SUMMARY
To advance the understanding of sleep regulation, we screened for sleep-promoting cells and identified neurons expressing neuropeptide Y-like short neuropeptide F (sNPF). Sleep-induction by sNPF meets all relevant criteria. Rebound sleep following sleep deprivation is reduced by activation of sNPF neurons and flies even experience negative sleep rebound upon cessation of sNPF neuronal stimulation, indicating that sNPF provides an important signal to the sleep homeostat. Only a subset of sNPF-expressing neurons, which includes the small ventrolateral clock neurons, is sleep-promoting. Their release of sNPF increases sleep consolidation in part by suppressing the activity of wake-promoting large ventrolateral clock neurons, and suppression of neuronal firing may be the general response to sNPF receptor activation. sNPF acutely increases sleep without altering feeding behavior, which it affects only on a much longer time scale. The profound effect of sNPF on sleep indicates that it is an important sleep-promoting molecule.
INTRODUCTION
Although animals need to coordinate sleep with food intake and/or metabolism, the relationship is quite complex. Hunger acutely suppresses sleep in flies and humans (Keene et al., 2010; MacFadyen et al., 1973), and sleep need is antagonistic to foraging/feeding behaviors. However, sleep is also essential for maintaining normal feeding patterns, body mass as well as metabolism (Howell et al., 2009; Knutson and Van Cauter, 2008). Neuropeptide Y (NPY) plays a central role in regulating both sleep and feeding in rats and humans. Although NPY receptors are potential drug targets for obesity treatment (Dyzma et al., 2010; Yulyaningsih et al., 2011), their regulation of sleep is not well understood and may be state-dependent. Moreover, injection of NPY into different brain regions led to either sleep promotion or suppression in rats, depending on the site of injection and dosage (Dyzma et al., 2010), while repetitive intravenous injection of NPY promoted sleep in young men (Antonijevic et al., 2000).
Flies express two NPY-like peptides, NPF and sNPF, which bind to NPFR1 and sNPFR, respectively (Garczynski et al., 2002; Mertens et al., 2002; Vanden Broeck, 2001). Both receptors are structurally similar to vertebrate neuropeptide Y2 receptors (Garczynski et al., 2002; Mertens et al., 2002). In the adult, NPF is expressed predominantly in two pairs of neurons (Wen et al., 2005), whereas sNPF is broadly expressed in multiple brain regions, including the mushroom body (MB), the pars intercerebralis (PI), the central complex (CC) and some clock neurons (Johard et al., 2009; Nassel et al., 2008). NPF has been shown to be important for feeding in larvae (Shen and Cai, 2001; Wu et al., 2003), alcohol sensitivity (Wen et al., 2005) and context-dependent memory retrieval in adults (Krashes et al., 2009). The major function of sNPF has been proposed to be the regulation of feeding and metabolism in adults (Hong et al., 2012; Lee et al., 2009; Lee et al., 2008; Lee et al., 2004; Root et al., 2011). Although it has also been shown to modulate the fine tuning of locomotion (Kahsai et al., 2010), a role for sNPF in sleep has not been identified.
In a screen to test the role of different peptidergic neurons in the adult brain we identified sNPF-expressing neurons as potently sleep-promoting. We found that the s-LNv clock neurons are part of this sNPF-expressing sleep promoting circuit, and the wake promoting l-LNvs are a postsynaptic target. sNPF has very different effects on feeding circuits, suggesting that the role of sNPF in feeding is more indirect and unrelated to its acute sleep-promoting effects.
RESULTS
Activation of sNPF neurons rapidly increases sleep independent of changes in locomotion
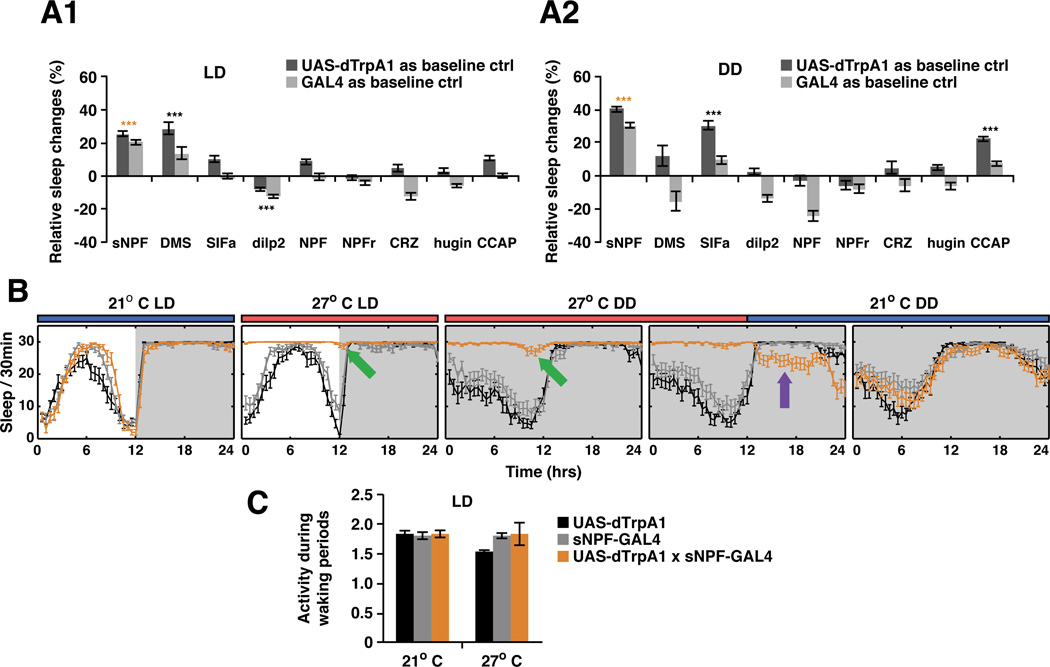
To test the role of different subsets of adult brain peptidergic neurons in sleep (Figure 1A), we used the warmth-activated dTRPA1 cation channel to acutely activate 9 different peptidergic neuron classes (Hamada et al., 2008). Since environmental light affects Drosophila sleep behaviors (Shang et al., 2011), we assayed activation under both light-dark (LD) and dark-dark (DD) conditions. To our surprise, most peptidergic neurons, including those that express NPF, do not affect sleep under either condition (Figure 1A). Neurons expressing DMS or DILP only affect sleep in LD conditions, and neurons expressing SIFa or CCAP only in DD conditions. This context-dependence may indicate that these neurons only affect sleep indirectly, by changing other internal physiological states.
Figure 1. Activation of sNPF expressing neurons promotes sleep independent of changes in locomotion.
A. A dTRPA1 screen identified neurons expressing sNPF as a potent sleep promoting system when activated in fly brains in both A1) LD and A2) DD. All experiments were repeated at least 3 times except for hugin-GAL4 (2 repeats). Calculation of relative sleep changes and statistical analyses are described in EXPERIMENTAL PROCEDURES. Most driver lines did not show significant effects on sleep pattern or sleep time. DMS expressing neurons promoted sleep in LD while sIFamide and CCAP neurons promoted sleep in DD. sNPF-GAL4 is the only driver line that dramatically increased total sleep and altered sleep pattern in both LD and DD. N = 52 for sNPF-GAL4:dTRPA1 flies. B. Sleep analysis showed that heat-induced firing of sNPF-expressing peptidergic neurons dramatically and rapidly (within 1 h) increased quiescent state in both LD and DD. Release from activation was followed by an immediate negative sleep rebound (purple arrow). The green arrows indicate flies still showed circadian related locomotor evening peak around ZT and CT12 (See Figure S1 for locomotor analyses). C. Waking activity between controls and experimental groups were compared using the DAM system. All the genotypes show similar activity at 21°C and heat activation of sNPF neurons led to no significant change of activity level while awake. Data are presented as mean ± SEM; *p < 0.01, **p < 0.001, and ***p < 0.0001 are significant difference from control group (ANOVA with Tukey post hoc test, described in EXPERIMENTAL PROCEDURES). See also Figure S1 and Movies S1 and S2.
The only condition-independent cell group was the short form NPY-like peptide sNPF, which led to a dramatic increase in quiescence in both LD and DD conditions (Figure 1A–B). High-resolution computer video tracking confirmed the results obtained with the DAM system (Donelson et al., 2012, Figure S6). Upon inactivation of dTRPA1 at 21°C, these effects were rapidly reversed, and the mean duration of quiescent episodes decreased (Figure 1B, Figure S6B). Remarkably, flies slept even less after reversal of dTRPA1 activation, i.e., they manifested negative sleep rebound (Figure 1B, purple arrow, Figure S6B green arrows), suggesting that the observed quiescence is really sleep and that this state can be homeostatically regulated in both directions (Hendricks et al., 2000; Shaw et al., 2000).
