Somatic mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 occur frequently in acute myeloid leukemia (AML). Chen et al. find that IDH2 mutants cooperate with oncogenic Flt3 or NRas alleles to drive leukemia in mice by impairing the differentiation of myeloid cells. Inhibiting the bromodomain-containing protein Brd4 triggers rapid differentiation and death of IDH2 mutant AML. These results demonstrate a critical role for mutant IDH2 in leukemogenesis and identify an IDH-independent strategy to therapeutically target these cancers.
Keywords: AML, Brd4 inhibition, IDH mutants, targeted therapy, tumor maintenance
Abstract
Somatic mutations in the isocitrate dehydrogenase (IDH) genes IDH1 and IDH2 occur frequently in acute myeloid leukemia (AML) and other cancers. These genes encode neomorphic proteins that produce the presumed oncometabolite 2-hydroxyglutarate (2-HG). Despite the prospect of treating AML and other cancers by targeting IDH mutant proteins, it remains unclear how these mutants affect tumor development and maintenance in vivo, and no cancer models exist to study the action of IDH2 mutants in vivo. We show that IDH2 mutants can cooperate with oncogenic Flt3 or Nras alleles to drive leukemia in mice by impairing the differentiation of cells of the myeloid lineage. Pharmacologic or genetic inhibition of IDH2 triggers the differentiation and death of AML cells, albeit only with prolonged IDH2 inhibition. In contrast, inhibition of the bromodomain-containing protein Brd4 triggers rapid differentiation and death of IDH2 mutant AML. Our results establish a critical role for mutant IDH2 in leukemogenesis and tumor maintenance and identify an IDH-independent strategy to target these cancers therapeutically.
Acute myeloid leukemia (AML) is a heterogeneous cancer involving the accumulation of immature cells of the myeloid lineage (Shih et al. 2012; The Cancer Genome Atlas Research Network 2013). Genomic and functional studies have identified two broad classes of mutations that cooperate during AML development (Kelly and Gilliland 2002; Takahashi 2011). Class I mutations confer a proliferative and/or survival advantage of hematopoietic stem and progenitor cells (HSPCs) and include activating mutations in NRAS or KRAS, loss of the Ras-GAP NF1, or upstream activation of RAS signaling through mutations affecting the FLT3 receptor tyrosine kinase. Class II mutations promote self-renewal and block the differentiation of HSPCs. Such mutations include translocations involving the MLL1 gene or the t(8:21) fusion involving AML1-ETO. Alone, class I mutations tend to trigger chronic myeloid leukemia-like diseases, whereas class II mutations lead to a myelodysplastic syndrome (MDS)-like state. Although an oversimplification, as a rule, class I and class II mutations cooperate to drive AML, leading to the aberrant proliferation and suppressed differentiation that are hallmarks of this disease.
Currently, most AML patients are treated with high-dose chemotherapy involving cytarabine (ara-C) and an anthracycline, a highly toxic drug combination to which >70% of patients develop resistance. However, the nature of certain genes mutated in AML suggests strategies for treating patients with molecularly targeted agents. For example, internal tandem duplication (ITD) mutations in Flt3 lead to constitutive activation of its tyrosine kinase activity, and drugs targeting these mutations are in clinical trials (Stirewalt and Radich 2003; Leung et al. 2013). Similarly, MEK inhibitors, which interfere with RAS effector mechanisms, show efficacy in certain preclinical models (Lauchle et al. 2009). Finally, inhibitors of DOT1L, a histone methyltransferase needed for the oncogenic activity of MLL fusion oncoproteins, are entering clinical trials (Bernt et al. 2011; Daigle et al. 2011). Such therapies require that leukemia remains dependent on the targeted oncogene or pathway, which is often difficult to predict a priori. Still, not all novel AML targets directly interfere with oncogenic drivers or their downstream effectors. For example, the BET family member Brd4, which is not mutated in AML, has been identified as a target in AML owing to its ability to sustain a Myc-dependent self-renewal program activated by certain AML drivers (Dawson et al. 2011; Zuber et al. 2011b). Regardless of their mode of action, genetically and pathologically accurate mouse models of AML have been important in informing target development (Bernt et al. 2011; Zuber et al. 2011b).
Recently, somatic mutations in the isocitrate dehydrogenase (IDH) genes IDH1 and IDH2 have been identified at high frequency in AML and other tumor types (Parsons et al. 2008; Mardis et al. 2009; Yan et al. 2009; Amary et al. 2011; The Cancer Genome Atlas Research Network 2013). These genes encode key metabolic enzymes that convert isocitrate to α-ketoglutarate (α-KG). IDH mutations mainly impact certain active site residues (e.g., IDH1R132, IDH2R140, or IDH2R172), resulting in loss of normal enzymatic function and the acquisition of a neomorphic activity that enables the mutant proteins to reduce α-KG to 2-hydroxyglutarate (2-HG) (Dang et al. 2009; Ward et al. 2010). The presumptive “oncometabolite” 2-HG can competitively inhibit multiple α-KG-dependent dioxygenases, including key epigenetic regulators such as histone demethylases and the DNA-demethylating TET proteins (Figueroa et al. 2010; Xu et al. 2011). Consequently, IDH mutants are associated with dramatic chromatin abnormalities, including globally altered histone and DNA methylation (Figueroa et al. 2010; Lu et al. 2012; Turcan et al. 2012). In the hematopoietic system and other cell types, these changes are associated with a differentiation block (Koivunen et al. 2012; Lu et al. 2012; Sasaki et al. 2012; Turcan et al. 2012).
The neomorphic action of IDH mutant proteins has created enthusiasm for targeting these enzymes with novel anti-cancer agents, and early studies using small molecules capable of inhibiting IDH1R132H and IDH2R140Q show some activity (Rohle et al. 2013; Wang et al. 2013). Still, there is a paucity of data documenting the oncogenic effects of IDH mutants on the development and maintenance of bona fide malignancies. On one hand, IDH2 mutants block the differentiation of cultured HSPCs (Figueroa et al. 2010), which accumulate in the hematopoietic compartment of mice expressing IDH1R132H (Sasaki et al. 2012). Also, enforced expression of this mutant can promote cytokine-independent growth and block the differentiation of an established erythroleukemic cell line in vitro (Losman et al. 2013). Still, in vivo models whereby IDH mutants drive a fully malignant disease have been lacking.
In this study, we describe a new mouse model in which IDH2 mutants cooperate with other lesions to drive an aggressive AML that accurately recapitulates features of the human disease. We further use this system to study the basis of IDH-mediated oncogenesis and as a preclinical model for testing novel therapies.
Results
IDH2 mutants cooperate with Flt3-ITD or NrasG12D to promote leukemia
Considering evidence that IDH mutations can block the differentiation of HSPCs (Figueroa et al. 2010; Sasaki et al. 2012), we hypothesized that they might act as canonical class II mutations and thus could cooperate with class I mutations to promote AML. We chose mouse models incorporating two common class I mutations observed in human AML: FLT3-ITD (Nakao et al. 1996) and NrasG12D (Schubbert et al. 2007). Flt3-ITD knock-in mice develop a chronic myelomonocytic leukemia that never progresses to AML (Lee et al. 2007; Chu et al. 2012), whereas Mx-1-mediated activation of a latent “lox–stop–lox” NrasG12D allele (Haigis et al. 2008) in hematopoietic cells results in a myeloproliferative disorder (Li et al. 2011; Wang et al. 2011).
