Abstract
Transcription factors IRF3 and NFκB, are activated by external stimuli, including virus infection, to translocate to the nucleus and bind genomic targets important for immunity and inflammation. To investigate RNA polymerase II (Pol II) recruitment and elongation in the human antiviral gene regulatory network, a comprehensive genome-wide analysis was conducted during the initial phase of virus infection. Results reveal extensive integration of IRF3 and NFκB, with Pol II and associated machinery, and implicate new partners for antiviral transcription. Analysis indicates that both de novo polymerase recruitment and stimulated release of paused polymerase work together to control virus-induced gene activation. In addition to known mRNA-encoding loci, IRF3 and NFκB, stimulate transcription at regions not previously associated with antiviral transcription, including abundant unannotated loci that encode novel virus-inducible RNAs (nviRNAs). These nviRNAs are widely induced by virus infections in diverse cell types and represent a previously overlooked cellular response to virus infection.
Keywords: IRF3, NF-κ, Transcription, Antiviral, RNA Polymerase II, NELF, Mediator, Innate, Immune Response
Introduction
Regulated gene expression is essential for most biological processes, from development to disease. Signal-responsive factors can modulate gene expression through the recruitment of co-activators to the promoter, which initiates a multitude of regulatory responses that include resolution of inhibitory chromatin configurations, modification of transcription regulators, engagement of Mediator complexes, and recruitment of RNA polymerase II (Pol II) and its associated machinery, ultimately resulting in increased rates of Pol II initiation and elongation at the target locus (Roeder, 1998; Kadonaga, 2004; Malik and Roeder, 2005; Fuda et al., 2009; Taatjes, 2010).
The traditional model of inducible transcription initiation facilitated primarily through transcription factor mediated de novo Pol II recruitment has been supplanted with evidence supporting pre-stimulation Pol II occupancy at gene promoters (Adelman and Lis, 2012). Genome-wide analysis of Pol II distribution has revealed that many metazoan genes, especially those associated with signal-responsive pathways including cell proliferation, development, stress, or damage responses, display higher levels of Pol II at their promoters than within the gene body (Guenther et al., 2007; Muse et al., 2007; Rahl et al., 2010; Cheng et al., 2012), indicating that the polymerase is paused at the promoter. Pol II is retained in a paused state in association with elongation-repressors, including DRB-sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) (Peterlin and Price, 2006). Activating stimuli lead to phosphorylation of the Pol II C-terminal domain (CTD), including phosphorylation of CTD serine 2 (S2) by the cyclin dependent kinase, P-TEFb (Marshall and Price, 1992, 1995; Wada et al., 1998; Sims et al., 2004). Pol II phosphorylation patterns and association with NELF can serve as diagnostic markers for determining the elongation status of an individual gene.
In addition to revealing novel aspects of Pol II activation, recent genome-wide RNA profiling experiments have demonstrated the production of a diversity of RNA transcripts beyond protein-coding genes (Djebali et al., 2012). RNAs produced from intergenic and unannotated loci account for a major subset of the total transcripts made in the human cell, and principles for long-range gene regulation, facilitated in part by Mediator, are beginning to be fully appreciated (Kagey et al., 2010; Sanyal et al., 2012). However, these studies have been carried out at steady-state rather than coupled with specific gene-activating stimuli, thereby inadvertently overlooking potential genomic targets relevant to biological processes.
In higher eukaryotes, transcriptional mechanisms are especially important for regulating appropriate immune functions, driving both adaptive and innate responses to pathogens. For viral pathogens, gene regulatory networks are activated to create initial barriers for virus replication and to shape subsequent innate and adaptive immunity. The inducible transcription of type I interferon (IFN) genes and other pro-inflammatory cytokines, as well as primary antiviral effectors, is initiated in response to virus-induced signal transduction (Taniguchi and Takaoka, 2002; Mogensen, 2009; Takeuchi and Akira, 2009), and secreted IFNs in turn can drive transcription of diverse target genes (de Veer et al., 2001; Stetson and Medzhitov, 2006). The regulation of cell autonomous antiviral responses has been the subject of intense investigation, and the virus-activated expression of the human IFNβ gene (Ifnb1) is among the best-characterized models of acute gene activation in mammals.
Transcriptional activation of human Ifnb1 requires the concerted actions of virus-activated and constitutive transcription factors to assemble an enhanceosome complex at a nucleosome-free region upstream of the transcriptional start site (TSS), which is obscured by a well-positioned nucleosome (Thanos and Maniatis, 1995; Lomvardas and Thanos, 2002). The assembled enhanceosome factors recruit co-activators including chromatin remodeling machinery that expose the TSS for de novo recruitment of Pol II and associated transcriptional machinery, ultimately initiating Pol II transcription (Agalioti et al., 2000; Lomvardas and Thanos, 2001; Freaney, 2013)
Two essential enhanceosome factors, the transcription factors IRF3 and NFκB, exist in latent cytoplasmic forms. Stimuli that evoke strong immune and/or inflammatory responses activate these factors to translocate to the nucleus and bind to their cognate recognition elements in target gene promoters (Chen and Greene, 2004; Honda and Taniguchi, 2006). These two proteins are critical regulators of immunity and inflammatory responses, control diverse normal cellular functions, and aberrations in their activity contribute to pathologies including inflammatory diseases and cancer.
Despite the wealth of information regarding the ability of IRF3 and NFκB, to regulate the expression of Ifnb1 and other individual target genes, their relative contributions to overall virus-activated transcription, their degree of overlap within the antiviral gene regulation network, and their breadth of target sites throughout the genome are under-investigated. The mechanisms used in regulating antiviral Pol II recruitment, initiation, and elongation at specific targets, as well as their abilities to regulate non-coding and protein-coding genes remain poorly understood. A comprehensive ChIP-seq study was carried out to directly address these questions, and provide a detailed and quantitative genome-wide analysis of transcriptional regulation of the cellular antiviral response. This study reveals extensive collaboration of IRF3 and NFκB, with Mediator throughout the genome, and implicates additional transcription factor partners for antiviral responses. Moreover, analysis of Pol II occupancy and elongation during virus infection indicates that IRF3 and NFκB, drive both de novo polymerase recruitment and mediate polymerase pause-release at their target sites, stimulating the expression of a variety of protein-coding, non-coding, and diverse unannotated loci.
Results
Genome-wide analysis of IRF3 and p65/RELA
IRF3 and p65/RELA are activated by virus infection and rapidly associate with target gene regulatory regions. To investigate the generality of this transcriptional paradigm in antiviral responses, chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) was used to determine the genome-wide occupancy of IRF3, p65/RELA, MED1, NELFA, total Pol II (All), which serves as a marker for transcription initiation and elongation, and Ser2-phosphorylated Pol II (S2P), an indicator of productive transcription elongation. In the human Namalwa B lymphocytic cell line, recruitment of IRF3 and p65/RELA to the Ifnb1 gene promoter occurs within 1h of Sendai virus infection (Figure S1A–D). Virus-induced recruitment of general transcriptional machinery to the Ifnb1 promoter, marked by occupation with Pol II, the negative elongation factor, NELFA, and a representative subunit of the Mediator complex, MED1, was observed at the promoter, and both Pol II and MED1 are later detected throughout the gene body (Figure S1E–N).
To capture initial antiviral transcriptional responses, ChIP-seq libraries were prepared from mock-infected cells or cells infected for 4 hours with Sendai virus. As an additional control, non-precipitated input DNA was isolated under the same conditions and subjected to sequencing. As a quality control measure for every ChIP DNA library, ChIP-qPCR was performed to evaluate an aliquot of the ChIP DNA to validate known protein binding sites (Figure S1O & P). The remainder of the library was used to produce paired-end sequence tags that were mapped to the human genome (hg19), producing 4.5–32 million mapped reads for the 6 factors in both uninfected and infected conditions (data for all libraries can be found in Table S1 ). Regions with statistically significant factor occupancy relative to input DNA were identified using the MACS program (p-value < 1e-5; Table S1 ) (Zhang et al., 2008).
In the absence of virus infection, few specific genomic loci were occupied with significant amounts of IRF3 or p65/RELA. After virus infection, the genome wide occupancy of IRF3 and p65/RELA, was dramatically elevated, giving rise to clear locus-specific binding relative to the surrounding genomic region ± 2.5 kb from the peak center (Figure 1A & E). Most sites occupied by virus-activated IRF3 or p65/RELA were not bound in the absence of infection (Figure S2). Furthermore, the loci occupied by virus-activated IRF3 or p65/RELA are sequences that exhibit high average conservation scores amongst vertebrates relative to the surrounding regions (Figure 1B & F) (Siepel et al., 2005), highlighting the importance of gene regulation by IRF3 or p65/RELA throughout the evolution of innate antiviral responses.