Importantly, excess quiescence is not due to a loss of ability to engage in locomotor activity. Quantitative analysis of single fly behavior showed similar or even slightly higher locomotor activity during periods when flies were awake (Figure 1C and Figure S6B). We also directly tested locomotion within 2 h after the temperature shift by tapping vials and assaying negative geotaxis behavior. Although at this time point sNPFGAL4:dTRPA1 flies already showed increased quiescence due to activation of sNPF-expressing neurons, they still rapidly climbed to the top of the vials when stimulated (movie S1). However, most flies were not responsive to the initial tap compared with controls three days after the shift (data not shown). Consistent with an increased sensory/arousal threshold, these flies required several taps but then climbed with a similar speed (movie S2). Moreover, flies with activated sNPF neurons still exhibited spontaneous circadian-related locomotor activity in both LD and DD conditions (Figure 1B, green arrows; see Figure S1 for locomotor analyses). This indicates that the locomotor circuitry is intact since it can be accessed by the circadian clock. Lastly, release from dTRPA1 activation reversed the increased locomotor activity observed during waking periods (Figure S6B, blue arrows). This indicates that the negative sleep rebound is not due to an increased intensity of locomotor activity.
sNPF is necessary for sleep maintenance
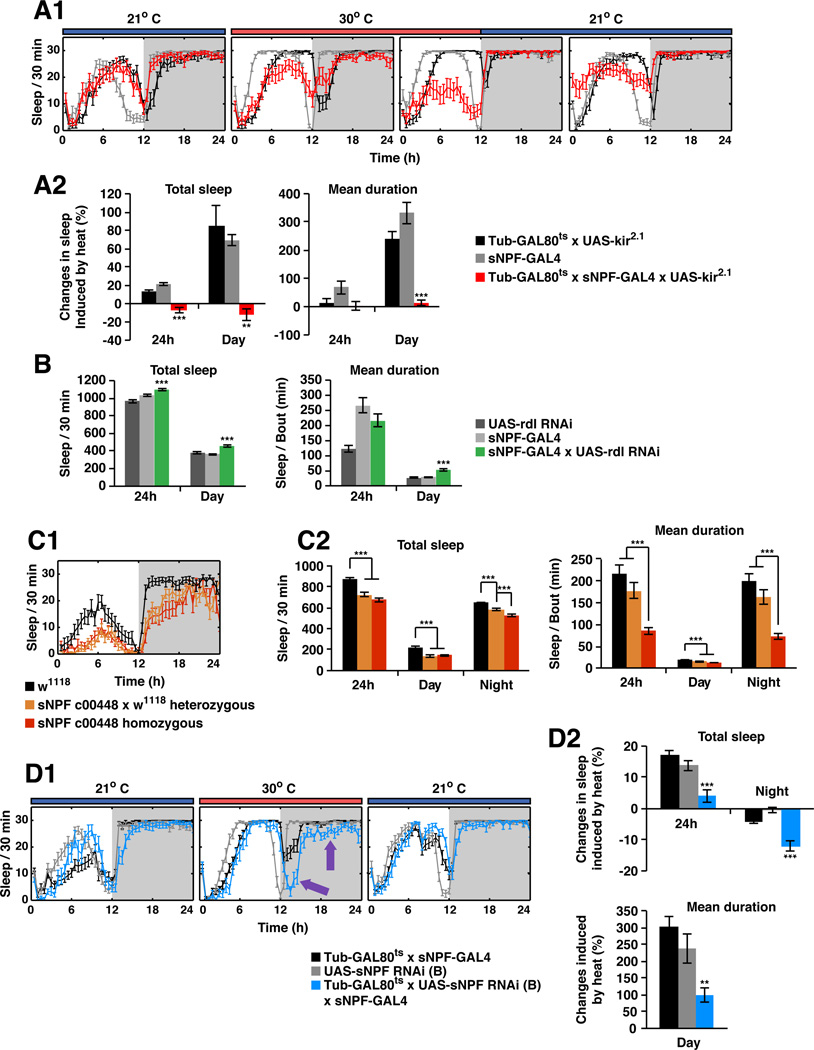
To investigate if the activity of sNPF neurons is required to maintain normal sleep, we silenced these neurons by co-expressing the Kir2.1 potassium channel (Nitabach et al., 2002). To restrict the silencing to adults, tubulin-GAL80ts was used to block expression of UAS-kir2.1 until adulthood (McGuire et al., 2003). After a temperature shift to release the GAL80 block, sleep levels as well as the mean duration of sleep bouts during the daytime were significantly reduced (Figure 2A). In contrast, control flies manifested a temperature-driven increase in daytime sleep.
Figure 2. sNPF is required for maintaining sleep.
A. sNPF-expressing neurons are required for maintaining sleep. A1) A tubGAL80ts transgene was used to block the expression of UAS-kir2.1 in sNPF-GAL4 neurons at 21°C. The GAL80 protein was inactivated at 30°C, allowing the expression of Kir2.1 mRNA driven by sNPF-GAL4 in adult brains. Temporally-controlled silencing of the sNPF neurons induced by heat led to a decrease of total as well as daytime sleep. The sleep loss was rapidly reversible once the GAL80 protein was reactivated at 21°C. A2) Quantitative data for the heat-induced sleep loss and changes in mean bout duration. The calculation for heat induced sleep changes is described in EXPERIMENTAL PROCEDURES. B. Sleep-promoting sNPF neurons are suppressed by GABA through Rdl GABAA receptors in the daytime. The total 24 h sleep time and daytime sleep for each genotype are shown, as well as mean bout duration. C. sNPF-deficient flies sleep less than genetic background control flies. C1) The sleep plot in LD for control w1118, heterozygous flies, as well as homozygous sNPFc00448 flies from one experiment is shown. The heterozygous flies were F1 progeny from w1118 (the genetic background line) crossed to sNPFc00448. C2) Quantitative analysis shows that reduction of sNPF led to less total sleep and decreased mean bout duration in both daytime and nighttime. The effect is dose dependent, i.e. the heterozygous flies slept less than the control flies but more than the homozygous flies. D. Transient knockdown of sNPF in the sNPF expressing neurons led to nighttime sleep loss. D1) A tubGAL80ts transgene was used to block the expression of UAS-sNPFRNAi in the sNPF-GAL4 neurons at 21°C. GAL80 protein was inactivated at 30°C, allowing the expression of UAS-sNPFRNAi driven by sNPF-GAL4. Transient knockdown of sNPF induced by heat led to significant decrease of total as well as nighttime sleep (purple arrows). The sleep loss was rapidly reversible once the GAL80 protein was reactivated at 21°C. D2) Quantitative data for the heat-induced sleep loss. The calculation for heat induced sleep changes is described in EXPERIMENTAL PROCEDURES. Data are presented as mean ± SEM; *p < 0.01, **p < 0.001, and ***p < 0.0001 are significant difference from control group (ANOVA with Tukey post hoc test, described in EXPERIMENTAL PROCEDURES). See also Figure S3.
Given the potency with which the sNPF-expressing neurons promote sleep, their activity should be regulated if they are part of normal sleep-promoting circuits. Since GABAergic neurons can function in wake-promotion by GABAA-mediated suppression of sleep-promoting regions (Y. Shang and M. Rosbash, unpublished observations; P. Haynes and L. Griffith, unpublished observations, Hassani et al., 2009), we knocked down the Rdl GABAA receptor in sNPF neurons. This indeed led to significant increases in both daytime and total sleep time as well as to longer sleep bouts, suggesting that sNPF neuron activation is normally attenuated by GABAA signaling (Figure 2B).
To determine if the sleep-promoting function of these neurons is due to the release of sNPF itself, we examined sNPFc00448 mutant flies, which have a more than 50% reduction of sNPF mRNA (Lee et al., 2008). These flies showed significant reductions of both daytime and nighttime sleep (Figure 2C). Sleep was also fragmented, as the mean duration of sleep bouts was reduced by more than 50% (Figure 2C2). Sleep was sensitive to gene dosage, as heterozygous flies had intermediate phenotypes compared to control and homozygous flies (Figure 2C), although statistical significance for the gene dosage effect was only reached in the case of nighttime sleep. Sleep was also assayed in animals with an adult-specific knockdown of sNPF mRNA in adult brains. Both total sleep and nighttime sleep were significantly reduced (Figure 2D). Daytime sleep was also more fragmented, although the amount of daytime sleep was unaffected (Figure 2D2). While the dTrpA1 activation strategy cannot rule out a role for a cotransmitter, these two independent methods of reducing sNPF levels also support a sleep-promoting function for the sNPF peptide in the adult fly.
