Abstract
The histone methyltransferase EZH2 is a central epigenetic regulator of cell survival, proliferation, and cancer stem cell (CSC) function. EZH2 expression is increased in various human cancers, including highly aggressive pancreatic cancers, but the mechanisms underlying for its biologic effects are not yet well understood. In this study, we probed EZH2 function in pancreatic cancer using diflourinated-curcumin (CDF), a novel analogue of the turmeric spice component curcumin that has antioxidant properties. CDF decreased pancreatic cancer cell survival, clonogenicity, formation of pancreatospheres, invasive cell migration, and CSC function in human pancreatic cancer cells. These effects were associated with decreased expression of EZH2 and increased expression of a panel of tumor-suppressive microRNAs (miRNA), including let-7a,b,c,d, miR-26a, miR-101, miR-146a, and miR-200b,c that are typically lost in pancreatic cancer. Mechanistic investigations revealed that reexpression of miR-101 was sufficient to limit the expression of EZH2 and the proinvasive cell surface adhesion molecule EpCAM. In an orthotopic xenograft model of human pancreatic cancer, administration of CDF inhibited tumor growth in a manner associated with reduced expression of EZH2, Notch-1, CD44, EpCAM, and Nanog and increased expression of let-7, miR-26a, and miR-101. Taken together, our results indicated that CDF inhibited pancreatic cancer tumor growth and aggressiveness by targeting an EZH2-miRNA regulatory circuit for epigenetically controlled gene expression.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies with worst prognosis. Annually, 37,000 patients are diagnosed with pancreatic cancer, and accounting for 34,000 deaths, which is the fourth leading cause of cancer-related deaths in the United States (1). Over the past 2 decades, numerous efforts have been made in the treatment strategies including chemotherapy, radiation therapy, gene therapy, and immunotherapy to improve the survival of patients diagnosed with pancreatic cancer; however, the outcome has been very disappointing. Because of the absence of specific early symptoms and the lack of early detection methods, pancreatic cancer is usually diagnosed at an advanced incurable stage (2, 3). Thus, the median overall survival is only 5 to 6 months after conventional therapies, resulting in less than 5 % overall 5-year survival rate (3). This disappointing survival outcome is, in part, due to late diagnosis as well as intrinsic (de novo) and extrinsic (acquired) therapeutic resistance of pancreatic cancer to conventional therapeutics.
A large number of studies have shown that the resistance to chemo radiation therapy and other currently available targeted therapies are, in part, due to the survival of tumor-initiating cells or cancer stem cells (CSC), a minor population of cells in the tumor mass (3–6), and their further enrichment after therapy. CSCs posses the ability of self-renewal and have the potential to regenerate into all types of differentiated cells within the tumor cell populations, which contribute to drive continued expansion of the population of malignant cells, which leads to increased invasion and metastasis (3–6). Thus, the failure to eliminate these special cells is considered to be one of the underlying causes of treatment failure, which is, in part, due to tumor cell resistance to conventional therapeutics, resulting in tumor recurrence and metastasis, suggesting that the identification of novel targets and targeted therapies are urgently needed for overcoming therapeutic resistance, which indeed could be achieved by targeted killing of CSCs.
To that end, enhancer of Zeste homologue 2 (EZH2) received increased attention in recent years. EZH2 is a histone methyltransferase enhancer of polycomb group complexes and it is a critical part of the cellular machinery involved in the epigenetic regulation of gene transcription (7). Polycomb group proteins are a class of chromatin-modifying enzymes that are capable of methylation of both DNA and core histones to regulate gene expression. Emerging evidence suggest that polycomb group proteins play important role in stem cell maintenance, X-inactivation, imprinting, development, differentiation, and proliferation. The altered expressions of many polycomb group proteins have been found to be associated with the development of cancers (8). Overexpression of EZH2 has been found in a variety of tumors such as breast cancer, prostate cancer, and PDAC (9–12) and found to be directly responsible for the de novo suppression in the expression of multiple genes involved in human cancers (13–15); however, the exact molecular mechanism(s) by which EZH2 plays a role in tumorigenesis is not clear. Emerging evidence suggest that the overexpression of EZH2 might cause normal cells to dedifferentiate back to the stem cell–like state by epigenetically repressing cell fate– regulating genes and tumor suppressor genes, leading to the development of tumor (8, 14, 16), suggesting that further mechanistic understanding of EZH2 regulation and finding novel agents that could inactivate EZH2 would become a novel strategy for the treatment of pancreatic cancer.
