for cytokine withdrawal-induced apoptosis of mast cells but dispensable for their functional and clonogenic death. Blood. 2006;107:1872–1877. doi: 10.1182/blood-2005-05-2160. [DOI] [PubMed] [Google Scholar]
- Baldi A, Santini D, Russo P, Catricala C, Amantea A, Picardo M, et al. Analysis of APAF-1 expression in human cutaneous melanoma progression. Exp Dermatol. 2004;13:93–97. doi: 10.1111/j.0906-6705.2004.00136.x. [DOI] [PubMed] [Google Scholar]
- Zucker RM, Hunter ES, III, Rogers JM. Apoptosis and morphology in mouse embryos by confocal laser scanning microscopy. Methods. 1999;18:473–480. doi: 10.1006/meth.1999.0815. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM. Development and tissue origins of the mammalian cranial base. Dev Biol. 2008;322:121–132. doi: 10.1016/j.ydbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Alappat S, Zhang ZY, Chen YP. Msx homeobox gene family and craniofacial development. Cell Res. 2003;13:429–442. doi: 10.1038/sj.cr.7290185. [DOI] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, Helms JA. A primary cilia-dependent etiology for midline facial disorders. Hum Mol Genet. 2010;19:1577–1592. doi: 10.1093/hmg/ddq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HJ, Young NM, Hu D, Jeong J, Mc Mahon AP, Hallgrimsson B, et al. Signaling by SHH rescues facial defects following blockade in the brain. Dev Dyn. 2012;241:247–256. doi: 10.1002/dvdy.23726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci. 2010;123:3209–3214. doi: 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia MJ, Eggenschwiler JT, Caspary T, Alcorn HL, Wyler MR, Huangfu D, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci USA. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci USA. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Belo JA, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, et al. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Dev. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]