Activation of sNPF neurons suppresses sleep homeostasis during mechanical deprivation
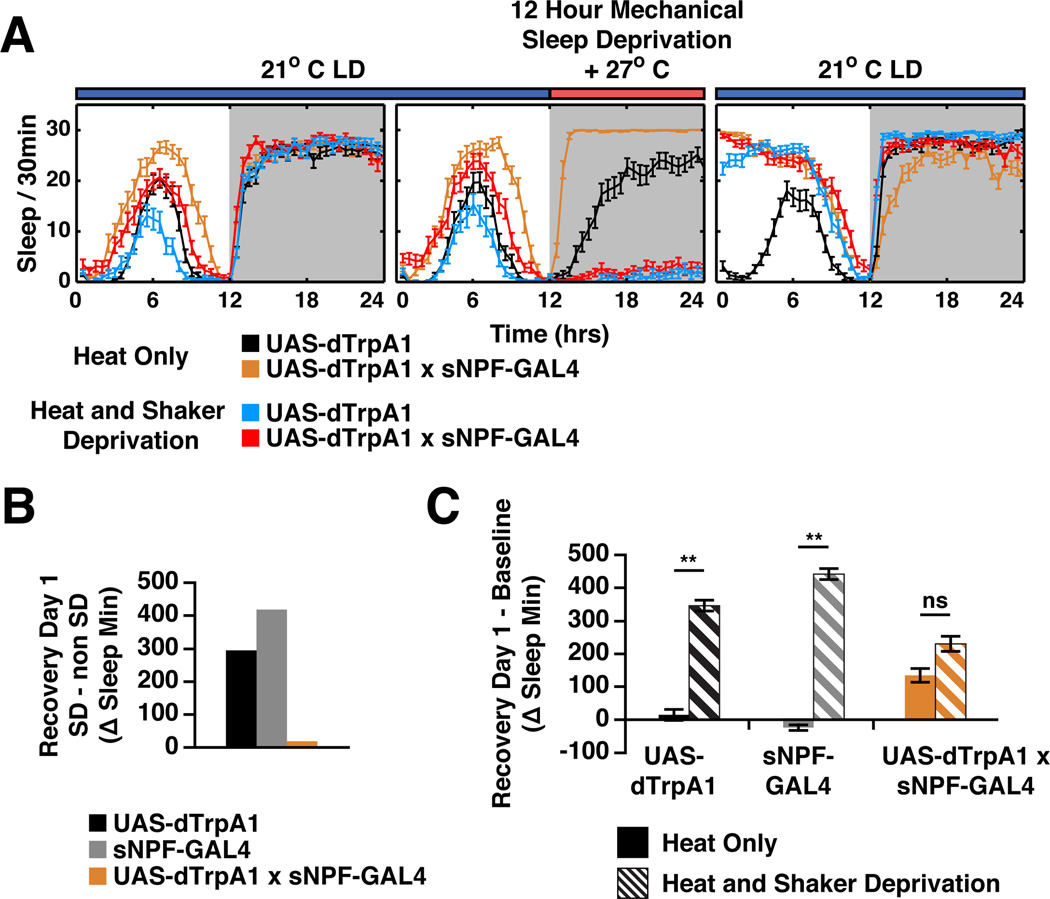
To test the idea that sNPF neuron activation might affect the sleep homeostat and therefore interfere with the effects of mechanical sleep deprivation (SD), we activated these neurons during traditional mechanical SD. The SD protocol was standard, with the exception that sNPFGAL4:dTRPA1 and control strains were heated to 27°C during the 12 h of deprivation to activate sNPF neurons and then returned to 21°C after the deprivation, at the end of the night (Figure 3). During the mechanical SD, the sNPFGAL4:dTRPA1 strain appeared indistinguishable from the control strains, i.e., it manifested no sleep during these 12 h as expected. This reflects true locomotor arousal, since DAM data from unconscious or dead flies can be distinguished from live moving animals (Figure S2). Remarkably however, this strain produced much less sleep rebound or recovery sleep after the deprivation than the control strains upon the return to 21°C. This can be seen both in comparisons of recovery day sleep levels of SD flies to non-SD flies (Figure 3B) and by comparison of the sleep of SD flies on the recovery day to their own pre-SD day sleep levels (Figure 3C). We conclude that activation of sNPF neurons during mechanical SD caused at least a partial sleep-like state, which was invisible with standard locomotor activity monitoring but could be interpreted by the sleep homeostat as genuine sleep. Another possibility is that sNPF provides a direct signal to the homeostat to indicate that sleep has occurred.
Figure 3. sNPF regulates the response to sleep deprivation.
A. Sleep plots of control and sNPF-GAL4 driven dTRPA1. Animals were maintained at 21°C and baseline sleep recorded. At ZT12 of the sleep deprivation (SD) day, temperature was increased to 27°C for 12 h ± mechanical SD. Flies were returned to 21°C to allow recovery sleep to occur. Animals with activated sNPF neurons that did not receive mechanical SD showed increased sleep, while controls showed a temperature-dependent decrease in sleep during the heat treatment. B. Amount of recovery sleep in SD flies was quantitated by comparison to sleep of siblings that were heated but did not receive SD. sNPF neuron activation significantly reduced recovery sleep. C. Recovery sleep was quantitated by comparison of each group (heat only, no SD and heat +SD) to its previous day’s sleep. Flies with activated sNPF neurons slept more than control flies even without SD, perhaps due to sleep inertia. Recovery sleep in SD flies was significantly different in flies with activated sNPF neurons (**p < 0.001, ANOVA, posthoc Tukey test shows only the UAS and GAL4 control heat alone are not significantly different from each other). Pairwise comparisons were all significantly different (p < 0.0001 for UAS and GAL4 controls, p = 0.0019 for the experimental cross). N ≥ 36 for each genotype and condition. See also Figure S2.
sNPF promotes nighttime sleep through the s-LNv-to-l-LNv circuit
sNPFGAL4 drives strong expression in many brain regions, including the MB, the PI and the FSB in the CC (Johard et al., 2008). We used multiple strategies to test the involvement of the MB, as it is a previously identified sleep-promoting region (Joiner et al., 2006; Pitman et al., 2006) Addition of a MB-GAL80 transgene blocked the expression of dTRPA1 in the MB without affecting the sleep promoting effect of sNPFGAL4:dTRPA1 flies (Figure S3B, E). The converse experiment, activation of just the MB-specific subset of sNPF-GAL4 cells using an intersectional strategy (Shang et al., 2008), also did not produce a sleep phenotype (Figure S3E). Although the PI has also been shown to be involved in sleep (Crocker et al., 2010; Foltenyi et al., 2007), direct activation of subsets of the PI did not significantly increase total sleep in a state-independent manner (Figure 1A: DILP2-, CRZ-, SIFa- and DMS-GAL4). Additionally, subdivision of the sNPF-GAL4 pattern with different GAL80s demonstrated no correlation of the sleep phenotype with PI or MB expression (Figure S3C–E). Therefore, most sleep-promoting sNPF neurons likely reside outside these two regions.
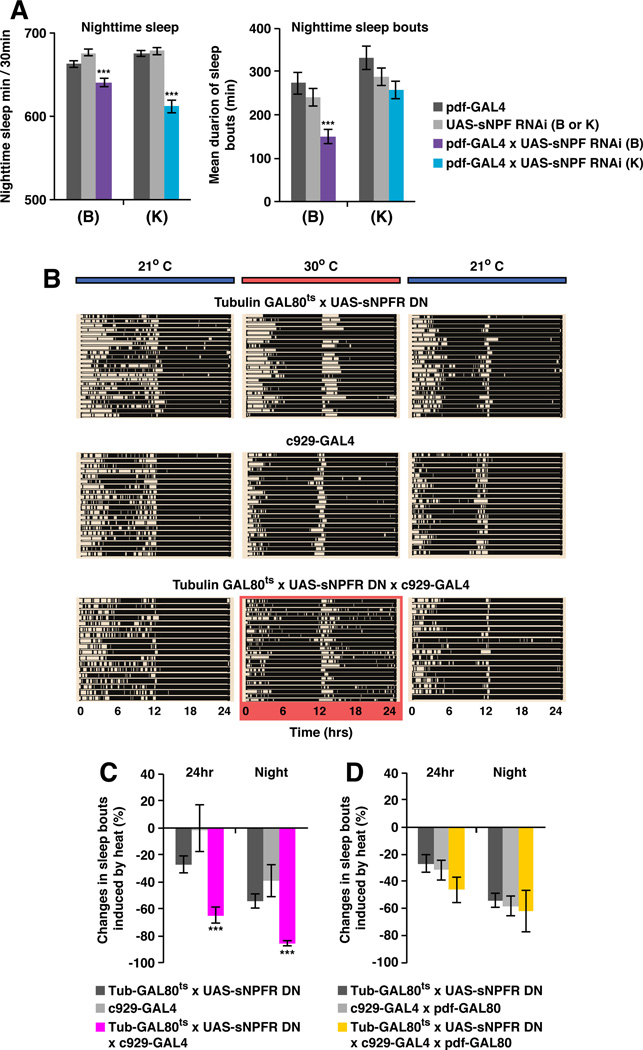
We then assayed PDF-expressing clock neurons. The 8 PDF+ small ventral lateral neurons (s-LNvs) are labeled by sNPF-GAL4 and have recently been shown to express sNPF (Johard et al., 2009). Moreover, sNPF mRNA is one of the most strongly cycling transcripts in the s-LNvs (Kula-Eversole et al., 2010). As the 10 neighboring cells, the PDF+ wake-promoting large ventral lateral neurons (l-LNvs), express little or no sNPF (Johard et al., 2009; Kula-Eversole et al., 2010), RNAi knockdown of sNPF using pdf-GAL4 should be specific for s-LNvs. Two independently generated RNAi lines against sNPF produced small but significant decreases in nighttime sleep without affecting daytime sleep (Figure 4A). Nighttime sleep bout length was also ~10%−50% shorter than control strains (Figure 4A). Therefore, sNPF within s-LNvs promotes normal nighttime sleep.
Figure 4. sNPF promotes nighttime sleep through the s-LNv-to-l-LNv circuit.