In this study, we examined the effect of diflourinated-curcumin (CDF), a novel synthetic derivative of curcumin and a known natural antitumor agent, on cell survival, clonogenicity, sphere-forming capacity, cell migration and invasion, CSC self-renewal capacity, and the expression regulation of EZH2 by miRNAs in pancreatic cancer cells in vitro. We also examined the role of EZH2 in the regulation of cell growth, clonogenicity, cell migration and invasion, and miRNA expression in pancreatic cancer cells and mechanistically investigated the regulation of EZH2 expression by siRNA transfection approach. Most importantly, we examined the role of miR-101 in the expression of EZH2, EpCAM, Notch-1, and Oct4 in pancreatic cancer cells in vitro and also in an orthotopic animal model of pancreatic cancer in vivo and their mechanistic correlation with antitumor activity of CDF.
Materials and Methods
Cell culture
Human pancreatic cancer cell lines AsPC-1 and MiaPaCa-2 were used for this study on the basis of their sensitivities to chemotherapeutic drug gemcitabine, as described previously (17, 18). Both cell lines were maintained in Dulbecco's Modified Eagle's Medium (DMEM; Invitrogen), supplemented with 5% FBS, 2 mmol/L glutamine, 50 units/mL penicillin, and 50 µg/mL streptomycin in a standard culture condition, as described previously (17, 18). The cell lines have been tested and authenticated in the core facility "Applied Genomics Technology Center" at Wayne State University, Detroit, MI, on March 13, 2009. The method used for testing was short tandem repeat (STR) profiling using the PowerPlex 16 System from Promega.
Reagents and antibodies
CDF, a synthetic derivative of curcumin, was synthesized as described in our earlier publications (17, 19). Antibodies against CD44, EpCAM, cleaved Notch-1, and SHH were purchased from Cell Signaling Technology. Antibodies against Notch-1, ABCG2, matrix metalloproteinase (MMP)-9, and Hes-1 were purchased from Santa Cruz. Antibodies against β-actin and EZH2 were acquired from Sigma Chemicals and BD Transduction.
Cell growth and survival assay
MTT assay was conducted to assess cell survival and growth. Both cell lines were exposed to different concentrations of CDF (0.1–1 µmol/L) for 3 days of treatment, and after 3 days, MTT assay was conducted as described previously (17).
Clonogenic assay
Clonogenic assay was conducted to examine the effect of CDF (0.5 µmol/L) on cell growth and proliferation of pancreatic cancer cells as described previously (17,20). Briefly, 5 × 10 cells were plated in a 6-well plate and after 72 hours of exposure to 0.5 µmol/L of CDF, 1,000 single viable cells were plated in 100-mm Petri dishes. The cells were then incubated for 10 to 12 days at 37° C in a 5% CO2/5% O2/90% N2 incubator. Colonies were stained with 2% crystal violet, washed with water, and counted.
Cell migration (wound-healing) assay
Cell migration (wound-healing) assay was conducted to assess the capacity of cell migration and invasion as described previously (21). Briefly, when the cells reached 90% to 95% confluency, the wound was generated by scratching the surface of the plates with a pipette tip. The cells were then incubated in the absence and presence of CDF (0.5 µmol/L) for 18 hours and then photographed with a Nikon Eclipse TS100 microscope.
Sphere formation assay
Sphere formation assay was conducted to assess the capacity of CSC self-renewal, as described previously (18). Briefly, single-cell suspensions of cells were plated on ultra-low-adherent wells of 6-well plate (Corning) at 500 to 1,000 cells/well in sphere formation medium (1:1 DMEM/F12 medium supplemented with B-27 and N-2; Invitrogen). After 7 days, the spheres termed as "pancreatospheres" were collected by centrifugation (300 × g, 5 minutes) and counted. The proportion of sphere-generating cells was calculated by dividing the number of cells seeded by the number of pancreatospheres with the diameter greater than 50 µm.
Protein extraction and Western blot analysis
Western blot analysis was conducted using whole-cell protein lysates of pancreatic cancer cells or tumor tissues. Total cell lysates from different experiments were obtained by lysing the cells in protein lysis buffer and 1× protease inhibitor cocktail, and Western blotting was conducted as described previously (17, 18).