We applied a mosaic mouse modeling approach in which HSPCs are isolated from 5-fluorouracil (5-FU)-treated Flt3-ITD mice, transduced with retroviral vectors expressing IDH2 mutants or a vector control, and then assessed for tumorigenic potential following transplantation into sublethally irradiated syngeneic recipient mice (Schmitt et al. 2002). For experiments involving NrasG12D, mice were pretreated with polyinosinic:polycytidylic acid (pIpC) to trigger cre-mediated oncogene activation (Supplemental Fig. 1A). Immunoblotting of protein extracts obtained from sorted GFP+ HSPCs confirmed expression of IDH2 wild-type and mutant proteins (Supplemental Fig. 1B). Gas chromatography-mass spectrometry (GC-MS) analysis revealed that 2-HG levels were elevated in HSPCs expressing IDH2R140Q and IDH2R172K but not wild-type IDH2, confirming that the mutant alleles function as expected (Supplemental Fig. 1C).
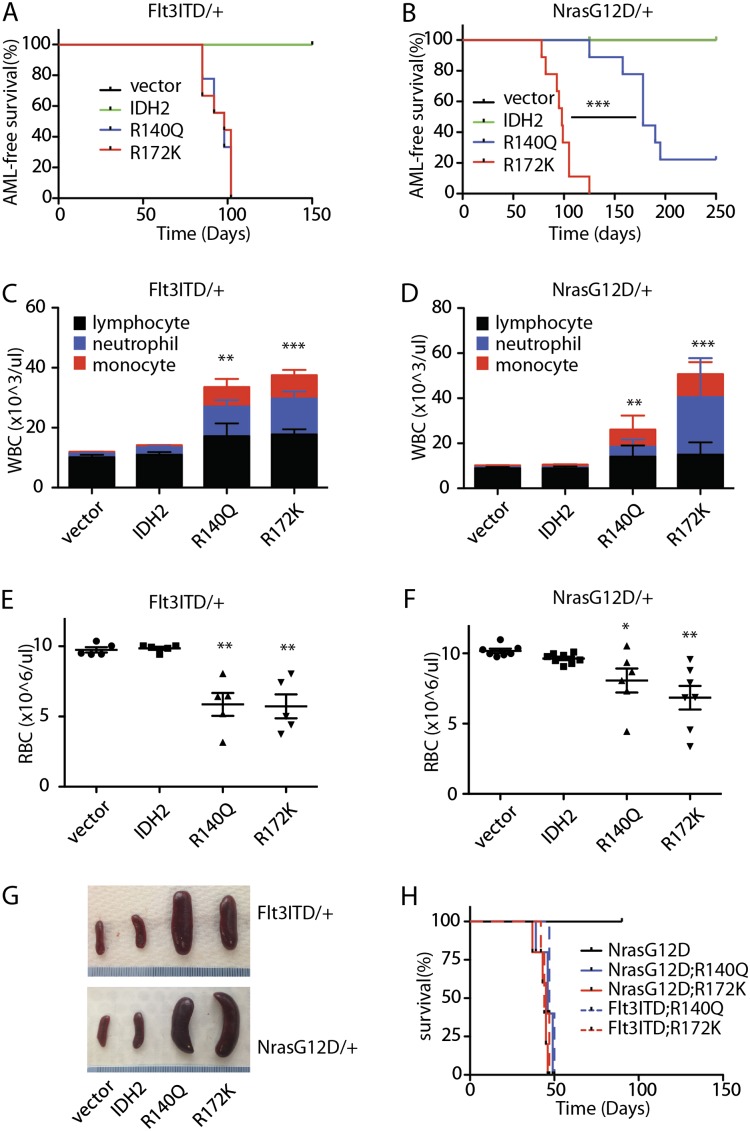
Recipients of HSPCs expressing Flt3-ITD or NrasG12D together with either IDH2R140Q or IDH2R172K displayed significantly reduced survival compared with recipients of HSPCs expressing Flt3-ITD or NrasG12D transduced with empty vector or wild-type IDH2 (Fig. 1A,B). Recipients of Flt3-ITD;IDH2R140Q- and Flt3-ITD;IDH2R172K-expressing HSPCs died with a similarly short latency after transplantation (median leukemia-free survival = 97 d); similarly, both NrasG12D;IDH2R140Q- and NrasG12D;IDH2R172K-expressing HSPC recipients also showed accelerated disease onset, although in this instance, the effect of IDH2R140Q was less potent than IDH2R172K (median leukemia-free survival = 178 d for IDH2R140Q vs. 98 d for IDH2R172K; P < 0.0001) (Fig. 1B). Complete blood counts (CBCs) showed that all IDH2R140Q and IDH2R172K recipients displayed leukocytosis (Fig. 1C,D), anemia (Fig. 1E,F), and gross splenomegaly (Fig. 1G). Importantly, disease could be transferred to secondary recipients by transplanting bone marrow (BM) cells derived from moribund mice, indicating that neoplastic cells arising in the presence of IDH2R140Q or IDH2R172K were fully malignant (Fig. 1H).
Figure 1.
IDH2 mutants cooperate with class I mutations to promote leukemia. (A) Kaplan-Meier survival curve of mice transplanted with Flt3-ITD HSPCs transduced with empty MSCV-IRES-GFP vector (pMIG), IDH2 wild type (WT), or mutants (IDH2R140Q and IDH2R172K). n = 9. (B) Kaplan-Meier survival curve of mice transplanted with NrasG12D HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). n = 9. (C) White blood cell (WBC) counts of mice transplanted with Flt3-ITD HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K) at 12 wk after transplantation. n = 5. (D) WBC counts of mice transplanted with NrasG12D HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K) at 12 wk after transplantation. n = 5. (E) Red blood cell (RBC) counts of mice transplanted with Flt3-ITD HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K) at 12 wk after transplantation. n = 5. (F) RBC counts of mice transplanted with NrasG12D HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K) at 12 wk after transplantation. n = 5. (G) Representative pictures of the spleens of recipient mice transplanted with Flt3-ITD or NrasG12D HSPCs transduced with empty vector (pMIG), IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (H) Kaplan-Meier survival curve of the secondary recipient mice transplanted with Flt3-ITD or NrasG12D HSPCs transduced with IDH2 wild type or mutants (IDH2R140Q and IDH2R172K). n = 5. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
IDH2 mutations drive aggressive AML
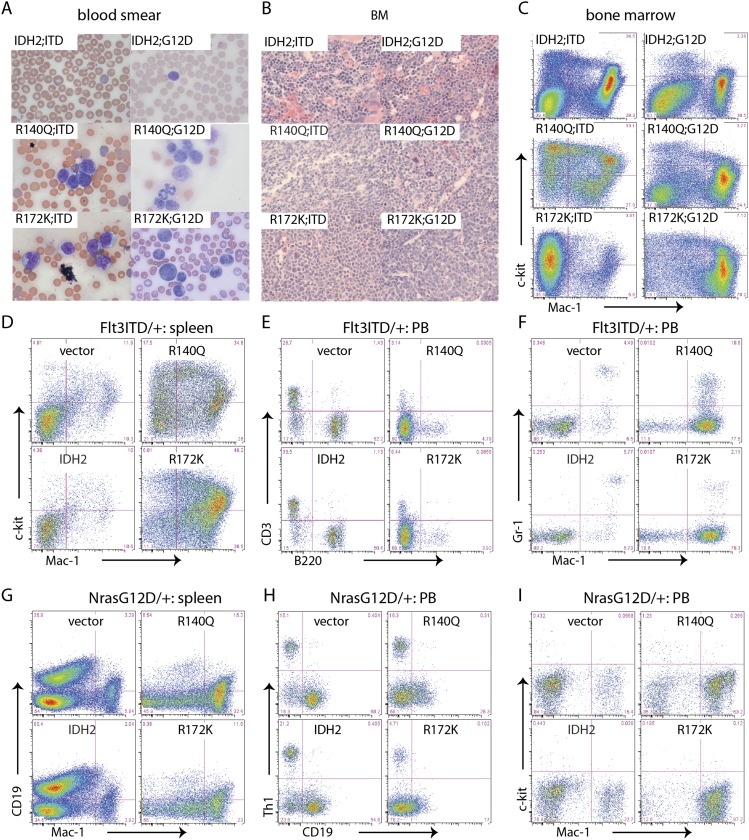
To further characterize the hematopoietic malignancy induced by each IDH2 mutant, we subjected moribund animals to histopathological analyses and immunophenotyping. At the time of sacrifice, all recipients of HSPCs harboring Flt3-ITD or NrasG12D and an IDH2 mutant contained numerous circulating blasts in the peripheral blood (Fig. 2A). Leukemic cells replaced normal hematopoietic cells within the BM (Fig. 2B) and spleen and disseminated to normally nonhematologic organs such as the liver (Supplemental Fig. 2A,B). Flow cytometry revealed that all leukemic cells expressed mutant IDH2, indicated by GFP expression (Supplemental Fig. 3). Whether obtained from the spleen or BM, they expressed the myeloid marker Mac-1 and/or c-kit, a marker of more immature myeloid progenitors (MPs) (Fig. 2C,D,G).