A. pdf-GAL4-driven knockdown of sNPF using two independently generated RNAi lines in s-LNvs led to sleep suppression at night. pdf-GAL4 only drives UAS expression in the l-LNvs and s-LNvs (Renn et al., 1999). Since the l-LNvs do not express sNPF (Johard et al., 2009; Kula-Eversole et al., 2010), the knockdown of sNPF mRNA using pdf-GAL4 should be specific for s-LNvs. More than 60 flies were tested in 3 trials. Nighttime sleep for the control and experimental lines are shown (left panel). Both RNAi led to significant decrease of sleep time at night. The mean duration for sleep episode at night for each genotype is shown (right panel). UAS-sNPF-RNAi (B) also affected sleep consolidation at night. B. Transient expression of UAS-sNPFR-DN in the c929+ cells caused sleep fragmentation. Raster plots of the sleep-wake pattern of individual flies in LD are shown for each genotype. Each row represents a single fly. Black bars are sleep episodes and white bars are wake episodes. A tubGAL80ts transgene was used to block the expression of UAS-sNPFR-DN in the c929-GAL4 neurons at 21°C. The tubulin-GAL80ts, UAS-sNPFRDN;c929-GAL4 flies showed sleep pattern similar to control genotypes (top two blocks of rasters). Expression of UAS-sNPFR-DN in the c929-GAL4 neurons was induced by shifting the temperature to 30°C (highlighted in red). Heat did not induce detectable changes in control strains, while severe sleep fragmentation was observed in the experimental flies. The fragmentation phenotype was rapidly reversed once the GAL80 protein was reactivated by reducing the temperature to 21°C. C. Quantitative analysis shows that the mean duration of 24 h sleep as well as nighttime sleep episodes were significantly reduced in the tubulin-GAL80ts, UAS-sNPFRDN;c929-GAL4 flies at 30°C. D. Defects in sleep consolidation are due to the reduction of sNPFR signaling in the l-LNvs. The sleep fragmentation phenotype was rescued by a pdf-GAL80 transgene, which blocks the expression of UAS-sNPFR-DN in the l-LNvs. Data are presented as mean ± SEM; *p < 0.01, **p < 0.001, and ***p < 0.0001 are significant difference from control group (ANOVA with Tukey post hoc test, described in EXPERIMENTAL PROCEDURES). See also Figure S3.
The wake-promoting l-LNvs express sNPFR (Kula-Eversole et al., 2010; Nitabach and Taghert, 2008). Because c929GAL4 expresses in the l-LNvs (and in multiple other peptidergic cells) but not in the s-LNvs (Shang et al., 2008), we used c929GAL4, tubulin-GAL80ts and UAS-sNPFR-DN to downregulate sNPF signaling (Lee et al., 2008) in adult cells. tubulin-GAL80ts; c929GAL4:UAS-DN-sNPFR flies had no detectable change in total sleep time or mean duration of sleep episodes at 21 °C. After shifting to 30°C for 3 days however, flies exhibited notable nighttime sleep fragmentation compared with parental controls (Figure 4B, for quantitative data, see Figure 4C). The effect on sleep consolidation was fully reversible after shifting back to 21°C (Figure 4B). Remarkably, addition of the pdf-GAL80 transgene to flies carrying c929GAL4 and UAS-sNPFR-DN strongly blocked the effects of UAS-sNPFR-DN (Figure 4D), indicating that most if not all of the sNPF effect on sleep from the diverse peptidergic neurons labeled by the c929GAL4 driver is due to the l-LNvs.
sNPF neuromodulation is predominantly inhibitory
We used functional imaging to address the cellular mechanisms by which sNPF affects neuronal function. Flies carrying pdf-GAL4:UAS-EPAC express the FRET-based cAMP reporter in both l-LNvs and s-LNvs (Shang et al., 2011). Because we previously showed that the l-LNvs receive synaptic inputs from dopaminergic neurons, with bath application of 100 µM DA evoking a dramatic increase in cAMP (Shang et al., 2011), we used subsaturating concentrations of DA and co-application sNPF. 20 µM DA with 20 or 80 µM sNPF suppressed cAMP responses in the l-LNvs compared with application of 20 µM DA alone (Figure S4A). Although these data suggest that the balance between sNPF and DA signaling in the l-LNvs affects nighttime sleep consolidation, the effect of sNPF did not always reach statistical significance. Moreover, sNPF alone did not alter FRET (Figure S4B).
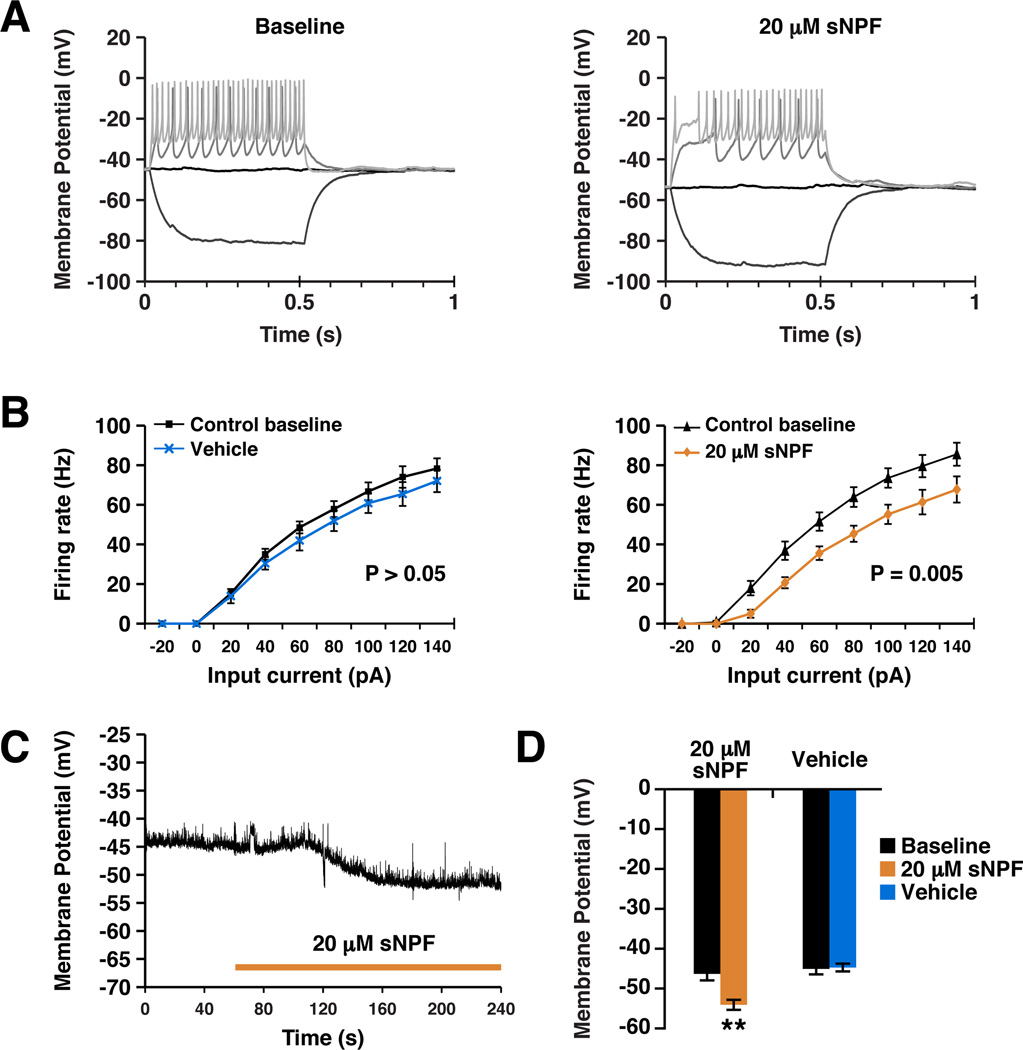
To address the mechanism by which sNPF regulates neuronal function in a more general way, we assayed the electrophysiological effects of the sNPF via its receptor, sNPFR in larval central neurons. The OK371-GAL4 driver was combined with UAS-sNPFR to ectopically express sNPFR in larval motor neurons. Perfusion of 20 µM sNPF reduced the firing response to current injection (Figure 5A; ANOVA, F(1,15) = 10.504, p = 0.005). The shift in the input-output function of the neuron was associated with a significant (p = 0.002) hyperpolarization in resting membrane potential, typically occurring within 1–2 minutes of treatment onset (Figure 5C). Vehicle had no effect (Figure 5B, 5D) and effects were completely dependent on expression of the sNPFR (Vecsey et al., submitted). Taken together with the strong sleep-promoting effect of sNPF firing, hyperpolarization and inhibition of firing may be the general response to sNPF. This is consistent with the fact that NPY, the mammalian analog of sNPF, is known to be primarily inhibitory (van den Pol, 2012).
Figure 5. sNPF reduces neuronal resting membrane potential and suppresses action potential generation.