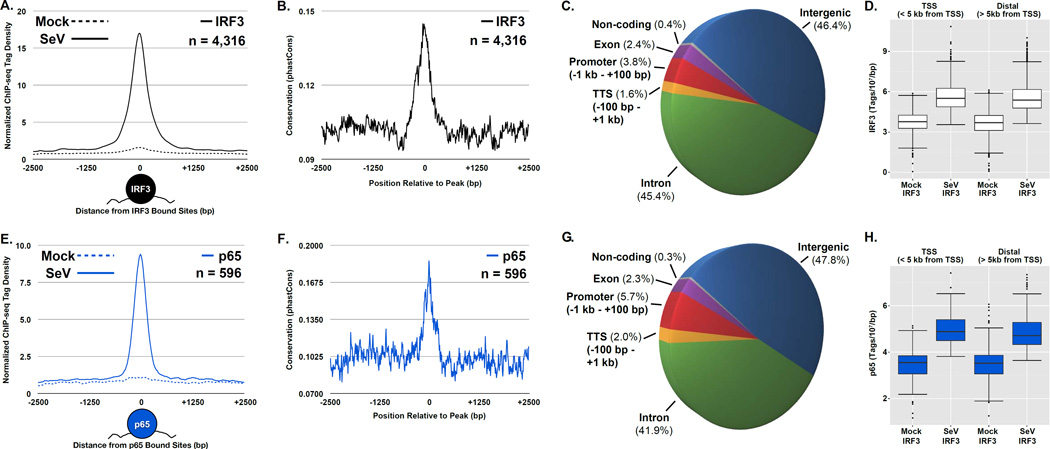
Figure 1. Virus activation of IRF3 and p65/RELA leads to widespread genome occupancy, see also Figure S2.
(A & E) Plot of the mean IRF3 (A) or p65/RELA (E) sequencing tag density at steady state (dashed) and following virus infection (solid) for 4,316 or 596 genomic loci that exhibit ≥ 2-fold increase in occupancy following Sendai infection, respectively (p-value < 1 × 10−5). ChIP-seq enrichment signals are grouped into 10 bp bins and graph illustrates 2.5 kb from the peak center. Graphical representation of factor binding displayed under the plot.
(B & F)Plot of the mean conservation scores at the differentially occupied IRF3 (B) or p65/RELA (F) regions defined in (A). Conservation score (grouped into 10 bp bins) represents the probability that a DNA sequence lies within a conserved element across all vertebrates (Siepel et al., 2005).
(C & G) Pie chart illustrating the annotation distribution of the differentially bound IRF3 (C) or p65/RELA (G) regions described in (A). Binding sites are mapped to one of six annotation categories: Promoter (−1kb to +100bp), TTS (−100bp to +1 kb), exon, intergenic, intron, and non-coding, with the percentage of sites corresponding to each category displayed in parentheses near the label.
(D & H) Boxplots of IRF3 (D) and p65/RELA (H) sequence reads ±1 kb centered at regions defined in (A), which are divided into TSS (< 5 kb from TSS) and distal (> 5 kb from TSS) regions. Changes between mean levels are significant for both IRF3 and p65/RELA (Welch’s two-tailed t test) at TSSs (p-value < 2.2 × 10−16) and distal regions (p-value < 2.2 × 10−16).
Virus-induced IRF3 or p65/RELA binding sites were analyzed with respect to the positions of annotated genomic features (Figure 1C & G). Unexpectedly, in light of the known activity of IRF3 and p65/RELA in driving the inducible expression of protein-coding genes, virus-activated binding within proximal promoter regions (defined as −1kb to +100bp from the nearest TSS) was the third most common area occupied by IRF3 or p65/RELA, comprising only 5.7% and 3.8% of all induced binding sites, respectively (Figure 1C & G). Intergenic regions (defined as greater than 1 kb from annotated genes) were the most frequently occupied by both IRF3 and p65/RELA, representing 46.4% and 47.8% of the inducibly bound loci, respectively. Intronic regions were the second most frequent binding site category representing 45.4% and 41.9% of the bound IRF3 and p65/RELA loci. Among the remaining inducibly bound sites for IRF3 or p65/RELA, 1.6% and 2.0% were located within −100bp to +1kb from transcription termination sites (TTS), 2.4% and 2.3% within exons, and 0.4% and 0.3% within non-coding regions. In addition, significant virus-induced IRF3 and p65/RELA sequence tag enrichment was observed in regions either close to (± 5 kb) or distant from (> 5 kb) the nearest TSS (Figure 1 D & H). This analysis demonstrates that a large proportion of virus-activated IRF3 or p65/RELA binding sites occur in poorly defined regions of the genome, that are likely to represent enhancer regions or previously uncharacterized sites of antiviral transcription initiation. These findings are highly reproducible, and the sequence tag density at the identified peaks is significantly correlated for each factor analyzed in biological replicate samples (Figure S2E).
Motif enrichment indicates cooperativity between IRF3 and p65/RELA, and reveals novel regulatory partners
Numerous studies have demonstrated that IRF and NFκB transcription factors have strong consensus binding sequences (Sen and Baltimore, 1986; Au et al., 1995; Chen et al., 1998; Escalante et al., 1998; Lin et al., 2000; Panne et al., 2007; Wong et al., 2011). DNA sequences within 50 bp from the peak center at 4,316 IRF3 or 596 p65/RELA bound regions that exhibit a greater than 2-fold increase in occupancy following virus infection (p-value < 1 × 10−5; Table S2) were analyzed with a motif discovery algorithm to reveal expected as well as novel motifs that could indicate potential antiviral regulatory partners (Heinz et al., 2010). As anticipated, IRF3 or p65/RELA recognition sequences were identified as the most significant motifs underlying their respective peaks (Figure 2A, top and bottom), and a variety of motif examples matching these canonical binding elements were found (Figure S3A & B). This unbiased approach validates the integrity and specificity of the ChIP-seq data.
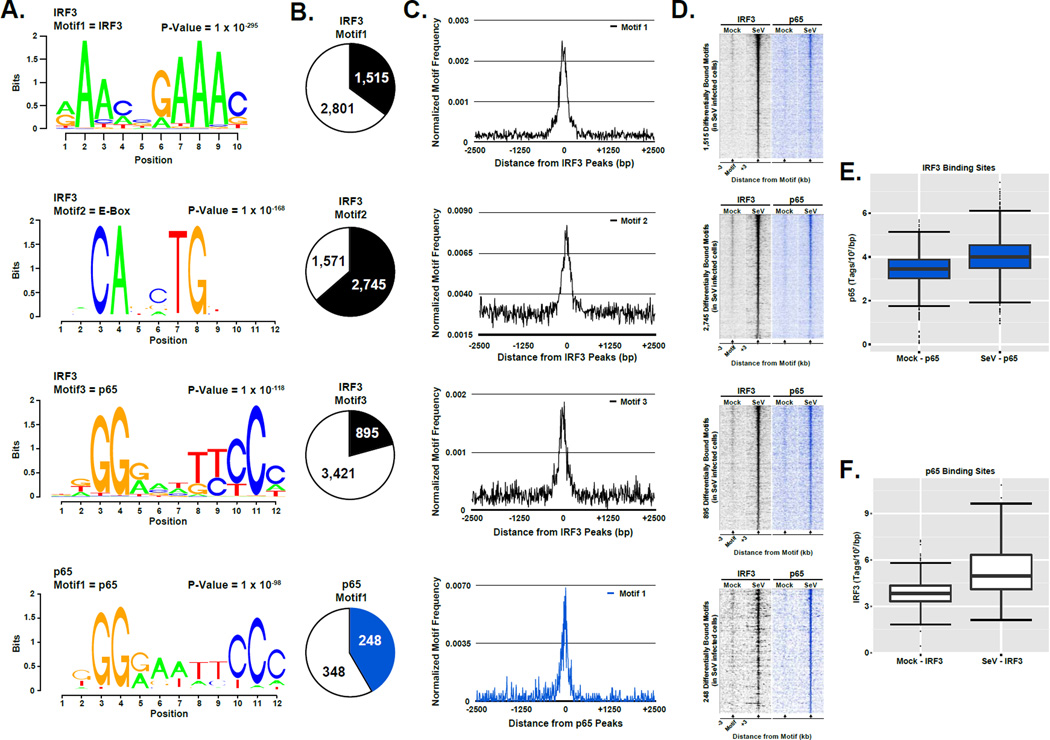
Figure 2. Enriched sequence motifs demonstrate IRF3 and p65/RELA co-occupancy and identify new potential regulatory partners, see also Figure S3.
(A) Graphical representations of the most frequent DNA sequence motifs identified within the 4,316 differentially occupied IRF3 or 596 p65/RELA regions determined by the de novo motif discovery algorithm in the software suite HOMER (Heinz et al., 2010). Each DNA logo represents the information content/bp by height, with the p-value indicating statistical significance for the motif enrichment. P-values and the representative motif name found in the databases (JASPAR, Transfac, HOMER) are displayed above their corresponding motif.