Figure 2.
IDH2 mutations result in AML. (A) Blood smear of recipient mice transplanted with Flt3-ITD or NrasG12D HSPCs transduced with IDH2 wild type (WT) or mutants (IDH2R140Q and IDH2R172K). (B) H&E staining of BM sections (400×) of recipient mice transplanted with IDH2 wild-type- or mutant-expressing cells. (C) Representative flow plots of BM cells from recipient mice transplanted with Flt3-ITD or NrasG12D HSPCs transduced with IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (D) Representative flow plots of splenocytes stained with Mac-1 and c-kit of recipients transplanted with Flt3-ITD cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (E) Representative flow plots of peripheral blood stained with B220 and CD3 of recipients transplanted with Flt3-ITD cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (F) Representative flow plots of peripheral blood stained with Mac-1 and Gr-1 of recipients transplanted with Flt3-ITD cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (G) Representative flow plots of splenocytes stained with Mac-1 and CD19 of recipients transplanted with NrasG12D cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (H) Representative flow plots of peripheral blood stained with CD19 and Thy1 of recipients transplanted with NrasG12D cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K). (I) Representative flow plots of peripheral blood stained with Mac-1 and c-kit of recipients transplanted with NrasG12D cells transduced with empty vector, IDH2 wild type, or mutants (IDH2R140Q and IDH2R172K).
Peripheral blood from recipient mice transplanted with HSPCs expressing the control vector or one encoding wild-type IDH2 contained mostly CD3+ or Thy1+ T cells and B220+ or CD19+ B cells, which is consistent with the cellular composition of blood from nonleukemic mice (Fig. 2E,H). In contrast, the peripheral blood from mice bearing leukemia driven by IDH2R140Q or IDH2R172K was overrun with CD3− B220− Mac1+ or Thy1− CD19− Mac1+ cells (Fig. 2F,I). Taken together, these features indicate that IDH2 mutants in combination with Flt3-ITD or NrasG12D drive disease resembling human AML.
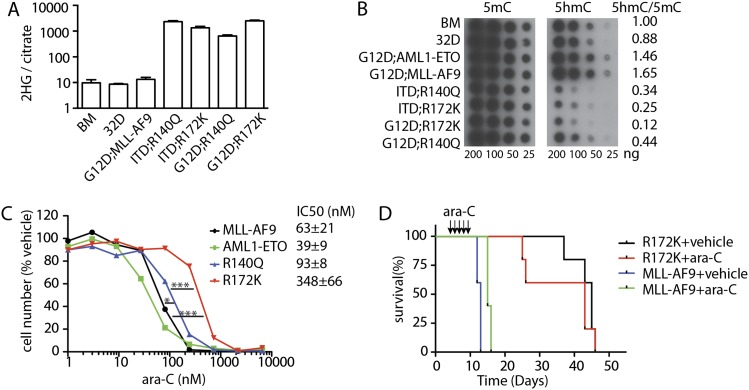
In vitro studies indicate that the high 2-HG levels produced by IDH2 mutants can impair the functions of TET proteins that convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), resulting in a decrease of 5hmC levels (Figueroa et al. 2010). Accordingly, murine AMLs triggered by either IDH2R140Q or IDH2R172K showed a marked increase in 2-HG production relative to nonleukemic BM, the immortalized myeloid cell line 32D, and AML induced by other oncogenes (Fig. 3A). Interestingly, although in vitro studies indicate that IDH2R172K produces more 2-HG than IDH2R140Q (Ward et al. 2013), the 2-HG levels present in Flt3-ITD;IDH2R140Q and Flt3-ITD;IDH2R172K AMLs were comparable, perhaps owing to the lower expression of IDH2R172K than that of IDH2R140Q (data not shown). Analysis of DNA methylation by dot blotting of normal and leukemia DNA showed no significant difference in the global levels of 5mC in AMLs expressing IDH2 mutants. However, consistent with their ability to inhibit TET proteins, IDH2 mutant leukemia displayed a massive reduction in 5hmC levels (Fig. 3B).
Figure 3.
IDH2 mutant-induced AMLs display high 2-HG levels and altered DNA methylation and are chemoresistant. (A) 2-HG levels in BM, 32D, and MLL-AF9 or IDH2 mutant-induced AMLs. n = 3–5. (B) Representative dot blotting with antibodies against 5mC and 5hmC displaying DNA methylation of nonleukemic (BM and 32D) and leukemic cells. (C) Dose response of NrasG12D;MLL-AF9, NrasG12D;AML1-ETO, NrasG12D;IDH2R140Q, and NrasG12D;IDH2R172K leukemic cells to ara-C. Leukemic cells were treated with vehicle or 1–6000 nM ara-C for 3 d. n = 4. (D) Kaplan-Meier survival curve of mice transplanted with NrasG12D;MLL-AF9 or NrasG12D;IDH2R172K leukemic cells treated with ara-C or vehicle. n = 5.
IDH2R172 mutations are associated with poor prognosis in AML patients, while the clinical effects of IDH2R140 mutations are controversial (Marcucci et al. 2010; Paschka et al. 2010; Patel et al. 2012). As underlying differences in survival of patients with different AML genotypes can be linked to treatment response (Zuber et al. 2009; Patel et al. 2012), we asked whether IDH2 mutant AMLs were intrinsically sensitive or resistant to ara-C chemotherapy. Consistent with previous reports (Zuber et al. 2009), NrasG12D;AML1-ETO leukemic cells were much more sensitive to ara-C than NrasG12D;MLL-AF9 leukemic cells in a 3-d treatment assay (IC50 = 39 vs. 63 nM; P = 0.03). In contrast, both NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K AML cells were highly resistant to ara-C in vitro (IC50 = 93 and 345 nM, respectively; P = 0.0001 for NrasG12D;IDH2R140Q vs. NrasG12D;AML1-ETO; P < 0.0001 for NrasG12D;IDH2R172K vs. NrasG12D;AML1-ETO). NrasG12D;IDH2R172K AML cells were even more resistant to ara-C than NrasG12D;IDH2R140Q AML cells (P = 0.0002), an intriguing observation in light of the poorer prognosis of patients with IDH2R172 mutations (Fig. 3C). This resistance was more striking in vivo, where some ara-C-treated mice harboring IDH2 mutant AMLs died even earlier than vehicle-treated controls (Fig. 3D). Therefore, IDH2 mutants can cooperate with class I mutations to promote AML with pathological, biological, and molecular features of the human disease.