A. Example current clamp recordings made from third instar larval motor neurons expressing sNPFR before (left panel) and after (right panel) treatment with sNPF. Current was injected to depolarize the neuron and elicit action potentials. Only selected sweeps of the current injection protocol are shown for clarity. B. Quantification of current clamp data, plotting firing rate as a function of input current (F/I curve). Vehicle treatment (N = 6) in flies expressing sNPFR did not cause a significant change in F/I curve (left panel). sNPF (N = 11) caused a significant rightward shift in the curve, showing that more current was required to elicit the same spike rate after sNPF treatment (right panel). Data are plotted as mean ± SEM and significance calculated by one-way ANOVA. C. An example trace showing a typical hyperpolarization response to sNPF treatment. Duration of treatment is indicated by bar. D. Quantification of the effects of sNPF and vehicle treatment on resting membrane potential. Numbers in parentheses represent the number of animals in each condition. Data are presented as mean ± SEM. ** represents p <0.001, t-test. See also Figures S4 and S5.
sNPF has different effects on sleep and feeding circuits
The inhibitory nature of sNPF action in non-feeding neurons contrasts sharply with its published role in feeding pathways. For example, sNPF enhances the responsiveness of olfactory receptor neurons (ORNs), which promotes foraging (Root et al., 2011), and sNPF has been shown to directly activate cyclase in the neuronal BG2-c6 Drosophila cell line (Hong et al., 2012). We therefore examined the effect of sNPF on DILP cells, which are reported to respond to octopamine (OA) and have functions in feeding as well as sleep (Crocker et al., 2010; Lee et al., 2008). Although sNPF alone did not evoke detectable changes in FRET (data not shown), co-application of 20 or 80 µM sNPF with subsaturating concentrations of OA (10 µM) evoked large increases of cAMP (Figure S5A and B), consistent with the excitatory effects of sNPF on the ORNs (Root et al., 2011). This is a direct effect since it was not blocked by TTX (Figure S5C).
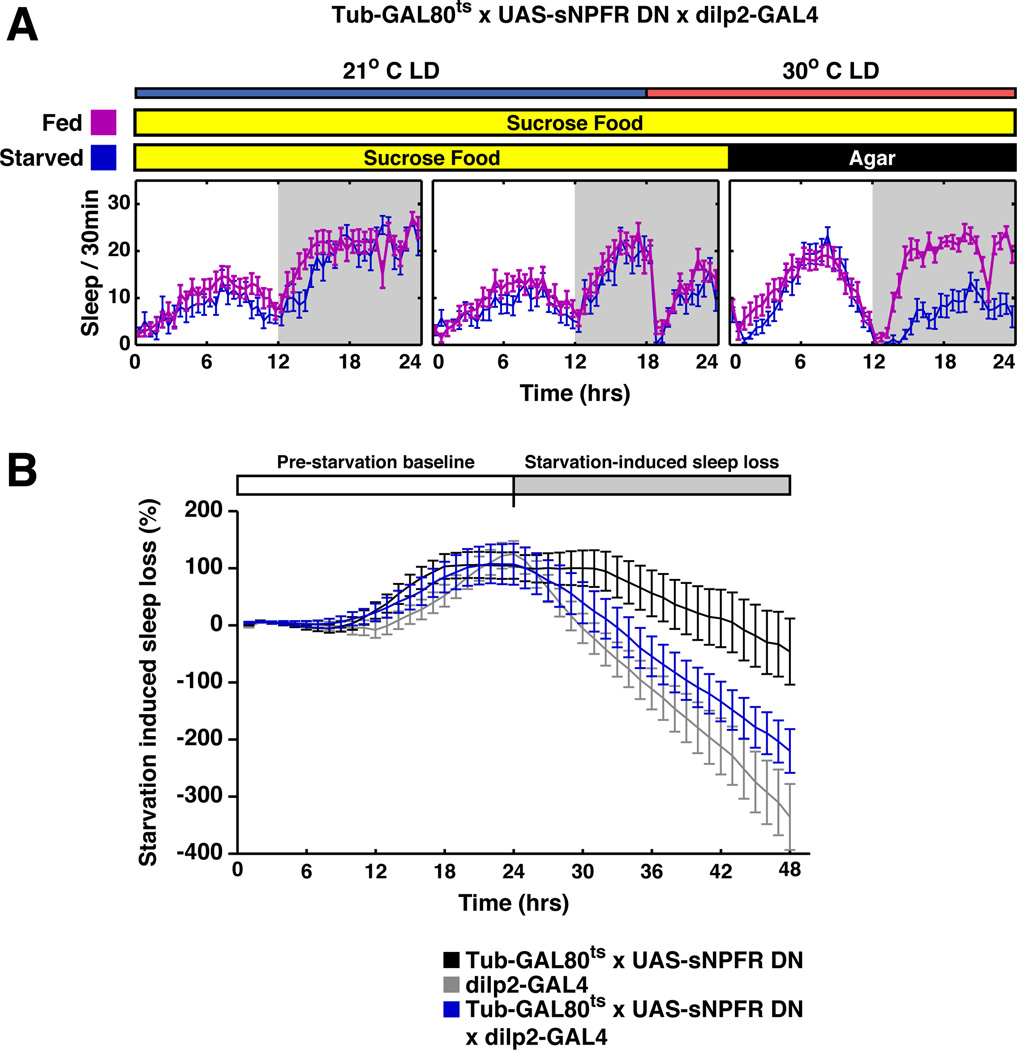
However, transient down regulation of sNPFR signaling in DILP cells did not affect sleep under starvation conditions, i.e., sleep was inhibited indistinguishably from control strains (Figure 6). This is despite the fact that dilp2-GAL4;UAS-sNPFR-DN flies show defects in metabolism and growth (Lee et al., 2008). We therefore suggest that the modest effect of DILP cell activation with dTRPA1 on sleep under LD conditions (Figure 1A) and the imaging results (Figure S5) may reflect a role of DILP cells in metabolism rather than a direct modulation of sleep circuitry (c.f. Erion et al., 2012).
Figure 6. Inhibition of sNPFR signaling in DILP cells does not affect starvation-induced sleep suppression.
A. Starved tubulin-GAL80ts, UAS-sNPFRDN;dilp2-GAL4 flies (blue curve) slept less compared to non-starved siblings (purple curve). GAL80 was inactivated for 6 h by shifting the temperature to 30°C at ZT18 and flies transferred onto agar or agar/sucrose food at ZT0. Sleep was monitored for 24 h. Starvation reduced sleep, indicating that sNPF signaling is not involved in starvation-induced wakefulness. B. The experimental flies (blue line) and the parental controls (black and gray lines) showed similar sleep loss in response to starvation. The baseline sleep for each genotype was collected for 3 days before heat inactivation of GAL80. The starvation induced sleep loss (%) = (Sleep during the starvation period – Baseline sleep)/Baseline sleep %.
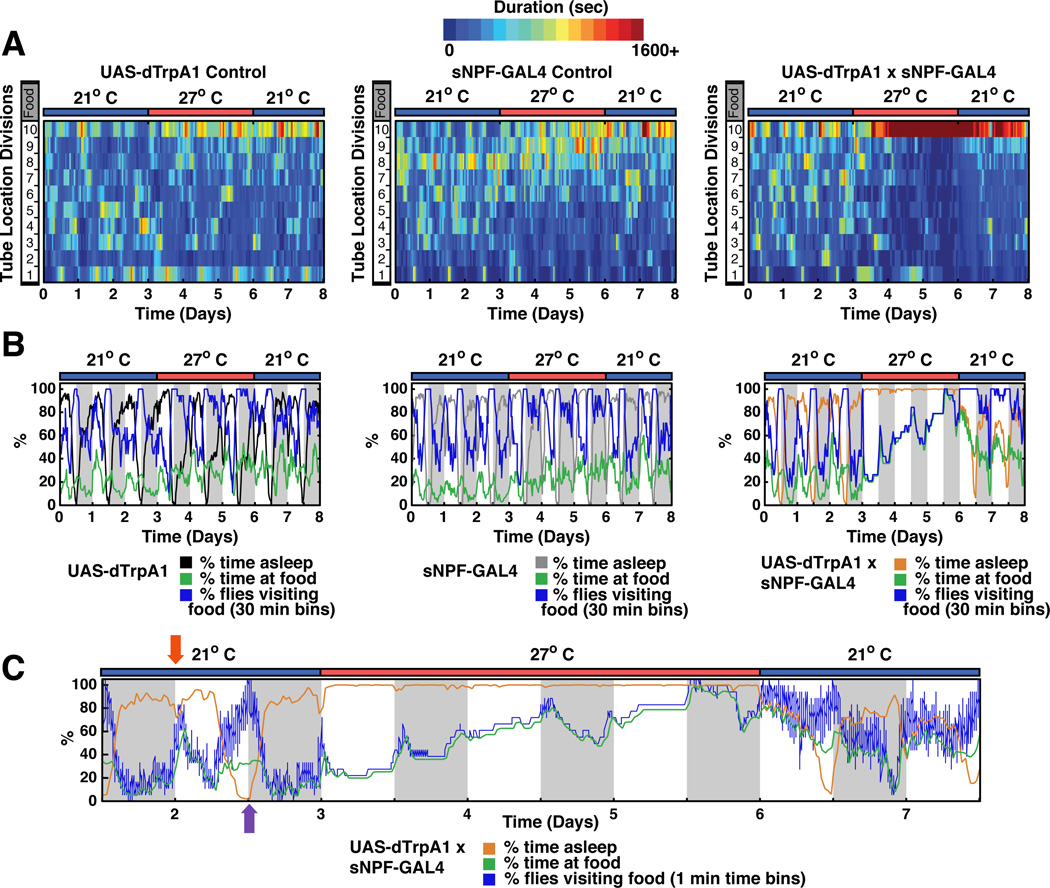
To further address the role of sNPF in fly feeding, we assayed the location of flies within behavior tubes subsequent to dTRPA1-mediated activation of sNPF cells. The temperature shift caused the flies to spend more time at the end of the tube containing food (Figure 7). Notably however, the onset of the location change or “food dwelling" was dramatically delayed from the onset of sleep by almost 12 h (compare orange and green lines in Figure 7C). This was due to a very slow accumulation of flies at the food (Figure 7C). Moreover, a temperature downshift led to rapid awakening, whereas food dwelling persisted for at least 2 days after dTRPA1 heat activation had ceased. In contrast to the slow effects on food dwelling, activation of dopaminergic neurons (THGAL4:UAS-dTRPA1) led to an immediate onset of food dwelling (Figure S7).