TaqMan miRNA real-time reverse transcriptase PCR
To determine the expression of miRNAs (let-7 family, miR-26a, miR-101, and miR-200 family) in the cells or tumor samples, we used TaqMan MicroRNA Assay kit (Applied Biosystems) following manufacturer's protocol. Five nanograms of total RNA was reverse transcribed and real-time PCR reactions were carried out in 10 µL of reaction mixture as described previously (17, 22) using AB StepOnePlus Real-Time PCR System (Applied Biosystems). Data were analyzed using Ct method and were normalized by RNU6B or RNU48 expression in each sample.
Levels of mRNAs by real-time reverse transcriptase PCR
To determine the mRNA expression of EZH2, Notch-1, EpCAM, Nanog, Sox2, and Oct4, 2 µg of total RNAs extracted from each sample (pancreatic cancer cells of xenograft tumor sample) were used for reverse transcription (RT) reaction in 20 µL of reaction volume using a reverse transcription system (Invitrogen) according to the manufacturer's instructions. SYBR Green Assay kit (Applied Biosystems) was used for real-time PCR reaction, following manufacturer's protocol. Sequences of PCR primers were described previously (17, 22). Data were analyzed using Ct method and were normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each sample.
Transfection of EZH2 siRNA
MiaPaCa-2 cells were plated in 6-well plate overnight and transfected with 10 µg of EZH2 siRNA or scramble control siRNA (Applied Biosystems) by ExGen 500 (Fermentas) following the manufacturer's protocol. After 48 hours of transfection, the cells were collected for the cell growth, clonogenic assay, wound-healing assay, Western blot analysis, or RT-PCR of miRNAs as described above.
Transfection of miRNA precursor miR-101
A total of 2 × 105 cells per well of MiaPaCa-2 cells were seeded in 6-well plates and transfected with pre-miR-101 or miRNA-negative control (Ambion) at a final concentration of 20 nmol/L using DharmaFECT transfection reagent (Dharmacon), following the manufacturer's protocol, and as described previously (21). After 3 days of transfection, the transfected cells were harvested for total RNA isolation or protein extraction. The relative levels of miRNA, mRNAs, and proteins were measured as described above.
Animal experiments
The animal protocol was approved by the Animal Investigation Committee, Wayne State University. Female CB17 severe combined immunodeficient (SCID) mice (4 weeks old) were purchased from Taconic Farms and fed Lab Diet 5021 (Purina Mills, Inc.). Initially, 1 × 106 MiaPaCa-2 cells or 5,000 pancreatospheres of MiaPaCa-2 were implanted in xenograft mouse model for comparison of protein expression between cells, xenograft tumor, and xenograft pancreatospheres. The tumors were removed and protein was extracted. For the actual therapeutic experiment, 1 × 106 MiaPaCa-2 cells were orthotopically implanted into the pancreas of the mice. One mouse was sacrificed after 2 weeks to confirm the growth of the tumor and were then randomly divided into 3 groups with 7 animals in each group: (i) untreated control; (ii) CDF (2.5 mg/mouse/d), intragastric once daily for 3 weeks; and (iii) CDF (5 mg/mouse/d), intragastric once daily for 3 weeks. The tumor tissue from all groups were rapidly frozen in liquid nitrogen and stored at −70° C for later use for RNA, protein extraction, and another portion was fixed in formalin for histopathologic and immunohistochemical studies.
Immunohistochemistry
Formalin-fixed tumor tissue sections were evaluated by hematoxylin and eosin staining and immunohistochemistry was carried out through the core facility of Department of Pathology, Karmanos Cancer Institute, Detroit, MI, as described previously (23). Briefly, immunohistochemical staining was carried out for the expression of Ki-67 and CD44 using specific antibodies followed by staining with appropriate horseradish peroxidase–conjugated secondary antibodies. The slides were developed in diaminobenzidine and counterstained with hematoxylin. The stained slides were dehydrated and mounted in Per mount solution and visualized using an Olympus microscope. Images were captured with an attached camera linked to the computer.
Statistical analysis
The data with mean and SD in this study were calculated using GraphPad Prism software (version 4.03). Comparisons of each treatment outcome were carried out for statistical difference by the paired t test. Statistical significance was assumed at a value of P < 0.05.