IDH2 mutants are sufficient to suppress hematopoietic differentiation and alter DNA methylation in vivo
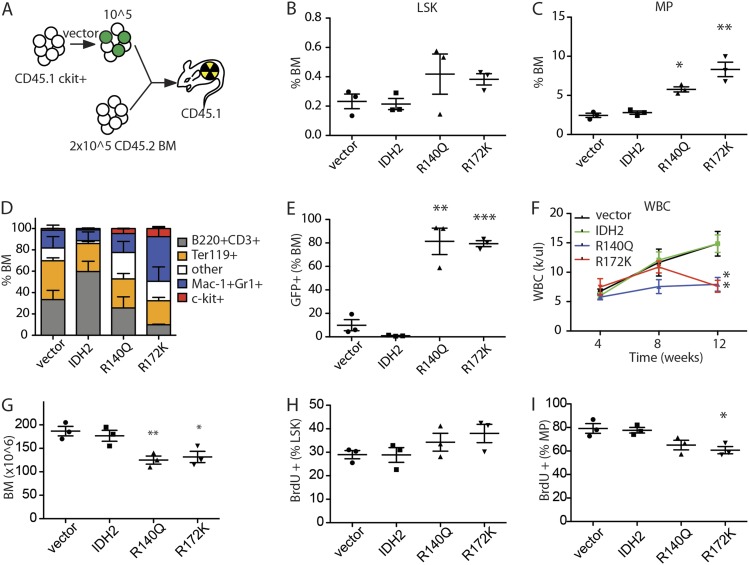
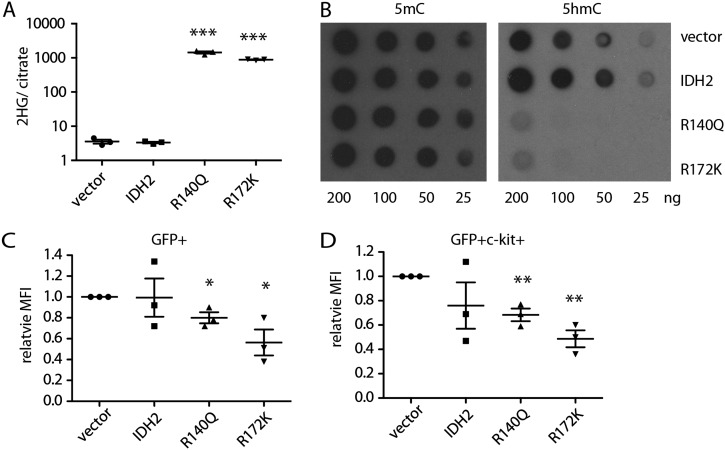
To determine whether the tumorigenic phenotype driven by mutant IDH2 might derive from their ability to impair differentiation in vivo, we examined the impact of IDH2 mutants on hematopoiesis using a competitive reconstitution assay. Here, CD45.1 HSPCs were transduced with a control vector expressing GFP alone or together with GFP vectors encoding either wild-type IDH2, IDH2R140Q, or IDH2R172K; the resulting populations were then mixed with CD45.2 BM cells as a competitive reference and transplanted into lethally irradiated CD45.1 mice (Fig. 4A). Peripheral blood, spleen, and BM from recipient animals were analyzed for cellular composition, 2-HG production, and changes in DNA methylation.
Figure 4.
IDH2 mutations are sufficient to block differentiation of HSPCs and alter DNA methylation. (A) Schematic diagram of competitive transplant assay. Vector refers to transduction of cells with empty MSCV-IRES-GFP vector or one coexpressing wild-type IDH2 or an IDH2 mutant. CD45.1 c-kit+ HSPCs (105) transduced with vectors were mixed with 2 × 105 CD45.2 BM and transplanted into lethally irradiated CD45.1 recipient mice. (B) Percentage of lin−Sca-1+c-kit+ (LSK) cells in the whole BM of recipient mice 12 wk after transplantation. n = 3. (C) Percentage of lin−Sca-1−c-kit+ (MP) cells in the whole BM of recipient mice 12 wk after transplantation. n = 3. (D) Percentage of c-kit+, Mac-1+ or Gr-1+, B220+ or CD3+, and Ter119+ cells in the whole BM of recipient mice 12 wk after transplantation. n = 3. (E) Percentage of GFP+ cells in the whole BM of recipient mice 12 wk after transplantation. n = 3. (F) WBC counts of recipient mice with IDH2 wild-type- or mutant-expressing cells. n = 4–5. (G) BM cellularities of recipient mice with IDH2 wild-type- or mutant-expressing cells at 12 wk after transplantation. n = 3. (H) BrdU incorporation rate of LSK cells after 1 d of labeling. n = 3. (I) BrdU incorporation rate of MP cells after 1 d of labeling. n = 3. Of note, in B–G, vector, IDH2, or IDH2 mutants refers to mice transplanted with HSPCs transduced with the indicated vector but does not necessarily indicate that all of the cells analyzed expressed the constructs. Specifically, while BM from mice transplanted with IDH2 mutant cells almost completely consisted of GFP+ IDH mutant-expressing cells (shown in E), empty vector and wild-type IDH produced a competitive disadvantage, and BM consisted entirely of GFP− normal competitors at this time point.
As expected, white blood cell (WBC) counts in mice receiving HSPCs transduced with empty or wild-type IDH2-expressing vector increased with time after transplantation (Supplemental Fig. 4A). Under these conditions, transplanted cells were able to reconstitute a normal hematopoietic compartment, which displayed the expected distributions of Lin− Sca+ c-Kit+ (LSK) HSPCs, Lin− Sca− c-Kit+ (MP), and other hematopoietic cells (Fig. 4B,D). If anything, empty vector and wild-type IDH2 appeared to confer a competitive disadvantage during hematopoietic reconstitution, as virtually no GFP+ cells remained after 12 wk, with most cells being derived from untransduced or competitor cells (Fig. 4E).
Mice reconstituted with HSPCs expressing IDH2R140Q and IDH2R172K showed a markedly different phenotype. Although at 4 wk post-transduction these mice retained WBC counts comparable with controls, by 12 wk, they showed significantly reduced WBC and had developed an anemia (Fig. 4F; Supplemental Fig. 4A) associated with significantly reduced BM cellularity (Fig. 4G). Nevertheless, IDH2 mutant cells (GFP+) predominated over controls (GFP−), suggesting that these cells outcompeted and excluded the nontransduced and cotransplanted CD45.2 cells (Fig. 4E). These changes were associated with an increased fraction of LSK+ and MPs in the BM (Fig. 4B–D). Although LSK cells expressing both IDH2 mutants showed slightly increased BrdU incorporation when compared with controls, the opposite occurred in the MP compartment, where IDH2 mutant MP cells displayed reduced BrdU incorporation (Fig. 4H,I). The reduced proliferation and increased percentage of MPs in recipients of IDH mutants were associated with a skewing of BM differentiation toward the myeloid lineage (Fig. 4D) comparable with phenotypes observed in IDH1R132H knock-in mice (Sasaki et al. 2012). Thus, IDH2 mutant proteins block the differentiation of MP cells in vivo, leading to the accumulation of more immature cell populations.