Figure 7. Activation of sNPF neurons alters sleep and food preference on different time scales.
A. Sleep, locomotor activity and position relative to food were monitored by computer video tracking (Donelson et al., 2012). In location heat plots, dark blue indicates flies spent no time at a particular location while dark red indicates flies spent more than 1,600 s at that location. The x-axis time and temperature and the y-axis indicates the location of each genotype within the behavioral tubes relative to food (location 10). Both parental control lines only showed slight increases in the time they spent on food upon heating, while the sNPFGAL4:UAS-dTRPA1 flies spent significantly more time on the food after heat activation. Sleep plots and sleep parameters for this data set are shown in Figure S6. B. Plots of % time asleep, % time at food and % flies visiting food for experiments in panel A. Control lines show a modest change in food dwelling with increased temperature. C. Expanded time scale for experimental fly data in panel B. During the morning peak of activity at 21°C when animals go to food they stay there (red arrow; blue line and green line overlap). During the evening peak of activity flies visit food often but do not stay. This likely reflects “patrolling behavior” (purple arrow; blue line higher than green line). Heat-induced acute firing of sNPF-expressing neurons dramatically and rapidly (within 1 h) increased sleep to maximal levels (orange line). The excessive sleep reversed within 1 h after inactivation by shifting temperature to 21°C. In contrast to the rapid effect on sleep, stimulation of sNPF expressing neurons caused a very slow accumulation of flies at the food which was not rapidly reversible. Flies remained closer to the food for at least 2 days after dTRPA1 N ≥ 17 for each genotype. See also Figure S7.
A predominant effect of sNPF neuronal activation on sleep rather than feeding was also observed in groups of flies housed in vials (Movie S1). At high temperature almost all of these flies avoided food and stayed at the top half of the vials, consistent with previous observations that sleep occurs preferentially away from food (Donelson et al., 2012). Parental control flies in contrast were frequently observed at the bottom of the vials, either near or on the food (movie S1).
Importantly, the total % time of the population at the food in the behavior tubes (green line) matched exactly the % of flies that have visited the food (blue line) during the period when sNPF neurons are active (Figure 7C). This indicates that the low level of locomotor activity of these flies (Figure S1) is used predominantly to go to the food, where they remain. The most parsimonious interpretation is that hypersomnolent flies are unable to feed properly and eventually become hungry or even starved, resulting in an increased drive to find food. This suggests that the observed changes in food-related behavior may be predominantly a result of the dramatic increase in sleep by sNPF.
DISCUSSION
We have presented several independent lines of evidence indicating that sNPF acutely increases sleep and alters sleep homeostasis. This is because release of animals from sNPF neuron activation after several days of hypersomnolence resulted in a transient decrease in sleep or negative sleep rebound. Moreover, activation of sNPF neurons during mechanical sleep deprivation blunted the rebound sleep following the deprivation. This suggests that sNPF might alter the internal perception of sleep state during the deprivation despite an apparently behaviorally awake state. It also suggests that sNPF might directly modulate the sleep homeostat.
The most potent in vivo manipulations of sNPF function, mutation of the sNPF gene and strong activation of sNPF neurons with dTRPA1, affect daytime as well as nighttime sleep levels. These manipulations also strongly alter sleep bout duration, a measure of consolidation, in the opposite direction to the sleep duration effects. More limited manipulations of sNPF signaling (cell-specific down regulation of sNPF levels or of sNPF signaling) indicate that sNPF is most important for promoting sleep at night. It also affects the structure of daytime sleep, a function of sNPF circuitry normally suppressed during the day by wake-promoting GABAergic neurons, acting via GABAA receptors. Suppression of excitability with Kir2.1 likely mimics this daytime GABAergic function. These results in aggregate suggest that sNPF action differs depending on the time of day, a result that supports the idea that daytime and nighttime sleep might be regulated by different circuitry.
The role of sNPF in promoting more consolidated sleep is consistent with a general antiarousal function. As in mammals, Drosophila arousal can be measured electrophysiologically (van Alphen et al., 2013; van Swinderen and Andretic, 2003), but the most straightforward measure of arousal state is behavioral, and sleep fragmentation is indicative of a less stable, more easily aroused state. The main neurochemical previously implicated in fly arousal is DA (Andretic and Shaw, 2005), and l-LNvs play a prominent role in the arousal circuitry (Lebestky et al., 2009; Parisky et al., 2008; Shang et al., 2008; Sheeba et al., 2008).
Although the imaging assays indicated that sNPF alone did not lead to significant cAMP changes in the l-LNvs, it mildly suppressed the activation effect of DA on the l-LNvs (Figure S4). As one subset of clock neurons in the sleep circuit releases sNPF and promotes sleep at night and an adjacent subset responds to sNPF and suppresses nighttime sleep, sNPF may be used by the s-LNv-to-l-LNv pathway to coordinate the timing of sleep with other circadian behaviors. Indeed, sNPF mRNA is a potent cycling mRNA in s-LNvs (Kula-Eversole et al., 2010). Importantly, the electrophysiological assays in larval central neurons suggest that inhibition of neuronal firing may be a general feature of sNPF function and relevant to other sleep centers in addition to the clock neurons.
sNPF and other sleep-relevant neuromodulators like DA are likely to act at multiple sites in the brain given the major state change effected by the sleep/wake transition. This expectation also reflects the modest effects of sNPFR manipulation within l-LNvs on total sleep time. Moreover, fan-shaped body neurons have recently been shown to be important for DA-mediated arousal (Liu et al., 2012; Ueno et al., 2012). The ability of these neuromodulators to act on many circuits may allow for more flexible integration of sleep with other behaviors and with other external and internal factors.
An important influence on sleep is metabolic state (Penev, 2007). Indeed, sNPF facilitated the OA-to-DILP circuit, which may reflect its role in sleep/wake, feeding and/or metabolic regulation (Figure S5). However, the wake-promoting effect of activating the DILP pathway is context-dependent, occurring only in LD (Figure 1A). Moreover, acute activation of octopaminergic neurons by dTRPA1 only mildly affects sleep and also in a condition-dependent manner (data not shown), and feeding animals with octopamine only significantly suppresses total sleep after 2–3 days of exposure (Crocker and Sehgal, 2008). Although long-term activation of octopaminergic neurons leads to long-lasting increases in food dwelling (N. C. Donelson and L.C. Griffith, unpublished results), these effects contrast sharply with the rapid and conditionindependent effects seen with acute increases in dopamine signaling (Shang et al., 2011). Dopaminergic neurons have also been shown to be a critical part of NPF-regulated changes in satiety and response to food (Inagaki et al., 2012; Krashes et al., 2009), and activation of these neurons indeed led to an immediate onset of food dwelling, which reversed rapidly upon dTRPA1 inactivation (Figure S7). As expected, tracker analysis shows that these food-dwelling flies also sleep very little, indicating that dopamine affects both sleep and feeding rapidly. These effects contrast with the slow effects on food dwelling by sNPF neuronal activation.
The simplest interpretation of this slow food-dwelling response is that it is secondary to a more primary effect of sNPF on sleep. Indeed, a slow buildup in hunger or even starvation as a consequence of too much sleep is a simple explanation consistent with most if not all of our data. Behavioral effects as a secondary consequence of some other more direct effect is also our interpretation of many of the sleep effects of activation of peptidergic neurons shown in Figure 1, in which only sNPF robustly increased sleep, i.e., under both LD and DD conditions. We therefore suggest that a necessary condition for serious consideration of a molecule as behaviorrelevant is a rapid response, which is also relatively condition-independent. Dopamine as a wake-promoting molecule and now sNPF as a sleep-promoting molecule meet these criteria.
EXPERIMENTAL PROCEDURES
Fly Stocks
Flies were raised on standard medium and 12 h light:dark cycles (we used fluorescent light and the light intensity was 1600 ± 400 lux). Flies carrying UAS-dTRPA1 or tubGAL80ts were raised at 21°C.