(B) Pie charts representing the frequency of each motif at the regions defined in (A) (± 100 bp from each peak center). Colored sections indicate the fraction of all differentially bound IRF3 (black) or p65/RELA (blue) genomic sites that contain at least one match to the indicated DNA sequence motif. The number of regions in which a match is present or absent is indicated.
(C) Plot of the mean motif density at the regions defined in (A) within 2.5 kb of the peak center. Motif density signals are grouped into 10 bp bins.
(D) Heatmap representation of IRF3 (black) and p65/RELA (blue) occupancy levels at the differentially occupied loci defined in (A) that encompasses the identified motif. Data are organized to illustrate 3 kb surrounding the motif center. Occupancy levels are rank ordered from most to least occupied according to the indicated factor.
(E & F) Boxplots of p65/RELA (E) and IRF3 (F) sequence reads ±1 kb centered on the regions defined in (A). Changes between mean occupancy levels are significant for both p65/RELA (p-value < 2.2 × 10−16) and IRF3 (p-value < 2.2 × 10−16; Welch’s two-tailed t test)
In addition to the consensus IRF3 recognition motif, a number of highly enriched (p-value < 1 × 10−50) DNA sequence motifs were identified underlying IRF3 occupied sites (Figure 2A, middle). The most frequently occurring motif in IRF3 targets matches the consensus sequence for an E-box element (p=1×10−168), which can be bound by transcription factors containing the basic helix-loop-helix protein structural motif (Massari and Murre, 2000). E-box proteins have not previously been linked to IRF3 and antiviral responses, but the E-box regulatory element has been previously described as a transcriptional partner for other members of the IRF family, specifically, IRF1 and IRF4, and this association is critical for CIITA gene activation by IFNγ and pre-B cell development, respectively (Muhlethaler-Mottet et al., 1998; Gobin et al., 2003; LeibundGut-Landmann et al., 2004; van der Stoep et al., 2004; Lazorchak et al., 2006). The E-box motif was found in 2,475 (57.3%) IRF3 bound regions within ±50bp proximity to the IRF3 binding sites (Figure 2B & 2C, 2nd row). The next most significant non-IRF3 motifs corresponded to p65/RELA binding sequences. Consideration of all p65-binding motif variants that lie within ±750bp of the nearest IRF3 binding site reveal over 76% cooccurrence. The most frequent p65 consensus motif (p=1×10−118; Figure 2, Row 3) is observed at 895 (20.7%) IRF3 bound sites within ±50bp proximity to the IRF3 occupied loci (Figure 2B & C, 3rd row).
Throughout the genome, a majority of IRF3 or p65/RELA bound loci investigated are co-occupied by the other antiviral transcription factor (Figure 2D) with a significant level of sequence tag enrichment (Figure 2E & F), indicating these factors work together globally to initiate the transcriptional regulatory network of the innate immune response. Significant enrichment of cyclic AMP response elements (CRE, p = 1 × 10−75) and consensus Ets-binding motifs (p = 1 × 10−52) (Figure S3C & D) suggests extensive cross-talk and coordination between the master antiviral factors and other essential cellular regulatory pathways during the onset of virus infection.
De novo assembly of transcriptional machinery at IRF3 and p65/RELA target sites
IRF3 and NFκB, are able to initiate recruitment of general transcription machinery at the Ifnb1 promoter in response to virus infection. To consider how these antiviral transcription factors and transcription machinery cooperate genome-wide to regulate antiviral target genes, ChIP-seq analysis focused on virus-induced target genes using strictly defined stringencies to determine differential binding sites for each factor ensuring that the investigation focused on genomic regions of high confidence (Tables S2 & S3). The number of occupied loci was highest for Pol II (S2P), Pol II (All), and NELFA (55,493, 28,182, and 12,769, respectively), presumably because of their more general roles in transcription regulation. MED1 occupied an intermediate number of loci (3,752), similar to the numbers observed for IRF3 and p65/RELA (6,141 and 1,107, respectively), though it is important to note that some variations in binding site frequency and signal strength may be due to the technical limitations resulting from differences in antibody efficiency, direct vs. indirect DNA binding, or other experimental idiosyncrasies.
To examine the overlap among all six factors, analysis was restricted to loci exhibiting a greater than 4-fold increase in signal following infection (IRF3 (n = 3,392), p65/RELA (n = 471), MED1 (n = 819), or Pol II (All; n = 3,449)). The occupancy of the other five factors relative to these differentially bound regions was determined (Figure 3). The results revealed that, in general, the inducibly occupied genomic sites are co-bound by all of the factors at precisely the same positions (Figure 3A & B). However, there are variances in the degree of factor recruitment at the differentially bound factor sites.
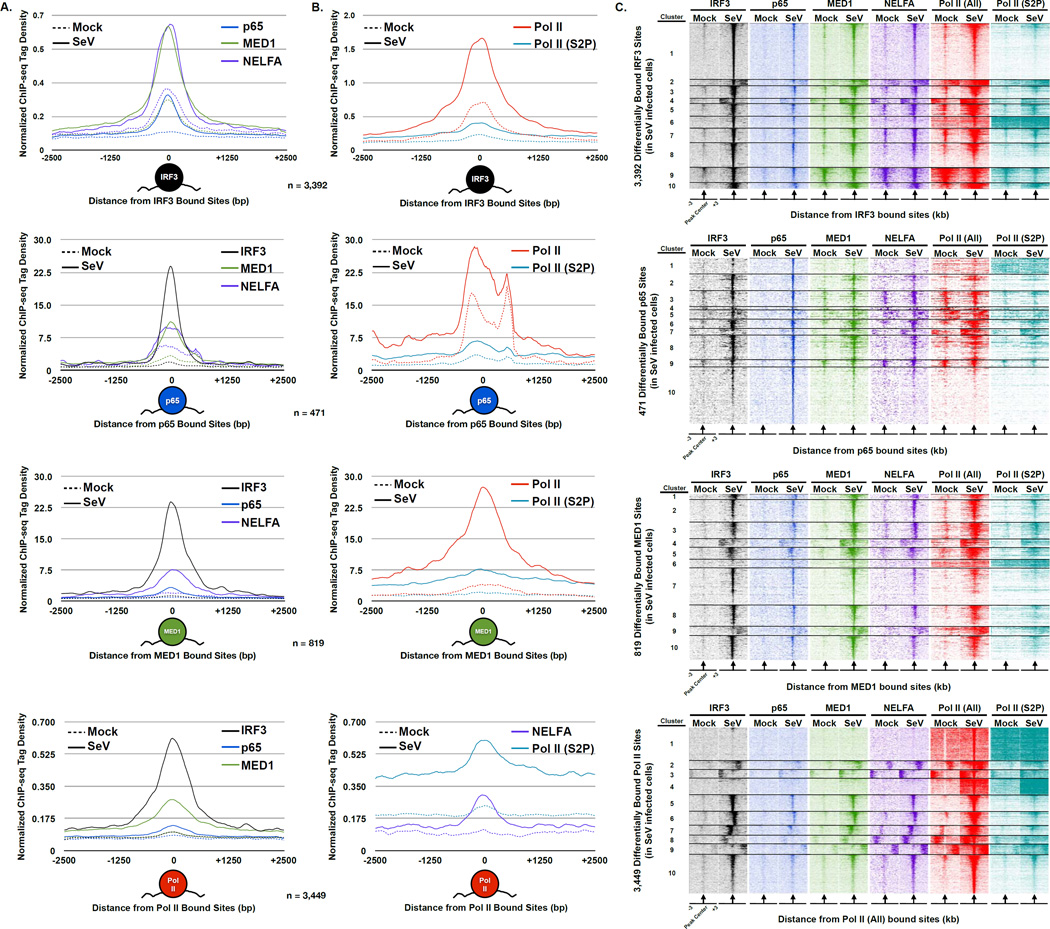
Figure 3. Coordinated de novo assembly of Pol II and general transcription machinery with IRF3 and p65/RELA.
(A) Plot of the mean IRF3 (black), p65/RELA (blue), MED1 (green), and NELFA (purple) sequence tag density at steady state (dashed lines) and following Sendai virus infection (solid lines). Only loci that exhibit ≥ 4-fold increase in occupancy following infection were included (p-value < 1 × 10−5); for IRF3 (n = 3,392), p65/RELA (n = 471), MED1 (n = 819), or Pol II (All; n = 3,449). ChIP-seq enrichment signals are grouped into 10 bp bins and graphs illustrate ± 2.5 kb. Graphical representation of bound factor displayed under plot.
(B) Plot of the mean NELFA (purple), Pol II (All; red), and Pol II (S2P; teal) sequence tag density at steady state (dashed) and following Sendai virus infection (solid) at differentially bound loci defined in (A). Graphical representation of bound factor displayed under plot.