As expected, BM cells derived from mice reconstituted with HSPCs expressing IDH2R140Q or IDH2R172K contained high levels of 2-HG (Fig. 5A). In contrast to what has been observed in other systems (Xu et al. 2011; Lu et al. 2012), no obvious changes in the global abundance of methylated histones (H3K4me3, H3K9me3, and H3K27me3) were observed in BM cells expressing IDH2 mutants (Supplemental Fig. 5) relative to vetor-only or wild-type IDH4-expressing cells, raising the possibility that IDH mutants can have tissue-specific effects on chromatin biology and cell fate. Nevertheless, these BM cells showed significantly decreased 5hmC levels without overt changes in global 5mC levels (Fig. 5B), an effect that was also observed in c-kit-positive cells subjected to flow cytometry (Fig. 5C,D; Supplemental Fig. 5B). Overexpression of wild-type IDH2 did not have significant effects on 5hmC levels (Fig. 5B,D; Supplemental Fig. 5B). Hence, IDH2 mutants are sufficient to drive 2-HG production and alter DNA methylation during leukemogenesis.
Figure 5.
IDH2 mutants lead to abnormal DNA methylation in hematopoietic cells. (A) 2-HG levels in the whole BM cells of recipient mice. n = 3. (B) Representative dot blots showing 5mC and 5hmC levels of whole BM genomic DNA. (C) Relative mean fluorescence intensity (MFI) of 5hmC in GFP+ BM cells. n = 3. (D) Relative MFI of 5mC in GFP+ c-kit+ BM cells. n = 3.
IDH2 mutants are required for leukemia maintenance
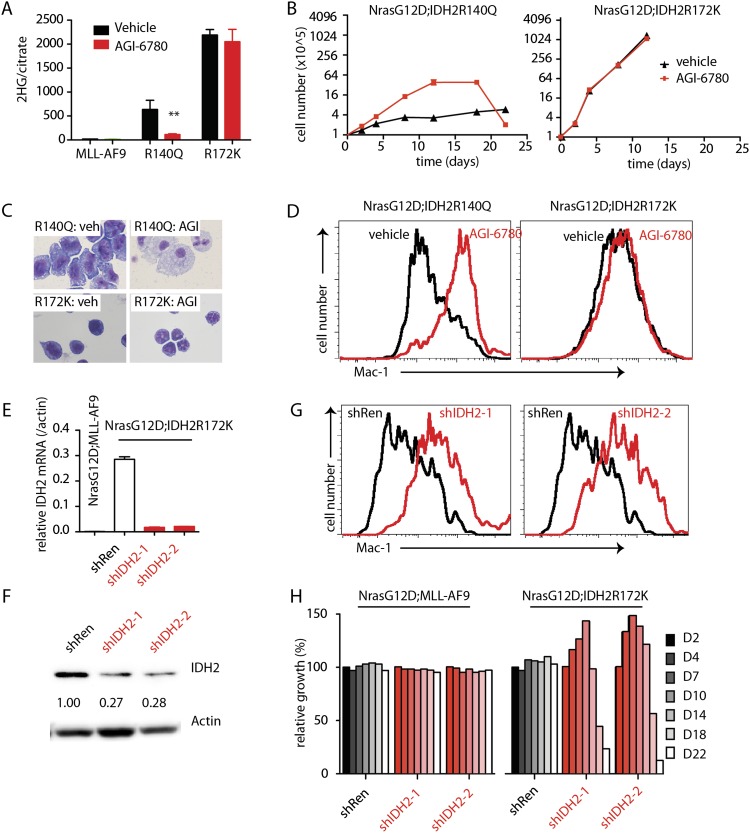
The availability of IDH mutant-driven leukemia models enabled us to assess whether ongoing expression of IDH2 mutants and their product, 2-HG, were required for tumor maintenance. AGI-6780 is a commercially available small-molecule inhibitor that targets IDH2R140Q but not IDH2R172K and has been shown to have anti-leukemic activity in human AML (Wang et al. 2013). To assess its activity against IDH2 mutant-driven murine AML, control NrasG12D;MLL-AF9 leukemia or NrasG12D leukemias coexpressing IDH2R140Q or IDH2R172K were treated with AGI-6780 in vitro, and 2-HG production, cellular proliferation, and differentiation were monitored. Within 2 d, AGI-6780 inhibited 2-HG production in AML cells expressing IDH2R140Q but not control or IDH2R172K-expressing leukemia cells (Fig. 6A). Remarkably, this effect was associated with a 2-wk proliferative burst that was unique to IDH2R140Q-expressing AML (Fig. 6B), after which these cells ceased to proliferate and underwent differentiation (Fig. 6C,D). This proliferative burst and eventual differentiation have also been seen following AGI-6780 treatment of a human AML explant harboring the IDH2R140Q allele (Wang et al. 2013).
Figure 6.
Inhibition of IDH2 mutants leads to loss of 2-HG and delayed differentiation. (A) 2-HG levels of leukemic cells treated with 5 μM AGI-6780 for 2 d. n = 3. (B) Growth of leukemic cells treated with 5 μM AGI-6780 or vehicle. n = 4. (C) Representative H&E staining of cytospin of NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 5 μM AGI-6780 at day 22. (D) Expression levels of Mac-1 by NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 5 μM AGI-6780 at day 22. (E) Transcript levels of hIDH2 in NrasG12D;IDH2R172K cells expressing shRen or shIDH2, normalized to actin mRNA. n = 4. (F) Western blot demonstrating hIDH2 knockdown in NrasG12D;IDH2R172K leukemic cells by shRNAs against mutant IDH2. (G) Relative growth of NrasG12D;MLL-AF9 and NrasG12D;IDH2R172K leukemic cells with shRen or shIDH2. n = 3. (H) Expression levels of Mac-1 by NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K leukemic cells treated with shRen or shIDH2.
Small-molecule inhibitors of the IDH2R172K protein are not currently available. To test its role in leukemia maintenance, we designed shRNA specifically targeting human IDH2, thus inhibiting only the exogenously transduced IDH2 mutant used in this model (Fig. 6E,F). Control (shRen, a potent shRNA targeting Renilla luciferase) and IDH2-specific shRNAs were cloned downstream from a tetracycline-responsive element and dsRed and retrovirally transduced into IDH2 mutant leukemia cells also expressing a rtTA transgene, thereby making the shRNAs doxycycline-inducible (Zuber et al. 2011a). Cells were treated with doxycycline and analyzed for the prevalence of the dsRed marker (and thus the competitive fitness of shRNA-expressing cells) over time.
As expected, IDH2 shRNAs had no effect on MLL-AF9-driven AML. However, similar to the effects of AGI-6780, inhibition of IDH2R172K by RNAi led to an initial enhanced proliferation and subsequent depletion of shRNA-expressing cells from the population (Fig. 6H), an effect associated with myeloid differentiation (Fig. 6G). Thus, ongoing expression of IDH2 mutants is required to block differentiation, although long periods of IDH2 inhibition are required to reverse these effects. In principle, the delay in response may reflect the requirement for cell division to renormalize the epigenetic changes produced by mutant IDH2.