The UAS-dTRPA1, UAS-rdlRNAi, and UAS-Epac1-cAMP (50A, II) flies were kindly provided by Drs. Paul Garrity (Brandeis University), Ron Davis (Scripps Florida), and Paul Taghert (Washington University). UAS-sNPFR-DN (X) and UAS-sNPFRNAi (K) were provided by Dr. Kweon Yu. The RNAi line was validated using immunoblotting (Lee et al., 2004) and immunohistochemistry (Kahsai et al., 2010). The dominant negative receptor competes with sNPFR for G protein binding and was functionally validated in multiple assays (Lee et al., 2008). UAS-sNPFRNAi (B), UAS-kir2.1, and tubGAL80ts (X) were ordered from Bloomington stock center. Dr. Paul Taghert also provided DMSGAL4, SIFaGAL4, CCAPGAL4, CRZGAL4, and huginGAL4 used in Figure 1. NPFGAL4 and NPFRGAL4 in Figure 1 were obtained from Dr. Ping Shen (University of Georgia). sNPFGAL4 flies (NP6301; order number 113901) were ordered from the Drosophila Genetic Resource Center (DGRC), Kyoto Institute of Technology, Kyoto, Japan. dilp2GAL4 were kindly provided by Dr. Ulrike Heberlein (Janelia Farm). pdf-GAL4/CyO or dilp2GAL4 flies were used to express the UAS transgenes in the PDF- expressing l-LNvs or DILP cells in fly brains, respectively.
Behavioral analysis
Individual flies were housed separately in 65 mm × 5 mm glass tubes (Trikinetics, Waltham, MA) containing 5% agarose with 2% sucrose. 2–5 day old flies were collected and entrained under standard light-dark conditions, with a 12hr light phase and followed by 12hr dark phase for 3–4 days. We first entrained adult flies at 21°C for 3–4 days. We then activated dTRPA1 or inactivated the GAL80 proteins by shifting the temperature to 27°C or 30°C for 3 days. This will either activate the neurons expressing the dTRPA1 or turn on the expression of UAS transgenes. Finally, we inactivate the dTRPA1 or reactivated the GAL80 proteins by shifting the temperature back to 21°C to test if the effects are reversible. For Figure 1B, the lights were turned off permanently upon the heat activation. The temperature was then returned to 21°C in DD to inactivate the dTRPA1 channel.
The sleep-like resting state is defined as no movement for 5 min (Hendricks et al., 2000; Shaw et al., 2000). Total sleep measures the amount of sleep per 24 h and the mean duration measures the average length of sleep episodes (Agosto et al., 2008). The behavioral pattern of each fly was monitored either by an automated method (DAM System, Trikinetics) or by a video-based tracking application (Donelson et al., 2012). The latter recorded the exact location of each fly every second for the 8 days of the experiment. Using this more direct method to record activity, we were able to gain a higher data resolution as well as analyze the preference for food location shown in Figures 7, S6 and S7. While DAM records beam breaks, the tracking system used in this experiment was able to detect movements of as little as 1.5 mm. In short, 3–5 day old female flies were loaded into the same sleep-tubes used for DAM. The tubes were capped with parafilm on both ends to prevent visual obstruction and were placed onto a piece of white paper, which afforded a high visual contrast to the fly. The flies were then placed under a video camera (Logitech Quickcam for Notebooks) connected to the computer running the tracking software. A red compact fluorescent light allowed for continuous recording during the flies 12 h dark period. The tracker data was transformed from coordinate data into DAM-style 1 and 30 min data files and analyzed as described previously. For sleep deprivation studies, DAM monitors were mounted on a Trikinetics plate attached to a VWR vortexer and shaken for 2 out of every 10 sec for 12 h.
Calculation of the relative sleep changes and statistical analysis
Sleep time as well as the effect of heat on sleep is highly sensitive to genotype. We therefore needed to subtract the heat induced changes occurring in the parental controls. We first calculated the heat induced percentage change in sleep (SI) for each genotype, which is SI = (sleep time 30°C – sleep time 21°C) / sleep time 21°C% (Figure 2A, 2D, 4C–D). To simplify the data presentation, we then calculated the relative sleep change (ΔSI), which is ΔSI = SIexp – SIctrl (Figure 1A). The SI of the experimental group was compared with the 2 control parental groups using the ANOVA with Tukey post hoc test. *p < 0.01, **p < 0.001, and ***p < 0.0001 are significant difference from both control groups. Error bar represents SEM.
Functional Imaging
For Figures S4 and S5A, live FRET imaging was performed as described in Shang et al 2011. Briefly, 3–6 day old entrained male flies were dissected in ice-cold adult hemolymph-like medium (AHL, Wang et al., 2003). 600 µl room temperature AHL was added to the imaging chamber. An individual brain was then placed in the chamber. We used an Olympus BX51WI microscope with a CCD camera (Hammamatsu Orca C472—80-12AG). The acquisition system for this set up allows for simultaneously recording both channels. The 86002v1 JP4 excitation filter (436, Chroma) as well as two-channel, simultaneous-imaging system from Optical Insights with the D480/30m and D535/40m emission filters were used. EPAC expressed in l-LNvs was excited with 50 ms pulses of light using CFP filters and fluorescent signals emitted by the l-LNvs or DILP cells were imaged every 5 s by an epifluorescent microscope using a 40X objective. The software Volocity (Perkin Elmer) was used for acquisition and the CFP and YFP images were recorded simultaneously. Under these conditions, we determined that the baseline fluorescent signal in l-LNvs stabilized after imaging the neurons for 150 frames. We were then able to obtain reliable responses induced by 10 µM forskolin (data not shown).
Octopamine and dopamine were purchased from Sigma and a stock solution (10 mM) was freshly prepared in H2O before the imaging (Cayre et al., 1999). sNPF was purchased from Polypeptide, San Diego, CA. 2mM stock solution in DMSO was vacuum dried and stored at −20°C. The baseline images were collected for 50 s before applying drugs to the brain. The mean intensity of CFP or YFP of a non-fluorescent brain region next to the l-LNvs or the DILP cells was first subtracted from that of l-LNvs or DILP cells. The YFP/CFP ratio for each time point was then calculated and normalized to the ratio of the first time point, before drug application. The relative cAMP changes were determined by plotting the normalized CFP/YFP ratio (%) over time. We also determined the average fluorescence change (area under the “relative cAMP change” curve) by calculating an average CFP/YFP ratio increase from 100 s to 200 s.
For Figure S5C, experiments were done in a different configuration with a different drug delivery method. This is the likely source of differences in OA effective dose and duration of effect. Optical signals from an Olympus BX51WI microscope were recorded using a back-illuminated CCD camera. A 45 ms exposure stimulated the FRET-based EPAC sensor, and CFP and YFP emissions were collected using a splitter (Photometrics). A 60x, 0.9 NA water immersion lens (Olympus LUMPlanFl) was used, and images were acquired at 1 Hz with the software Volocity (PerkinElmer, Inc.), with 4x binning. Filters used for cAMP imaging were: excitation 86002v1 JP4 filter (436, Chroma), and emission D480/30m and D535/40m (Optical Insights). Off-line data analysis was performed using ImageJ (U.S. National Institutes of Health) and Matlab (Mathworks). To limit bleaching, a 25% neutral density filter (Chroma) was used for all experiments, and brains were pre-exposed to 436 nm blue light twice, for 90 seconds followed by another 60 seconds, with a 5 second off period between. Imaging experiments were performed in male progeny from crosses of w;dilp-GAL4 x w;;UAS-EPAC-55A. After collection, flies were raised to 20–23 days old at 25°C on a 12:12 light/dark cycle with lights on at 9 am. All imaging was carried out during the light period. Each brain was dissected in ice-cold 0 mM Ca2+ Modified A solution containing (in mM) 118 NaCl, 2 NaOH, 2 KCl, 4 MgCl2, 22.3 sucrose, 5 trehalose, 5 HEPES, pH 7.15, mOsm 281, and was transferred to an RC-26 chamber on a P1 platform (Warner Instruments, LLC) and pinned in Sylgard (Dow Corning). The brain was then perfused with AHL by gravity feed at 3–4 mL/min. Switching between solutions was achieved using a three-way valve solenoid (Cole-Parmer) under manual control. All Recordings lasted 240 seconds, with 30 seconds of baseline before 60 seconds of drug treatment before washout with control AHL. TTX (Tocris) was stored as a 100µM stock at −20°C and was used at 1µM. TTX was added to all AHL solutions so that brains were TTX-treated throughout the light preexposure and the entire recording. Octopamine (Sigma) was made fresh daily to 50 mM in water, and was kept wrapped in foil on ice until dilution to 200 nM in AHL. sNPF was prepared as described above, and was used at 40 µM. The FRET signal (CFP/YFP ratio) for each time point was calculated and normalized to the ratio of the first baseline time point. The relative cAMP changes were determined by plotting this normalized CFP/YFP ratio (%) over time. Relative cAMP values were averaged from 90–150 seconds, and resulting response averages were compared between OA+TTX and OA+TTX+sNPF groups using a t-test assuming unequal variances.
Electrophysiology
Flies were raised at 25°C. To drive expression of transgenes in larval motor neurons, the OK371-GAL4 driver line was used (Bloomington stock 26160). The UAS-sNPFR line was generated in the Yu lab (Lee et al., 2008). sNPF-1 (H-AQRSPSLRLRF-NH2) was commercially synthesized (PolyPeptide). sNPF was stored as powder at room temperature, and as then dissolved in DMSO at 20 mM. Aliquots were desiccated using a SpeedVac (Savant) and were stored at −20C.