(C) Heatmap representation of IRF3 (black), p65/RELA (blue), MED1 (green), NELFA (purple), Pol II (All; red), and Pol II (S2P; teal) occupancy levels at the differentially occupied loci defined in (A). Occupancy levels are rank ordered from most to least occupied according to the indicated factor and then divided into 10 sections by k-means clustering analysis. 3 kb surrounding the peak center (arrow) are illustrated.
For example, virus-induced IRF3-bound sites were co-occupied by p65/RELA and associated with dramatic increases in MED1, NELFA, Pol II (All), and Pol (S2P) occupancy (Figure 3A & B – top). Similarly, sites of newly bound p65/RELA were co-occupied by IRF3 and associated with increases in transcriptional machinery occupancy, albeit to a lesser degree than IRF3 target loci (Figure 3 A & B – 2nd row). These data suggest that an array of genomic sites bound by both virus-activated factors can recruit regulatory machinery to initiate transcription. Regions of newly bound MED1 are co-occupied by IRF3 and p65/RELA, and also correspond to elevated NELFA, Pol II (All), and Pol II (S2P) occupancy, verifying through an unbiased approach that newly bound MED1 sites are inextricably linked to loci bound by virus-activated IRF3 and p65/RELA (Figure 3A & B – 3rd row). Similarly, sites of newly bound Pol II showcase the co-occupancy of IRF3 and p65/RELA and are marked by dramatic increases in MED1, NELFA, and Pol II (S2P) occupancy. This analysis reveals extensive genome-wide IRF3 and p65/RELA collaboration and suggests that they can recruit Mediator to direct the transcriptional response of infected cells.
Organizing the variety of binding patterns at highly induced loci through k-means clustering provided even greater insights into co-regulation of antiviral gene expression. IRF3 and p65/RELA co-occupy virtually every inducible IRF3 or p65/RELA region, but virus-inducible recruitment of transcriptional machinery is not always apparent at these sites (Figure 3C – 1st row (Cluster 9) and 2nd row (Cluster 1, 4, 9, & 10)). Inducibly-bound MED1 regions reveal a small subset that was not occupied by IRF3 or p65/RELA (Figure 3C – 3rd row (Cluster 6)). These regions do not exhibit increased occupancy of Pol II and its co-associated pausing factor, NELFA, suggesting that a subset of MED1 is reorganized during infection without the support of IRF3 and/or p65/RELA, and may function in a context independent of the general transcription machinery. Clustering of the inducibly-bound Pol II (All) sites revealed only a small portion of loci that are not co-occupied by IRF3, p65/RELA, MED1, or NELFA (Figure 3C – bottom row (Cluster 1 & 4)). These loci can be explained by at least two distinct mechanisms, including: (i) Pol II recruitment independent of IRF3, p65/RELA, and MED1 or (ii) IRF3, p65/RELA, and MED1 participate in Pol II recruitment, but either transiently or distal to the defined region.
Genome-wide and contextual co-localization of virus-activated transcription machinery
To examine the spatial relationship between all six factors throughout the infected cell genome, the total binding sites of each protein were evaluated relative to annotated genes to reveal similarities and differences in their binding sites (Figure 4A). While IRF3, p65/RELA, MED1, and Pol II (S2P) were largely bound outside of proximal promoters in intergenic and intronic regions (91.7%, 92.5%, 86.1% and 84.9% of sites, respectively), Pol II (All) and NELFA were more commonly associated with promoters (18.0% and 42.1% of sites, respectively). These data demonstrate that all of the factors do not solely bind to promoters, but rather a variety of genomic locations. A majority of Pol II (All) and NELFA is found within 5kb of the nearest TSS, while Pol II (S2P) binding sites largely lie downstream of the nearest TSS (Figure 4B; Table S4 ). Similar distributions of IRF3, p65/RELA, and MED1 were found with a large fraction of occupied sites greater than 10 kb from an annotated TSS.
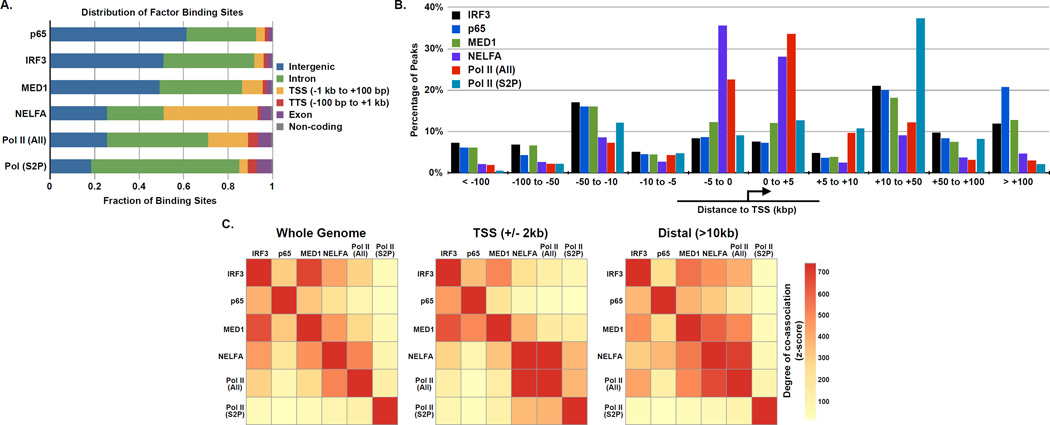
Figure 4. Genome-wide occupancy and co-localization of virus induced transcription factors with general transcription machinery.
(A) The distribution of genomic sites bound by IRF3, p65/RELA, MED1, NELFA, Pol II (All), and Pol II (S2P) in Sendai virus infected cells.
(B) Distribution of virus-induced factor binding sites relative to the nearest annotated TSS.
(C) Graphical representation of the co-association of virus-induced factors. The color strength represents the extent of association from red (strongest) to yellow (weakest). Left: whole genome analysis reveals factors have nonrandom associations with each other. Oriented analysis of promoter proximal (i.e., within 2 kb from the nearest TSS) or distal associations (i.e. > 10 kb from the nearest TSS) reveals more specific relationships (Center and Right, respectively).
Pairwise analysis of inducibly bound peaks in every dataset demonstrates that all of the tested factors share significant levels of co-association to one another genome-wide (Figure 4C, left chart). This co-association reflects both known interactions, such as that between Pol II (All) and NELFA, as well as unexpected novel associations, like that of IRF3 and p65/RELA with MED1. An oriented co-association analysis focused on promoter regions (defined as +/−2 kb surrounding the TSS) and distal regions (defined as > 10 kb from an annotated TSS). Distinct and specific patterns of co-association were identified for a subset of factors. For example, at distal regions, increased association between IRF3 or MED1 with NELFA and Pol II (All) were detected, suggesting that these factors function together outside of annotated regions of the genome to initiate antiviral transcription. Additionally, IRF3 and p65/RELA share a stronger relationship around the promoters of genes than they do at distal regions, suggesting that while they occupy and regulate a similar set of genes, they bind to distinct targets in unannotated genomic regions during the early innate antiviral response.