IDH mutant AMLs rapidly differentiate in response to Brd4 inhibition
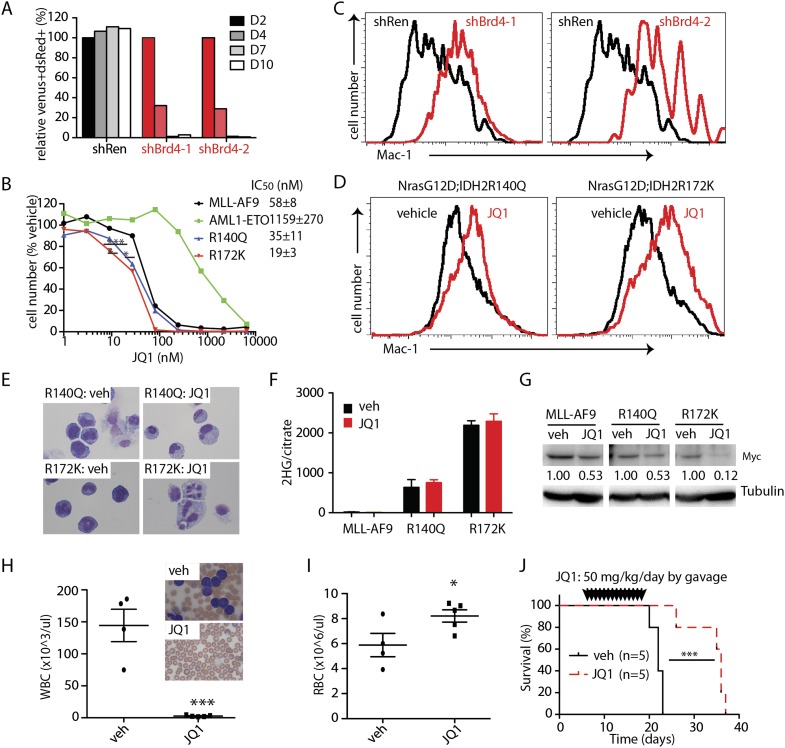
The BET family protein BRD4 has been identified as a therapeutic target in AML produced by MLL fusion oncoproteins owing to its ability to sustain an aberrant self-renewal circuit controlled by Myc (Dawson et al. 2011; Zuber et al. 2011b). To test whether the aberrant self-renewal program produced by IDH2 mutants is also sensitive to Brd4 inhibition, we examined the impact of two independently validated Brd4 shRNAs (Zuber et al. 2011b) or the small-molecule Brd4 inhibitor JQ1 on leukemic cell proliferation, viability, differentiation, and Myc levels in IDH2 mutant AML. Cultured cells were infected with the doxycycline-inducible construct used above expressing either shRNAs targeting Brd4 or Renilla as a control. Brd4 suppression acutely reduced the competitive fitness of cultured NrasG12D;IDH2R172K cells compared with controls, suggesting that Brd4 is essential to maintain IDH2 mutant leukemia (Fig. 7A).
Figure 7.
IDH mutant leukemias rapidly differentiate in response to Brd4 inhibition. (A) Relative growth of NrasG12D;IDH2R172K leukemic cells with Brd4 knockdown. n = 3. (B) Dose response of NrasG12D;MLL-AF9, NrasG12D;AML1-ETO, NrasG12D;IDH2R140Q, and NrasG12D;IDH2R172K to JQ1. Leukemic cells were treated with vehicle or 1–6000 nM JQ1 for 3 d. n = 4. (C) Representative flow plots showing Mac-1 expression levels in NrasG12D;IDH2R172K leukemic cells with shRen or shBrd4 3 d after doxycycline induction. (D) Representative flow plots showing Mac-1 expression levels in NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 50 nM JQ1 for 2 d. (E) Representative cytospin staining of NrasG12D;IDH2R140Q and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 50 nM JQ1 for 2 d. (F) 2-HG levels of NrasG12D;MLL-AF9, NrasG12D;IDH2R140Q, and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 50 nM JQ1 for 2 d. n = 3. (G) Western blotting of Myc levels in NrasG12D;MLL-AF9, NrasG12D;IDH2R140Q, and NrasG12D;IDH2R172K leukemic cells treated with vehicle or 50 nM JQ1 for 2 d. The numbers indicate normalized Myc levels by densitometry. (H) WBC counts of NrasG12D;IDH2R172K recipient mice treated with vehicle or JQ1 at day 20 after transplant. Inserts show the representative blood smear of vehicle- or JQ1-treated mice. Vehicle, n = 4; JQ1, n = 5. (I) RBC counts of NrasG12D;IDH2R172K recipient mice treated with vehicle or JQ1 20 d after transplant. Vehicle, n = 4; JQ1, n = 5. (J) Kaplan-Meier survival curve of NrasG12D;IDH2R172K recipient mice treated with vehicle or JQ1. The mice were treated with vehicle or 50 mg/kg per day JQ1 by gavage from day 5 to day 18 after transplant. n = 5.
In parallel, we tested response to the small-molecule JQ1 in vitro. As expected, NrasG12D;MLL-AF9 AML cells were highly sensitive to JQ1 (IC50, 58 nM) (Zuber et al. 2011b), while NrasG12D;AML1-ETO AML cells were more resistant (IC50, 1159 nM). IDH mutant AMLs were even more sensitive to JQ1 than NrasG12D;MLL-AF9-expressing cells (IC50, 34 nM and 19 nM for NrasG12D;IDH2R140Q and NrasG12D IDH2R172K, respectively; P = 0.01 for NrasG12D;IDH2R140Q vs. NrasG12D;MLL-AF9; P < 0.0001 for NrasG12D;IDH2R172K vs. NrasG12D;MLL-AF9) (Fig. 7B).
The hypersensitivity of IDH mutant AMLs to Brd4 inhibition was associated with rapid differentiation after either shRNA Brd4 knockdown or JQ1 treatment (Fig. 7C–E). Nevertheless, JQ1 treatment did not affect 2-HG levels in either IDH2R140Q or IDH2R172K cells (Fig. 7F), perhaps indicating that Brd4 inhibition acts downstream from mutant IDH2 proteins and mediates its effects without directly affecting its neomorphic activity. However, as has been reported in MLL-AF9-driven AML, immunoblotting of IDH2 mutant AML lysates treated for 48 h with JQ1 revealed reduced levels of Myc (Fig. 7G). Therefore, IDH2 mutant AMLs are addicted to a Brd4-driven, Myc-dependent, self-renewal program that can be inhibited by JQ1 treatment.
Although the pharmacologic properties of JQ1 are not ideal for in vivo studies (Matzuk et al. 2012), we tested its activity against IDH2 mutant AML in vivo. Mice were transplanted with IDH2R172K AML and, upon engraftment, treated (day 5) with vehicle or 50 mg/kg per day JQ1 by gavage for 2 wk (Filippakopoulos et al. 2010; Zuber et al. 2011b). Leukemic cells were apparent in the peripheral blood in vehicle-treated mice at day 19 but completely absent in JQ1-treated animals (Fig. 7H). JQ1-treated mice also displayed significantly improved erythropoiesis, leading to reduced anemia (Fig. 7I). While all vehicle-treated mice died within 3 d post-treatment, JQ1 treatment extended median survival to 18 d (Fig. 7J). Taken together, IDH mutant-driven AML is susceptible to Brd4 inhibition both in vitro and in vivo.
Discussion
A mouse AML model driven by IDH2 mutants suggests therapeutic approaches for AML treatment
In vivo cancer models are essential for understanding genetic alterations that contribute to oncogenesis and can provide preclinical systems for testing novel therapies. Here we demonstrate that the IDH2R140Q and IDH2R172K mutants observed in human cancers can be potent oncogenes in mice, acting as class II driver mutations that cooperate with the class I mutations Flt3-ITD and NrasG12D to promote aggressive AML. Murine AMLs expressing IDH2 mutants display the histopathological and molecular features that are characteristic of the human disease and show a marked chemoresistance phenotype that may underlie the association between IDH2R172 mutations and poor patient survival.