Third instar larvae were dissected and pinned in Sylgard (Dow Corning) in 0 mM Ca2+ Modified A solution (see above). The brain was removed and pinned, and the preparation was perfused with adult hemolymph (AHL), containing (in mM) 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4, 5 trehalose, 10 sucrose, 5 HEPES, pH 7.5, mOsm 265 (based on Wang et al. (2003)). Protease XIV (Sigma-Aldrich) treatment (0.5–1% w/v) was used to dissolve the glial sheath, allowing access to the motor neurons. Patch electrodes were (in mm) 1.2 OD × 0.9 ID × 100 L (Friedrick & Dimmock, Inc.), and were pulled and fire-polished to achieve a resistance of 3–7 MQ. Pipettes were loaded with internal solution as per Choi et al (2004), containing (in mM) 20 KCl, 0.1 CaCl2, 2 MgCl2, 1.1 EGTA, 120 K-Gluconate, 10 HEPES, pH 7.2, mOsm 280. Signals were acquired using the Axopatch 200B amplifier and Clampex (Molecular Devices). Current-clamp pulses were 500 ms in duration, and stepped in 20 pA increments from −20 to +140 pA, with a 10-second inter-pulse interval. sNPF was bath-applied at 20 µM, and current-clamp recordings and effects on resting membrane potential were carried out after 5 mLs of treatment.
Statistical analysis was carried out using JMP, Version 7 (SAS Institute, Inc.) and SPSS (IBM), Version 19. The change in resting membrane potential from baseline to post-treatment was calculated for each recording, and this value was compared between sNPF and vehicle treatments using a t-test assuming unequal variances. A one-way repeated-measures ANOVA was used to analyze the change in firing rate following drug treatment, with treatment as a between-subject factor and input current as a within-subject factor. No significant interaction between drug treatment and input current was observed, but drug treatment caused a significant overall effect on firing rate across all input currents.
Immunocytochemistry
The protocol is described in (Shang et al., 2008). Briefly, fly heads were fixed in 4% paraformaldehyde/0.008% PBS-Triton X100 for 1hr at 4°C. Paraformaldehyde fixed samples were washed for 1hr in 0.1% PBS- TritonX100 at RT and then dissected in PBS. Fixed brains were washed twice, 10 min each, in 0.5% PBS-Triton X100 at RT and then blocked in 10% goat serum with 0.5% PBS-TritonX100 for 1hr at RT. Brains were incubated with primary antibody (anti-GFP) at 4°C overnight then washed 3 times and incubated in secondary antibodies (Molecular Probes) at 1:500 dilution for 1hr at RT. Brains were washed 3 times and resuspended in mounting solution (Vectashield, Vector Laboratories, Inc.). Brain samples were visualized by a Leica TCS SP2 confocal microscope.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Paul Garrity, Ron Davis, Paul Taghert, Kweon Yu, Ping Shen, and Ulrike Heberlein for kindly providing transgenic flies. We also obtained fly lines from Bloomington Stock Center and DGRC. We thank E. Dougherty for assistance in confocal microscopy and Kristyna Palm and Patricia Parmenter for administrative assistance. The work was supported by grants from the NIH: R01 MH067284 to LCG and P01 NS044232-06 to MR. NCD was supported by post-doctoral training grant T32 NS007292 and CGV was supported by F32 MH090711.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: Y.S. conceived the project. Y.S., N.C.D., C.G.V. and F.G. performed the experiments. Y.S., M.R., and L.C.G. wrote the paper.
REFERENCES
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nature neuroscience. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods in enzymology. 2005;393:759–772. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- Antonijevic IA, Murck H, Bohlhalter S, Frieboes RM, Holsboer F, Steiger A. Neuropeptide Y promotes sleep and inhibits ACTH and cortisol release in young men. Neuropharmacology. 2000;39:1474–1481. doi: 10.1016/s0028-3908(00)00057-5. [DOI] [PubMed] [Google Scholar]
- Cayre M, Buckingham SD, Yagodin S, Sattelle DB. Cultured insect mushroom body neurons express functional receptors for acetylcholine, GABA, glutamate, octopamine, and dopamine. Journal of neurophysiology. 1999;81:1–14. doi: 10.1152/jn.1999.81.1.1. [DOI] [PubMed] [Google Scholar]
- Choi JC, Park D, Griffith LC. Electrophysiological and morphological characterization of identified motor neurons in the Drosophila third instar larva central nervous system. J Neurophysiol. 2004;91:2353–2365. doi: 10.1152/jn.01115.2003. [DOI] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:9377–9385. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65:670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson N, Kim EZ, Slawson JB, Vecsey CG, Huber R, Griffith LC. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the "tracker" program. PloS one. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyzma M, Boudjeltia KZ, Faraut B, Kerkhofs M. Neuropeptide Y and sleep. Sleep medicine reviews. 2010;14:161–165. doi: 10.1016/j.smrv.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Erion R, DiAngelo JR, Crocker A, Sehgal A. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. The Journal of biological chemistry. 2012;287:32406–32414. doi: 10.1074/jbc.M112.360875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nature neuroscience. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides. 2002;23:773–780. doi: 10.1016/s0196-9781(01)00647-7. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani OK, Lee MG, Henny P, Jones BE. Discharge profiles of identified GABAergic in comparison to cholinergic and putative glutamatergic basal forebrain neurons across the sleep-wake cycle. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:11828–11840. doi: 10.1523/JNEUROSCI.1259-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, Kwon OY, Song WJ, Tatar M, Yu K. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS genetics. 2012;8:e1002857. doi: 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MJ, Schenck CH, Crow SJ. A review of nighttime eating disorders. Sleep medicine reviews. 2009;13:23–34. doi: 10.1016/j.smrv.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Inagaki HK, Ben-Tabou de-Leon S, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, Anderson DJ. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell. 2012;148:583–595. doi: 10.1016/j.cell.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HA, Enell LE, Gustafsson E, Trifilieff P, Veenstra JA, Nassel DR. Intrinsic neurons of Drosophila mushroom bodies express short neuropeptide F: relations to extrinsic neurons expressing different neurotransmitters. The Journal of comparative neurology. 2008;507:1479–1496. doi: 10.1002/cne.21636. [DOI] [PubMed] [Google Scholar]
- Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Forster C, Nassel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. The Journal of comparative neurology. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Kahsai L, Martin JR, Winther AM. Neuropeptides in the Drosophila central complex in modulation of locomotor behavior. The Journal of experimental biology. 2010;213:2256–2265. doi: 10.1242/jeb.043190. [DOI] [PubMed] [Google Scholar]
- Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J. Clock and cycle limit starvation-induced sleep loss in Drosophila. Current biology : CB. 2010;20:1209–1215. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson KL, Van Cauter E. Associations between sleep loss and increased risk of obesity and diabetes. Annals of the New York Academy of Sciences. 2008;1129:287–304. doi: 10.1196/annals.1417.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang JS, Dankert H, Zelnik L, Kim YC, Han KA, Wolf FW, Perona P, Anderson DJ. Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron. 2009;64:522–536. doi: 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Hong SH, Kim AK, Ju SK, Kwon OY, Yu K. Processed short neuropeptide F peptides regulate growth through the ERK-insulin pathway in Drosophila melanogaster. FEBS letters. 2009;583:2573–2577. doi: 10.1016/j.febslet.2009.07.024. [DOI] [PubMed] [Google Scholar]
- Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nature cell biology. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. The Journal of biological chemistry. 2004;279:50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Current biology : CB. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFadyen UM, Oswald I, Lewis SA. Starvation and human slow-wave sleep. Journal of applied physiology. 1973;35:391–394. doi: 10.1152/jappl.1973.35.3.391. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochemical and biophysical research communications. 2002;297:1140–1148. doi: 10.1016/s0006-291x(02)02351-3. [DOI] [PubMed] [Google Scholar]
- Nassel DR, Enell LE, Santos JG, Wegener C, Johard HA. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC neuroscience. 2008;9:90. doi: 10.1186/1471-2202-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Currentbiology : CB. 2008;18:R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penev PD. Sleep deprivation and energy metabolism: to sleep, perchance to eat? Current opinion in endocrinology, diabetes, and obesity. 2007;14:374–381. doi: 10.1097/MED.0b013e3282be9093. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145:133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008; 105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pirez N, Harrington KI, Guo F, Pollack J, Hong P, Griffith LC, Rosbash M. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nature neuroscience. 2011;14:889–885. doi: 10.1038/nn.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Current biology : CB. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. Journal of neurobiology. 2001;47:16–25. doi: 10.1002/neu.1012. [DOI] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nature neuroscience. 2012;15:1516–1523. doi: 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- van Alphen B, Yap MH, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den, Pol AN. Neuropeptide transmission in brain circuits. Neuron. 2012;76:98–115. doi: 10.1016/j.neuron.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Swinderen B, Andretic R. Arousal in Drosophila. Behavioural processes. 2003;64:133–144. doi: 10.1016/s0376-6357(03)00131-1. [DOI] [PubMed] [Google Scholar]
- Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. 2001;22:241–254. doi: 10.1016/s0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- Yulyaningsih E, Zhang L, Herzog H, Sainsbury A. NPY receptors as potential targets for anti-obesity drug development. British journal of pharmacology. 2011;163:1170–1202. doi: 10.1111/j.1476-5381.2011.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.