IRF3 and p65/RELA participate in diverse Pol II regulatory paradigms
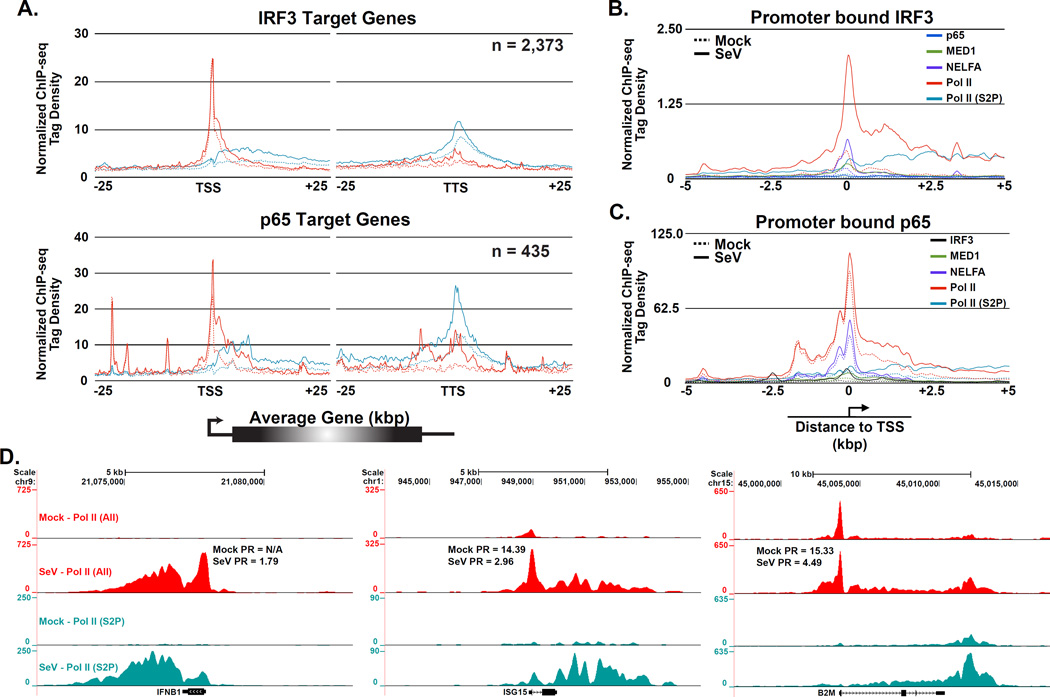
The virus activation of Ifnb1 gene expression is well known to feature recruitment of Pol II de novo. The recent appreciation that Pol II regulatory mechanisms involving pause and release are widespread in diverse metazoans led to comparison of virus-induced Pol II pause release versus de novo Pol II recruitment genome-wide and at specific target genes. To determine if virus-activated transcription factors contribute to pause release of their target genes, ChIP-seq data were analyzed to determine the fraction of genes bound by IRF3 or p65/RELA that were associated with increased levels of Pol II (All) and Pol II (S2P), as an indication of active transcription. Nearly all IRF3-occupied sites and a majority of all p65/RELA occupied loci show evidence of transcription elongation, indicating that most of the regions associated with these transcription factors are transcriptionally active (Figure 3A & B). Both IRF3 and p65/RELA target genes exhibit a substantial virus-induced increase in total Pol II and Ser-2 phosphorylated Pol II within the gene body while the promoter proximal Pol II (All) remained largely unaffected (Figure 5A). The similar overall magnitude of evidence for pausing phenomena at both IRF3 and p65/RELA targets suggests they may share similar abilities to promote Pol II elongation. In addition, a global increase in Pol II elongation following virus infection was observed, most likely due to the dramatic increases in transcriptional load at the virus-activated target genes (Figure S4A). Calculated pausing ratio values (PR; see Experimental Procedures) for IRF3 and p65/RELA target genes decreased as a result of virus infection (Figure S4B). The similarity in Pol II pause release found between the two data sets is, in part, a result of the overlap in target genes between the two factors.
Figure 5. Activated IRF3 participates in diverse Pol II regulatory paradigms, see also Figure S4.
(A) Pol II pause release is a general feature of virus-induced gene expression. Plot of mean Pol II (All; red) and Pol II (S2P; teal) sequence tag density at steady state (dashed lines) and following Sendai virus infection (solid lines). Data are displayed in a 50 kb window surrounding the TSSs and TTSs of all IRF3 (top) or p65/RELA (bottom) target genes that exhibit ≥ 4-fold increase in factor occupancy following Sendai virus infection (p-value < 1 × 10−5). ChIP-seq enrichment signals are grouped into 150 bp bins.
(B & C) De novo Pol II recruitment to IRF3 (B) and p65/RELA (C) target promoters. Plot of mean IRF3 (black), p65/RELA (blue), MED1 (green), NELFA (purple), Pol II (All; red), and Pol II (S2P; teal) sequence tag density at steady state (dashed lines) and following Sendai virus infection (solid lines). Data are displayed in a 10 kb window surrounding the TSSs of 200 promoters containing the greatest increases in IRF3 (B) or p65/RELA (C) occupancy after virus infection. ChIP enrichment signals are grouped into 25 bp bins.
(D) Gene browser views of specific examples of virus-dependent recruitment of transcriptional machinery with distinct Pol II regulatory mechanisms. Displayed are the virus induced Ifnb1 (left), Isg15 (middle), and B2m (right) genes with their corresponding PR values.
Despite this evidence for release of paused Pol II, IRF3 and p65/RELA both have been associated with promoting the de novo recruitment of Pol II and general transcription machinery to activated genes (Lenardo et al., 1989; Wathelet et al., 1998; Sato et al., 2000; Wietek et al., 2003; Leung et al., 2004; Ogawa et al., 2005). To determine the extent of IRF3 or p65/RELA-mediated de novo recruitment, 200 TSSs demonstrating the greatest increase in factor occupancy following virus infection (ranked by factor levels within 2 kb of annotated TSSs) were examined (Figure 5B and C). The proximal promoter regions of virus-activated IRF3 target genes were largely devoid of Pol II in the uninfected cells, but exhibited dramatically increased levels of Pol II (All) and Pol II (S2P) occupancy after virus infection. In stark contrast, Pol II occupied the proximal promoters of p65/RELA target genes to similar levels irrespective of virus infection. Both sets of target genes were marked by virus-induced increases of all forms of Pol II in the gene bodies. These data indicate that IRF3 is more competent than p65/RELA in recruiting the Pol II transcription machinery to the promoters of genes not previously bound by Pol II and its associated factors, while p65/RELA is an effector of pause-release.
Detailed examination of three target genes (Ifnb1, Isg15, and B2m) provides clear examples of both pause-release and de novo Pol II recruitment mechanisms underlying antiviral target gene regulation (Figure 5D). Ifnb1 and Isg15 display varying levels of de novo Pol II recruitment and pause release, with the Ifnb1 gene harboring no Pol II and the Isg15 gene containing limited Pol II occupancy prior to infection. B2m shows all the hallmarks of pause release after infection, with increased Pol II levels in the gene body, similar Pol II levels in the promoter, and a significantly decreased PR value after activation. Together, the association of IRF3 or p65/RELA with a substantial fraction of actively transcribed genes containing promoter-bound NELF and virus-induced decreases of PR values, is consistent with the model that both IRF3 and p65/RELA contribute to Pol II pause release at a large portion of genes in virus-infected cells.
IRF3 and p65/RELA are primary drivers of innate antiviral transcription
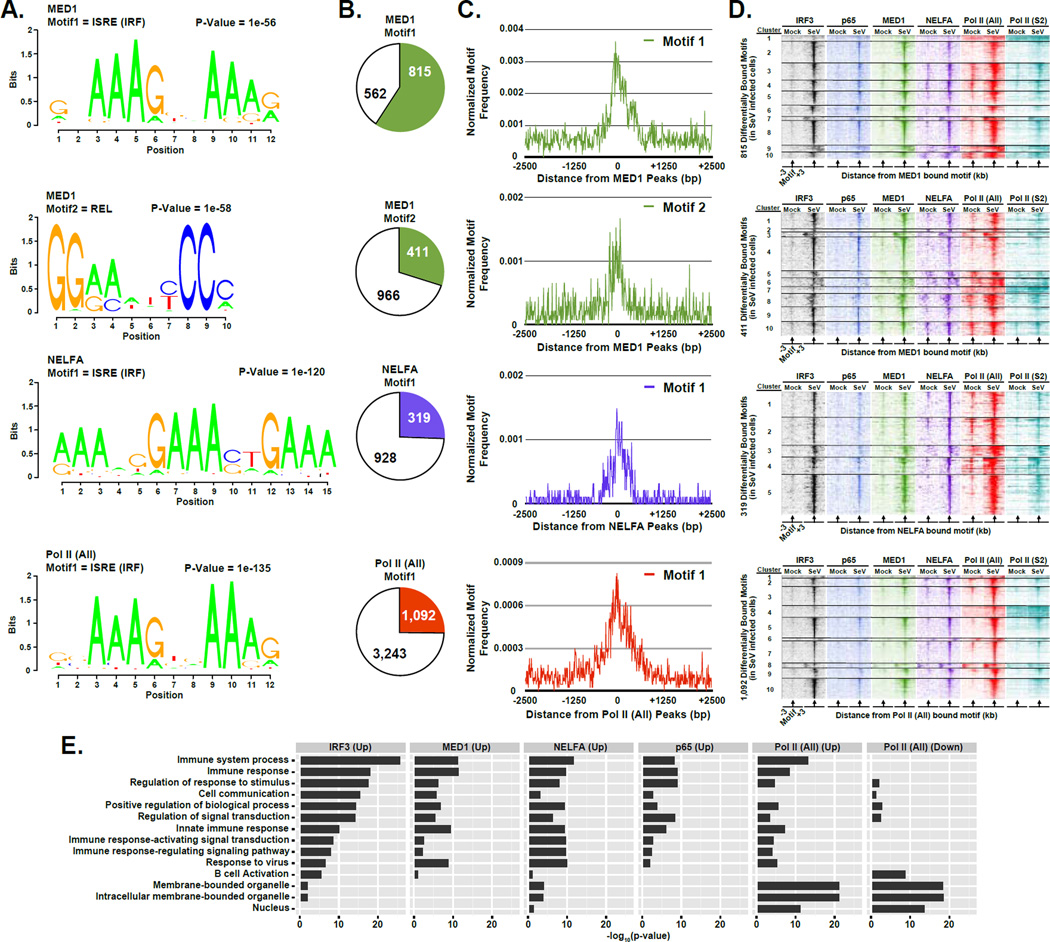
Analysis of DNA sequences underlying virus-induced occupancy of MED1, NELFA, and Pol II sites was carried out to reveal the primary transcription factor motifs that underlie antiviral transcription regulation. The DNA sequences within 100 bp from the peak center at 1,377 MED1, 1,247 NELFA, or 4,335 Pol II (All) regions with > 2-fold increase in occupancy following Sendai virus infection (p-value < 1 × 10−5; Table S2 ) were subjected to de novo motif discovery. For MED1, two motifs were recognized. Motif 1 is a consensus IRF-binding ISRE element (Honda and Taniguchi, 2006), and was identified in 815 (59.2%) MED1-bound loci. Motif 2 represents a consensus p65-binding REL element (Chen and Greene, 2004), and was identified in 411 (29.8%) MED1-bound loci. The IRF binding motifs were also significantly enriched in all the differentially bound data sets (Figure 6A) and was found in 319 (25.6%) NELFA sites, which also display a second IRF half site motif, and 1,092 (25.2%) Pol II bound loci. In all cases, the motif is found in close association to the ChIP-seq peak center (Figure 6B & C – top and bottom 2 rows).