IDH2 mutant proteins promote AML by blocking the differentiation of HSPCs; in the absence of a cooperating event, this produces an MDS-like state, a disorder in which IDH2 mutations are frequent in humans (Patnaik et al. 2012). While these effects correlate with the ability of IDH2 mutants to produce 2-HG and alter DNA methylation, our experiments do not prove a causal role for these downstream changes and disease etiology or maintenance. Still, in a parallel study (Lu et al. 2013), IDH2R172K acts similarly to drive the malignant conversion of mesenchymal progenitor cells into sarcoma in vivo, which also correlates with its ability to produce 2-HG, block differentiation, and alter DNA methylation. Irrespective of the precise mechanism, these data imply a broad and potent action of IDH mutant oncogenes across diverse disease states.
Owing to their similarity to the human disease, the AML models described herein may be useful for evaluating therapies to target IDH2 mutant leukemia. Using genetic and pharmacological tools to manipulate IDH2 activity, we show that IDH2 mutants are required for sustained 2-HG production and leukemia maintenance. Suppression of IDH2 mutant levels and its neomorphic activity triggered myeloid differentiation, albeit requiring prolonged IDH2 inhibition. This delayed response may reflect the requirement for multiple rounds of proliferation to restore normal epigenetic states but might also present a confounding factor when treating patients with acute leukemia. In contrast, shRNA or small-molecule-mediated inhibition of Brd4 causes rapid terminal differentiation and elimination of IDH2 mutant leukemia cells even in the presence of sustained 2-HG levels. This effect is associated with loss of a Myc-dependent self-renewal circuit and triggers substantial anti-leukemic effects in vivo. Notably, IDH2R172K mutant AML, which is not inhibited by AGI-6780 (Wang et al. 2013) and is associated with poor prognosis (Marcucci et al. 2010; Paschka et al. 2010; Patel et al. 2012), displays the highest sensitivity to Brd4 inhibition.
While high-dose chemotherapy can be highly effective against AML, these agents produce substantial toxicity, and patients frequently relapse with resistant disease. One notable exception involves the treatment of acute promyelocytic leukemia (APL), which can be cured without substantial toxicity by a combination of all trans retinoic acid (ATRA) together with arsenic or other molecularly targeted agents (Shen et al. 1997). In contrast to conventional chemotherapy, which triggers leukemia cell death, ATRA acts more directly by reversing the differentiation block produced by the driving PML-RARα oncoprotein (Zhang et al. 2000). As shown here, similar effects can be achieved in murine IDH2 mutant AMLs following IDH2 or Brd4 inhibition, raising hope that improved agents will produce sustained anti-leukemic responses in patients. In any event, these observations validate IDH2 as a therapeutic target in AML and point to an alternative approach to IDH2 inhibition for treatment of IDH2 mutant cancers.
Materials and methods
Mice
All mouse experiments were conducted in accordance with institutional guidelines at Memorial Sloan-Kettering Cancer Center (MSKCC). Recipient C57Bl/6 mice (National Cancer Institute) were irradiated at a dose of 4.5 Gy (Cs137) for sublethal irradiation or two doses of 5.5 Gy for lethal irradiation before transplantation. Mice were transplanted with 1 × 106 or the indicated number of cells by tail vein injection. Mice were monitored for leukemogenesis by complete blood cell count (Hemavet, Drew Scientific), spleen palpation, and blood smear.
Plasmid construction
Human IDH2 wild type and mutants R140Q and R172K were cloned into a pMIG vector (Lu et al. 2012). shRNAs against Renilla (TGCTGTTGACAGTGAGCGCAGGAATTATAATGCTTATCTATAGTGAAGCCACAGATGTATAGATAAGCATTATAATTCCTATGCCTACTGCCTCGGA), IDH2 (shIDH-1, TGCTGTTGACAGTGAGCGCCAAGCTGAAGAAGATGTGGAATAGTGAAGCCACAGATGTATTCCACATCTTCTTCAGCTTGATGCCTACTGCCTCGGA; shIDH2-2, TGCTGTTGACAGTGAGCGCTAAGACCGACTTCGACAAGAATAGTGAAGCCACAGATGTATTCTTGTCGAAGTCGGTCTTATTGCCTACTGCCTCGGA), and Brd4 (shBrd4-1, TGCTGTTGACAGTGAGCGCCCCATGGATATGGGAACAATATAGTGAAGCCACAGATGTATATTGTTCCCATATCCATGGGTTGCCTACTGCCTCGGA; shBrd4-2:,TGCTGTTGACAGTGAGCGACACAATCAAGTCTAAACTAGATAGTGAAGCCACAGATGTATCTAGTTTAGACTTGATTGTGCTGCCTACTGCCTCGGA) (Zuber et al. 2011b) were cloned into a TRIN (Tre-dsRed-mir30-PGK-Venus-IRES-Neo) vector (Zuber et al. 2011a).
Cell culture and transduction
BM cells were isolated from the indicated young adult donor mice, and c-kit-positive HSPCs were separated by autoMACS (Miltenyi Biotec, Inc.). HSPCs were cultured with stem cell medium as described (Schmitt et al. 2002; Zuber et al. 2009). IDH2 mutant AMLs were cultured with IMDM medium supplemented with 20% FBS and 0.2 ng/mL IL-3. NrasG12D;MLL-AF9 AML was cultured with RPMI-1640 plus 10% FBS. Retroviruses were made by calcium phosphate-mediated transfection of Plat-E (Morita et al. 2000) packaging cells. HSPCs and AML cells were transfected by spinoculation.
Measurement of 2-HG
Frozen cell pellets were extracted with 1 mL of ice-cold 80% methanol containing 20 μM deuterated 2-HG as an internal standard (D-hydroxyglutaric-2,3,3,4,4-d5). Methanol extracts were incubated at −80°C for 30 min and centrifuged at 21,000g for 20 min at 4°C to remove precipitated protein. Nine-hundred microliters was evaporated to dryness under a nitrogen gas stream. Dried organic acids were derivatized by the sequential addition of 50 μL of 40 mg/mL methoxyamine hydrochloride in pyridine with incubation for 90 min at 30°C followed by 80 μL of MSTFA + 1% TMCS (Thermo Scientific) and 70 μL of ethyl acetate with incubation for 30 min at 37°C using an automated sample preparation platform (Gerstel). One microliter of the trimethylsilyl-derivatized organic acids was analyzed by GC-MS using an Agilent 7890A gas chromatograph with an HP-5MS capillary column connected to an Agilent 5975 C mass selective detector operating in splitless mode with electron impact ionization. Relative quantitation of 2-HG was determined from extracted ion chromatograms (EICs) for 2-HG (m/z: 349 or 247) normalized to the EICs of intracellular citrate (m/z: 465).
Histology and pathology assay
Bone, spleen, and liver were fixed in 10% formalin. Embedding, sectioning, and H&E staining were performed by IDEXX RADIL. Blood smears were stained with Hema-Quik II stain solution (Fisher Scientific). Cytospin was performed on Shandon Cytospin 4 and then stained with JorVet DipQuick stain.