Figure 6. IRF3 acts as a master regulator of innate immunity activated transcriptional networks.
(A) Graphical representations of the most frequent DNA sequence motifs identified by HOMER within the 1,377 MED1-, 1,247 NELFA-, or 4,335 Pol II (All)-bound regions with ≥ 2-fold occupancy increase following virus infection (p-value < 1 × 10−5). Each DNA logo represents the information content/bp by height, with p-value indicating statistical significance for the motif enrichment. P-values and the representative motif name found in the databases (JASPAR, Transfac, HOMER) are displayed above their corresponding motif.
(B) Pie charts representing the genomic DNA sequences of the bound regions described in (A) (± 100 bp from each peak center) scanned for matches to each of the motifs. Pie charts representing the distribution of motifs present in differentially bound MED1, NELFA, or Pol II (All) peaks. Colored sections indicate the fraction of bound MED1, NELFA, or Pol II (All) genomic sites that contain at least one match to the indicated DNA sequence motif. The number of regions in which a match is present or absent is indicated.
(C) Plot of the mean motif density at bound regions described in (A) within 2.5 kb of the peak center. Motif density signals are grouped into 10 bp bins.
(D) Heatmap representation of IRF3 (black), p65/RELA (blue), MED1 (green), NELFA (purple), Pol II (All; red), and Pol II (S2P; teal) occupancy levels at the differentially occupied loci defined in (A) that encompasses the identified motif. Data are organized to illustrate 3 kb surrounding the motif center. Occupancy levels are rank ordered from most to least occupied according to the indicated factor and divided into 5 or 10 sections by k-means clustering analysis
(E) Enrichment of gene ontology (GO) categories among genes nearest IRF3, p65/RELA, MED1, NELFA, or Pol II (All) bound regions demonstrating ≥ 4-fold increase or decease (as noted) in occupancy following Sendai virus infection (p-value < 1 × 10−5). Representative GO categories are shown; the complete set of enriched GO categories is listed in Supplemental Table 5.
To test whether the identified sequence motifs were occupied by their cognate transcription factor, the occupancy of each factor was plotted within 3 kb of the motif (Figure 6D). In general, the identified sequence motif-containing loci were all found to be associated with the corresponding transcription factor and were also associated with surrounding increases in occupancy by transcriptional machinery (Figure 6D – all rows), validating that these sequences play a vital role in recruiting transcription factors to facilitate transcription. These findings support the overall conclusion that IRF3 and p65/RELA are master transcription factors that drive the innate antiviral gene regulatory response.
To understand biological pathways that are affected by gene transcription during the early stages of virus infection, genes inducibly bound by each factor were functionally categorized using gene ontology (GO) (Harris et al., 2004), and this information was used to plot enriched categories (Figure 6E). Results indicate that each factor converges on regulation of immunological and signaling pathways; for example, genes involved in the innate immune response and related processes were preferentially occupied by each factor (Figure 6E).
Novel sites of virus-induced transcription
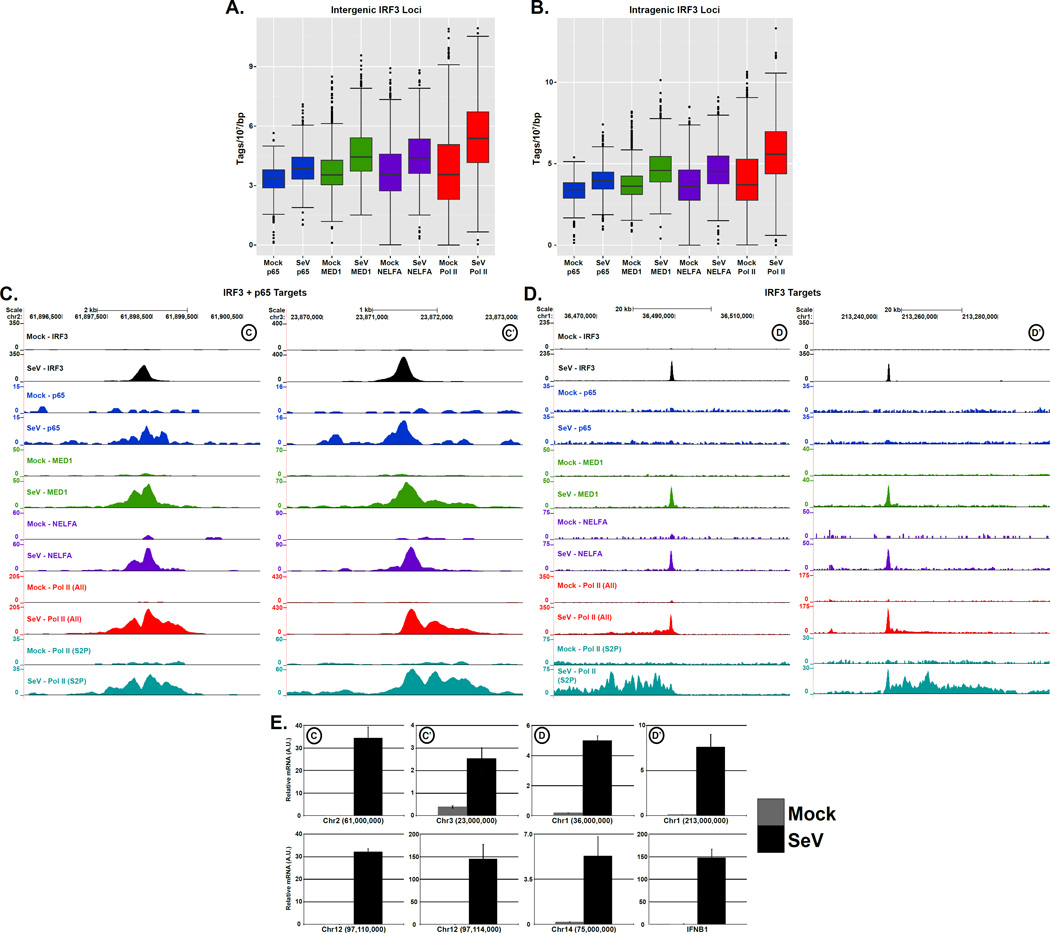
Pol II transcribes all protein-coding RNAs as well as an assortment of non-coding RNAs, including those associated with uncharacterized “intergenic” and “intragenic” regions as well as enhancer-associated transcripts (Kim et al., 2010; Djebali et al., 2012). The identification of virus-induced binding sites that are outside of previously annotated regions prompted the use of ChIP-seq data to identify novel sites of transcription initiated in response to virus infection. Inducible IRF3 targets in intergenic and intragenic loci experienced a concomitant increase in p65/RELA, MED1, NELFA and Pol II (All) occupancy, which suggests that these sites are transcriptionally active (Figure 7A & B). Detailed examination of both IRF3+p65/RELA and IRF3-only unannotated target loci indicates that transcription factor binding promotes the recruitment of transcription machinery and activation of Pol II (Figure 7C & D). These unannotated regions generate RNA transcripts in response to virus infection as detected by specific RT-qPCR (Figure 7E). This demonstrates that the antiviral response is capable of activating transcription to produce distinct classes of previously unrecognized RNAs. For reference, we refer to these RNA species as novel virus-induced RNAs, or nviRNAs.
Figure 7. Virus-activated IRF3 and p65/RELA initiate transcription at novel genomic locations, see also Figure S5.
(A & B) Boxplots of indicated sequence reads ±1 kb centered at IRF3 intergenic (A) or intragenic (B) regions demonstrating ≥ 4-fold increase in occupancy following SeV infection (p-value < 1 × 10 −5). Changes between mean levels are significant for all factors (Welch’s two-tailed t test) at both intergenic (p-value < 2.2 × 10−16) and intragenic sites (p-value < 2.2 × 10−16).
(C & D) Examples of virus-dependent occupancy of IRF3+p65/RELA (C) or IRF3 (D) targets at unannotated loci. The IRF3, p65/RELA, MED1, NELFA, Pol II (All), and Pol II (S2P) occupancy at basal state and 4 h.p.i. are illustrated in genome browser views of virus-induced regions in chromosome 1, 2, and 3.
(E) Induction of nviRNAs by Sendai virus infection. Cells were mock infected or infected (5 pfu/cell), and 5 × 106 cells were harvested for RNA isolation, and random-primed RT-PCR was performed with primers for the indicated locus and GAPDH for normalization. Error bars denote standard deviation for triplicate PCR reactions. The top 4 graphs illustrate loci from panels C & D, and the bottom 4 graphs show additional regions, including those displayed in Figure S6 , and Ifnb1 as a control.
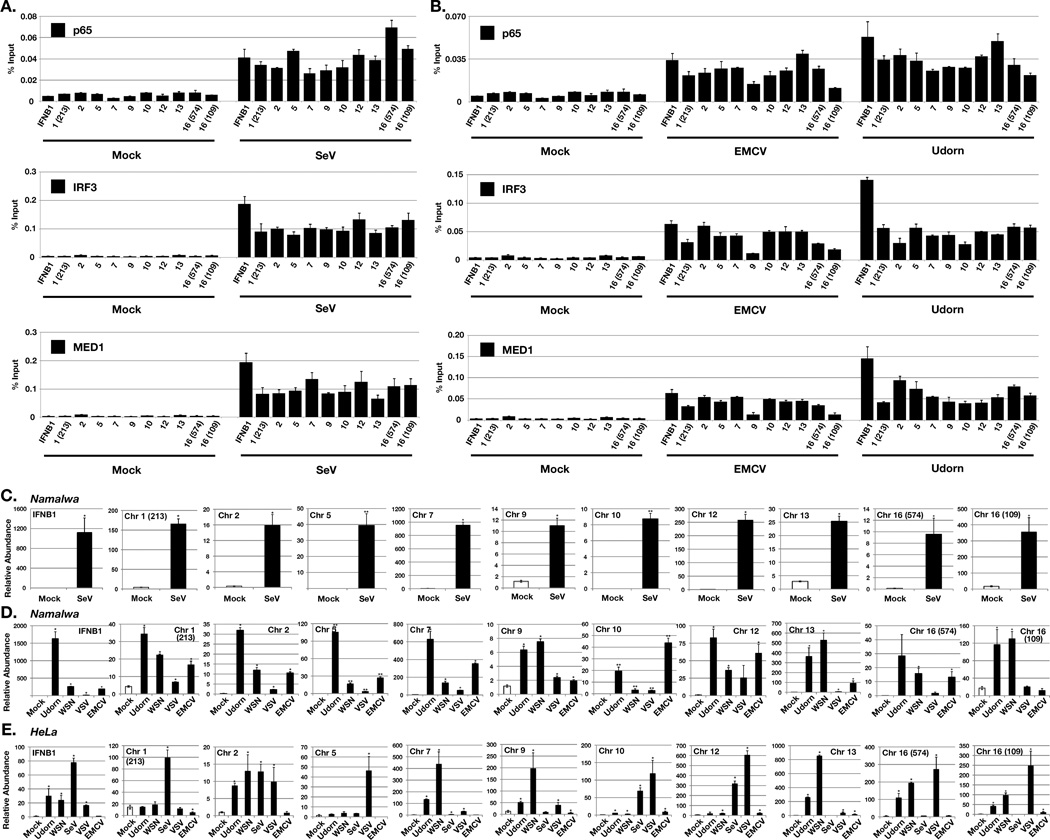
The behavior of the identified nviRNA loci was further tested by direct analysis, and independently validated the Sendai virus-inducible occupancy of IRF3, p65/RELA, and MED1 at every investigated nviRNA locus, confirming the accuracy of the ChIP-seq datasets (Figure 8A). Further, most of the tested nviRNA loci exhibited inducible IRF3, p65/RELA, and MED1 recruitment in response to distinct viruses tested, including encephalomyocarditis virus (EMCV), and influenza A virus (A/Udorn/72) (Figure 8B). Each of the nviRNA loci generated an inducible RNA transcript detected by RT-qPCR in response to Sendai virus (Figure 8C), influenza A viruses (A/Udorn/72 and A/WSN/33), vesicular stomatitis virus (VSV), and EMCV, albeit with some locus-specific differences in absolute activation levels reflective of activation kinetics or other parameters (Figure 8D). To test the generality of nviRNA induction, parallel experiments were carried out in HeLa cells. Direct analysis by RT-qPCR reveals that a large majority of the nviRNA loci generate inducible transcripts in response to diverse virus infections (Figure 8E). These findings solidify the conclusion that these novel sites of transcription are widely activated by virus infections and represent an overlooked feature of the innate antiviral response.
Figure 8. Novel sites of virus-induced transcription factor recruitment result in novel innate antiviral RNA transcription.
(A & B) Namalwa cells were infected with (A) Sendai virus, (B) EMCV, or influenza A/Udorn/72 or mock infected for 10 hours and analyzed by ChIP (IRF3, p65/RELA, and MED1). Numbers on the x-axis refer to the nviRNA locus chromosome number, with numbers in parentheses to distinguish loci that are located on the same chromosome. Error bars denote standard deviation for triplicate PCR reactions.
(C, D & E) nviRNA induction by Sendai virus, influenza A/Udorn/72 or A/WSN/33, VSV, and EMCV infection. (C & D) Namalwa and (E) HeLa cells were mock infected or infected (5 pfu/cell), and 5 × 106 cells were harvested 10 h.p.i. for RNA isolation, and RT-PCR was performed with primers for the indicated nviRNA locus and GAPDH for normalization. Error bars denote standard deviation for triplicate PCR reactions. * indicates p≤0.05; ** indicates RNA was not detected in mock-infected cells.
In addition to virus-induced transcription, several intergenic regions were identified that featured virus activated IRF3+p65/RELA but were constitutively occupied by elongating Pol II and did not recruit additional transcription machinery de novo (Figure S5, top right). This behavior is not uncommon and is reflected in clusters 9 and 10 in the differentially bound IRF3-occupied loci (Figure 3C, IRF3-bound heat maps), and in clusters 1, 2, 4, 9, and 10 in differentially-bound p65/RELA-occupied loci (Figure 3C, p65/RELA-bound heat maps). These data strongly support a role for IRF3 and NFκB, as regulators of a wide range of known and novel RNA classes that contribute to the cellular response to virus infection.
Discussion
Signal-activated transcription factors function independently or in concert to ensure rapid and specific induction of gene expression programs. During the transcriptional response to virus infection, Pol II is recruited to new sites and activated for efficient elongation by the transcription factors IRF3 and NFκB. Examination of the occupancy of Pol II and its associated co-activating partners across the genome revealed that IRF3 and p65/RELA are responsible for regulating the vast majority of transcriptional responses to virus infection. Both IRF3 and p65/RELA were identified within close proximity to one another’s ChIP-seq peak centers throughout the human genome, and were also identified within close proximity to the ChIP-seq peak centers of loci with infection-induced occupation by general transcriptional machinery (MED1 and NELFA), and Pol II. These transcription factors act in close association with each other to regulate antiviral target genes, and also act independently to regulate specific loci, in agreement with the designation of IRF3 and NFκB, as master regulators of innate antiviral responses (Honda and Taniguchi, 2006). In addition, subordinate regulators that had not been previously recognized in the antiviral system were identified, in the form of E-box, CREB, and ETS family transcription factors whose cognate recognition motifs were highly represented within master regulator ChIP-seq peaks.
Virus-activated transcription is achieved through two means: pause-release mechanisms that enhance the elongation rate of promoter-stalled Pol II and de novo recruitment of Pol II and associated transcriptional machinery. The virus-activated loci that feature a paused polymerase associated with NELF in the absence of stimulation undergo a quantifiable increase in polymerase elongation after virus infection, as indicated by their decreased PR values. During the antiviral response, both IRF3 and p65/RELA are recruited to NELFA-occupied loci, and these loci are heavily enriched for IRF-binding consensus motifs. The inducible transcription factors set the stage to alleviate the constraints of the negative elongation machinery, and activate Pol II for productive transcription.
In addition to activation of paused Pol II, a significant number of inducible genes were observed to have little or no Pol II present prior to infection, with substantial increases in Pol II following exposure to virus. These genes have a high or incalculable PR at steady state, but exhibit high levels of Pol II at the promoter and throughout the gene body after infection. The newly recruited Pol II is associated with NELFA and Mediator, and rapidly becomes Ser-2 phosphorylated, resulting in productive transcription.