Flow cytometry
Flow cytometry was performed on LSR II or Fortessa machines (BD Bioscience). All of the cell surface marker antibodies were from eBioscience. 5-hmC antibodies were from Active Motif (no. 39769). Cell surface marker staining was performed with Hank's balanced salt solution plus 2% FBS. For 5-hmC staining, cells were fixed and permeabilized with BD fix/perm buffer and then treated with 0.3 mg/mL DNase I for 30 min at 37°C. For the BrdU incorporation assay, mice were intraperitoneally (i.p.) injected with 100 mg/kg BrdU and then fed with 1 mg/mL BrdU in water for 24 h (Chen et al. 2008). BrdU staining was performed with a BD BrdU kit (BD Bioscience).
Dot blotting
Dot blotting was performed as described (Ko et al. 2010). Briefly, purified genomic DNA was quantified on NanoDrop and denatured by 0.1 M NaOH. Serial-diluted DNA was spotted on a nitrocellulose membrane. 5mC and 5hmC antibodies (Active Motif) were diluted at 1:1000 and 1:5000, respectively.
Western blotting
For IDH2 and Myc detection, cells were lysed in RIPA buffer, and lysates were separated with SDS-PAGE gel electrophoresis. For histone acid extraction, cells were lysed in hypotonic lysis buffer. The primary antibodies were anti-IDH2 (Abcam, ab55271), anti-c-Myc (Santa Cruz Biotechnology, sc-764), anti-actin (Sigma, T6074), anti-tubulin (Sigma, T9026), anti-H3K4me3 (Active Motif, 39916), anti-H3K9me3 (Active Motif, 39765), anti-H3K27me3 (Millipore, 07-449), and anti-H3 (Cell Signaling Tech, 4499).
Quantitative real-time PCR
mRNA was extracted from FACS-sorted cells with Trizol (Invitrogen), and then cDNA was made with SuperScript III (Invitrogen) according to the manufacturer's manual. Quantitative PCR was performed on a 7900HT Fast Real-Time system (Applied Biosystems). The sequences of PCR primers were mouse Actin forward (TGGTGATAAGTGGCCTTGGAGTGT) and reverse (ATGCAAGGAGTGCAAGAACACAGC) and hIDH2 forward (AGACCGACTTCGACAAGAATAAG) and reverse (GACTGCACATCTCCGTCATAG).
Two-color shRNA competitive proliferation assay
Cells were transduced with shRNAs and then selected with G418 (US Biological). Infected cells were mixed with uninfected cells at a 1:1 ratio for the IDH2 knockdown experiment and 4:1 for the Brd4 knockdown experiment. Next, the shRNAs were induced by 1 mg/mL doxycycline (Sigma). The percentage of shRNA-expressing cells (Venus+dsRed+) was measured by a Guava easyCyte flow cytometer (Millipore) at the indicated time points.
Drug treatment
In vitro, cells were treated with vehicle, ara-C (Bedford Laboratories), AGI-6780 (Xcess Biosciences), or JQ1 for 3 d or the indicated times. Viable cells were counted by a Guava easyCyte flow cytometer. For in vivo treatment, JQ1 was suspended in vehicle (0.5% [w/v] hydroxy-propyl-methylcellulose [Sigma], 0.2% [v/v] Tween 80 [Sigma]) at a concentration of 10 mg/mL. The suspension was sonicated before use. Mice with AMLs were treated with vehicle, 100 mg/kg per day ara-C by i.p. injection for 5 d, or 50 mg/kg per day JQ1 by gavage for 2 wk.
Acknowledgments
We thank C.C. Sherr, Dr. Z. Zhao, Dr. C. Miething, Dr. C. Chen, and other members of the Lowe laboratory for suggestions and/or technical help; S. Kogan for histopathological analysis; C. Sherr, L. Dow, and S. Mayack for editorial assistance; and T. Jacks for the NrasG12D mice. C.C. was supported by a career development fellowship from the Leukemia and Lymphoma Society (LLS). This work was supported by a Specialized Center of Research (SCOR) grant from the LLS (to S.W.L.) and a U01 CTDD award from the National Cancer Institute (to S.W.L.). S.W.L. is the Goeffrey Beene Chair of Cancer Biology at MSKCC and an Investigator in the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226613.113.
References
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T, et al. 2011. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Zheng P 2008. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 205: 2397–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu SH, Heiser D, Li L, Kaplan I, Collector M, Huso D, Sharkis SJ, Civin C, Small D 2012. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell 11: 346–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. 2008. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet 40: 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LM, Gilliland DG 2002. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet 3: 179–198 [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, et al. 2012. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchle JO, Kim D, Le DT, Akagi K, Crone M, Krisman K, Warner K, Bonifas JM, Li Q, Coakley KM, et al. 2009. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature 461: 411–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Tothova Z, Levine RL, Anderson K, Buza-Vidas N, Cullen DE, McDowell EP, Adelsperger J, Frohling S, Huntly BJ, et al. 2007. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell 12: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AY, Man CH, Kwong YL 2013. FLT3 inhibition: A moving and evolving target in acute myeloid leukaemia. Leukemia 27: 260–268 [DOI] [PubMed] [Google Scholar]
- Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, Wong JC, Braun BS, Wolff L, Jacks T, et al. 2011. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117: 2022–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG Jr 2013. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339: 1621–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Venneti S, Akalin A, Fang F, Ward PS, DeMatteo RG, Intlekofer AM, Chen C, Ye J, Hameed M, et al. 2013. Induction of sarcomas by mutant IDH2. Genes Dev (this issue). doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, et al. 2010. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol 28: 2348–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. 2012. Small-molecule inhibition of BRDT for male contraception. Cell 150: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T 2000. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther 7: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S 1996. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia 10: 1911–1918 [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, et al. 2010. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol 28: 3636–3643 [DOI] [PubMed] [Google Scholar]
- Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, et al. 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik MM, Hanson CA, Hodnefield JM, Lasho TL, Finke CM, Knudson RA, Ketterling RP, Pardanani A, Tefferi A 2012. Differential prognostic effect of IDH1 versus IDH2 mutations in myelodysplastic syndromes: A Mayo Clinic study of 277 patients. Leukemia 26: 101–105 [DOI] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, et al. 2013. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340: 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. 2012. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1: 289–298 [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G 2007. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer 7: 295–308 [DOI] [PubMed] [Google Scholar]
- Shen ZX, Chen GQ, Ni JH, Li XS, Xiong SM, Qiu QY, Zhu J, Tang W, Sun GL, Yang KQ, et al. 1997. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL): II. Clinical efficacy and pharmacokinetics in relapsed patients. Blood 89: 3354–3360 [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 12: 599–612 [DOI] [PubMed] [Google Scholar]
- Stirewalt DL, Radich JP 2003. The role of FLT3 in haematopoietic malignancies. Nat Rev Cancer 3: 650–665 [DOI] [PubMed] [Google Scholar]
- Takahashi S 2011. Current findings for recurring mutations in acute myeloid leukemia. J Hematol Oncol 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu Y, Li Z, Wang Z, Tan LX, Ryu MJ, Meline B, Du J, Young KH, Ranheim E, et al. 2011. Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood 118: 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, et al. 2013. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340: 622–626 [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Lu C, Cross JR, Abdel-Wahab O, Levine RL, Schwartz GK, Thompson CB 2013. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 288: 3804–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Wang JY, Chen SJ, Chen Z 2000. Mechanisms of all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. J Biosci 25: 275–284 [DOI] [PubMed] [Google Scholar]
- Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, McCurrach ME, Yang MM, Dolan ME, Kogan SC, et al. 2009. Mouse models of human AML accurately predict chemotherapy response. Genes Dev 23: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW 2011a. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011b. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]