Evidence for both pause-release and de novo Pol II recruitment was found at IRF3 and p65/RELA target genes, but examination of highly-inducible individual targets of these factors revealed a clear difference in the predominant mode of Pol II activation between the two regulators. The p65/RELA target genes were primarily regulated by pause-release, with little change in Pol II occupancy at the promoter following infection. In contrast, the IRF3 target genes exhibit strong de novo recruitment, with low levels of Pol II found prior to infection that dramatically increase after virus-mediated activation. The difference in principal mechanisms used for target gene activation may in part explain why these factors have evolved to co-regulate gene expression. IRF3 is able to organize promoter-specific recruitment of Pol II, while NFκB, provides the ability to stimulate its efficient and processive elongation through its recruitment of P-TEFb (Barboric et al., 2001; Brasier, 2008; Nowak et al., 2008; Hargreaves et al., 2009). Gene-specific effects are certain to regulate the degree and quality of cooperation in this system, and this phenomenon has been documented for several NFκB, target genes. Specific sequences within κB elements have been identified that can determine whether or not IRF3 is used as an essential co-activator for NFκB-dependent activation (Leung et al., 2004), and the p65/RELA component of NFκB, was demonstrated to be required as a co-activator of IRF3 target genes in response to TLR4 activation (Wietek et al., 2003). The strong enrichment of IRF3 and NFκB, targets in association with MED1 suggests that the Mediator complex is a likely vehicle for coordinate organization of Pol II activation by these two regulators. In addition, the identification of E-box, CRE, and Ets sequence elements among activated IRF3 targets suggests additional co-activators in antiviral responses.
The prevalence of paused Pol II at genes involved in immunological signaling, environmental stress, and developmental processes has several implications for the transcriptional regulation of the human innate immune response (Muse et al., 2007; Adelman et al., 2009). Unlike the coordinated sequences of molecular events that are required for Pol II recruitment at genes like Ifnb1, pre-association of Pol II at an antiviral gene promoter can enable efficient and rapid control of gene expression with virus-activated transcription factor(s). Bypassing the need for more complex regulatory steps is likely to enable immediate changes in transcriptional responses to virus infection. Pol II pause-release can provide a means to fine-tune the expression of key targets that are integral to the cellular response to infection by regulating the intensity of inducible transcription beyond the “all or nothing” effects characteristic of de novo Pol II recruitment. More complex mechanisms that remodel chromatin and recruit Pol II de novo may represent a more secure system that prevents inappropriate activation of signaling pathways and biological responses in the absence of suitably strong stimulation. Consequently, this may grant antiviral transcription factors, like IRF3 and p65/RELA, the opportunity to work together with transcriptional regulatory partners, such as Mediator and NELF, to allow for collaborative regulation of a wider range of antiviral transcription responses.
Annotated mRNA-coding and non-coding genes account for only a small fraction of the genomic loci with virus-inducible transcription factor and Pol II association. Unexpectedly, most virus-activated factor deposition and activation of Pol II-mediated transcription was found to occur at regions distal from known targets, many of which remain without annotation in the human genome. These novel sites of antiviral transcription feature virus-inducible occupation by IRF3 and/or p65/RELA, as well as Pol II and its transcriptionally active Ser2-phosphorylated form. They encode RNA transcripts, termed nviRNAs, potentially ranging from hundreds to thousands of bases, throughout the genome. Direct evaluation of transcription at a sample of these loci confirms that they encode transcribed RNAs that respond to virus infection (Figure 7E). Analysis of nviRNAs revealed their common activation in diverse cell lines by several viruses tested, including EMCV, Sendai virus, and influenza A virus. The potential roles for these RNAs during the antiviral response will be revealed by further investigation, but as IRF3 and p65/RELA play central roles in promoting their transcription we speculate that they participate in the regulation of the human innate immune response by unique mechanisms. The number of identified nviRNA loci suggests that a large portion of virus-inducible transcription responses have been overlooked, and these may represent new targets for use in diagnostic or therapeutic applications.
Experimental Procedures
Cell culture and virus infection
The human immortalized Namalwa B cell line (ATCC CRL-1432) was cultured in RPMI 1640 medium (Life Technologies) and the cervical carcinoma line HeLa was cultured in DMEM. Media were supplemented with 10% cosmic calf serum (Hyclone), glutamine, 1 mM sodium pyruvate (Cellgro), and 1% penicillin/streptomycin (Life Technologies). Infection of cells with Sendai virus (Cantell), influenza A virus (A/Udorn/72 and A/WSN/33), vesicular stomatitis virus (VSV; Indiana), or encephalomyocarditis virus (EMCV; Mengo) was performed at 5 plaque-forming units per cell (pfu/cell) in serum-free media. After 1 hr, cells were lightly centrifuged, washed, and re-suspended in growth media supplemented with 2% CCS. For mock-infection, cells were subjected to all manipulations with PBS substituted for virus inoculum. Cells were centrifuged at 4°C at indicated time points.
Expression analysis
For mRNA analysis by RT-PCR, total RNA was extracted from 5 × 106 cells using TRIzol Reagent (Invitrogen). Samples were treated with DNase I (Life Technologies), and 1 ug of RNA was random primed and subjected to reverse transcription with Superscript III (Life Technologies) for cDNA synthesis. Quantitative real time PCR (qPCR) was carried out using the MX3000P SYBR Green real time PCR system (Agilent) and analyzed using the ΔΔCT method. Expression was normalized against GAPDH and displayed as arbitrary units (A.U.). The primers used for this analysis are listed in Table S7 .
Chromatin immunoprecipitation (ChIP) assays
ChIP analysis was carried out as described in (Lee et al., 2006)
ChIP-qPCR analysis
ChIP DNA was analyzed using SYBR Green real-time PCR analysis (Agilent). ChIP signal was normalized to total input. The oligos used for this analysis can be found in Table S7 .
SOLiD ChIP-seq data analysis
Data analysis was performed using HOMER, a software suite for ChIP-Seq analysis. Each ChIP-Seq experiment was normalized to a total of 107 uniquely mapped tags by adjusting the number of tags at each position in the genome to the correct fractional amount given the total tags mapped. This normalization was used for all downstream analysis (See Extended Experimental Procedures).
Supplementary Material
Highlights.
Comprehensive genome-wide analysis of virus-induced gene regulation.
Antiviral transcription features de novo pol II recruitment and release of paused pol II.
Vast majority of virus-induced transcription outside of known genes.
Identification of numerous novel RNAs induced by diverse virus infections.
Acknowledgments
We thank Dr. John Crispino and Dr. Jason Brickner for helpful discussions and critical reading of this manuscript, Dr. Nadereh Jafari and the Northwestern University Genomics Core for help with SOLiD sequencing, and members of the Horvath laboratory for helpful discussions and support. J.F. was supported by a Dr. John N. Nicholson Fellowship and a Malkin Scholars Award. Supported by NIH grant R01A1073919 and U01AI082984 to C.M.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Access
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE44939 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44939).
References
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci U S A. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- Freaney JEZQ, Yigit E, Kim R, Widom J, Wang JP, Horvath CM. High Density Nucleosome Occupancy Map of Human Chromosome 9p21-22 Reveals Steady-State and Virus-Induced Chromatin Organization of the Type I Interferon Gene Cluster. Submitted for BMC Genomics. 2013 [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobin SJ, Biesta P, Van den Elsen PJ. Regulation of human beta 2-microglobulin transactivation in hematopoietic cells. Blood. 2003;101:3058–3064. doi: 10.1182/blood-2002-09-2924. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol Cell Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Waldburger JM, Krawczyk M, Otten LA, Suter T, Fontana A, Acha-Orbea H, Reith W. Mini-review: Specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Fan CM, Maniatis T, Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Lin R, Genin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Nucleosome sliding via TBP DNA binding in vivo. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Di Berardino W, Otten LA, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr Opin Immunol. 2002;14:111–116. doi: 10.1016/s0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- van der Stoep N, Quinten E, Marcondes Rezende M, van den Elsen PJ. E47, IRF-4, and PU.1 synergize to induce B-cell-specific activation of the class II transactivator promoter III (CIITA-PIII) Blood. 2004;104:2849–2857. doi: 10.1182/blood-2004-03-0790. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. Embo J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Wietek C, Miggin SM, Jefferies CA, O’Neill LA. Interferon regulatory factor-3-mediated activation of the interferon-sensitive response element by Toll-like receptor (TLR) 4 but not TLR3 requires the p65 subunit of NF-kappa. J Biol Chem. 2003;278:50923–50931. doi: 10.1074/jbc.M308135200. [DOI] [PubMed] [Google Scholar]
- Wong D, Teixeira A, Oikonomopoulos S, Humburg P, Lone IN, Saliba D, Siggers T, Bulyk M, Angelov D, Dimitrov S, et al. Extensive characterization of NF-kappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 2011;12:R70. doi: 10.1186/gb-2011-12-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.