Results
Effect of CDF on cell survival of pancreatic cancer cells
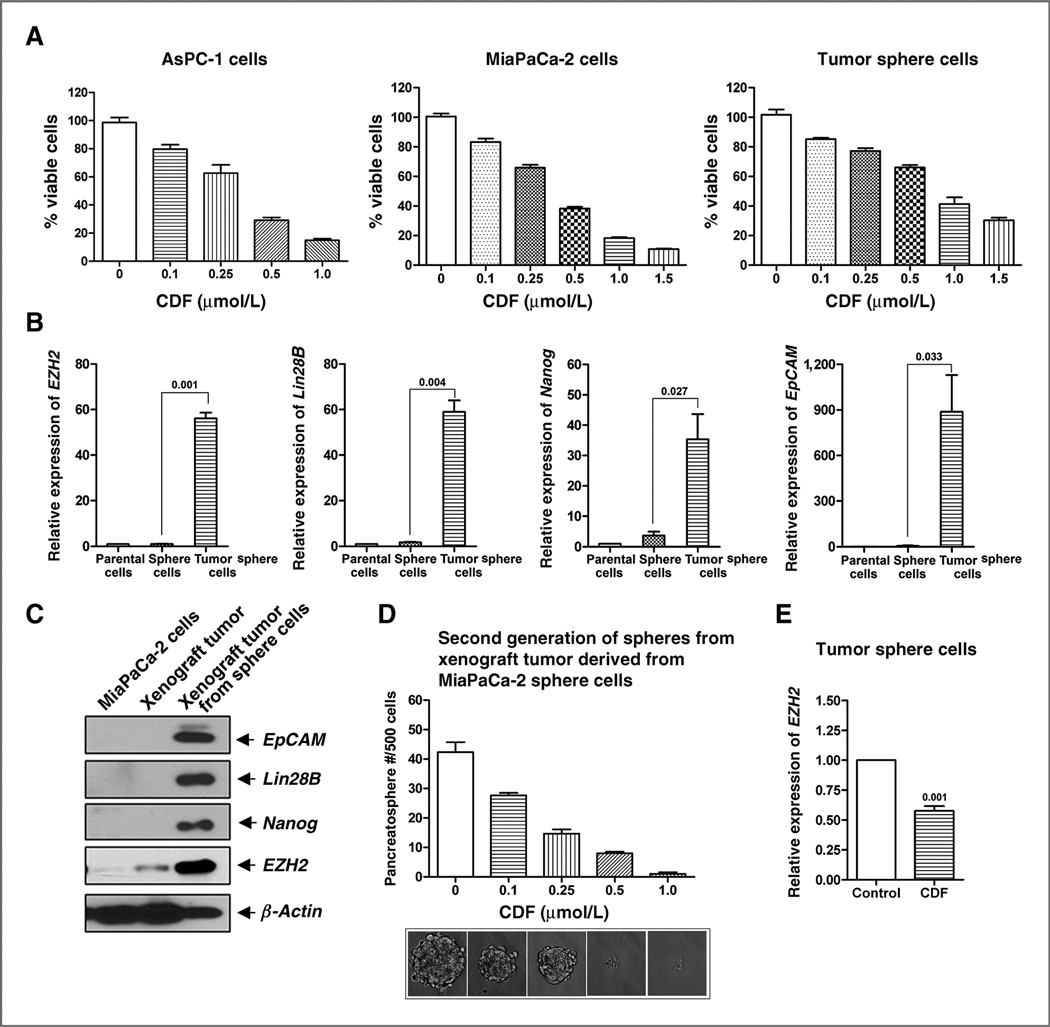
To investigate the effect of CDF on cell survival of pancreatic cancer cells, we conducted MTT assays using AsPC-1, MiaPaCa-2 cells, and MiaPaCa-2 tumor sphere cells. The results indicate that CDF significantly inhibited cell survival in a dose-dependent manner in AsPC-1, MiaPaCa-2 cells, and MiaPaCa-2 tumor sphere cells (Fig. 1A). These results suggests that CDF could exert a pronounced cell growth–inhibitory effect on pancreatic cancer cell growth and proliferation, which is consistent with our previously published reports (17, 18).
Figure 1.
A, CDF decreased cell survival of AsPC-1, MiaPaCa-2 cells, and MiaPaCa-2 tumor sphere cells as assessed by MTT assay. B, differential mRNA expression of CSC marker genes in the parental MiaPaCa-2 cells, sphere cells, and tumor sphere cells. C, differential protein expression of CSC marker genes in MiaPaCa-2 cells, xenograft tumor from MiaPaCa-2 cells, and xenograft tumor derived from MiaPaCa-2 sphere cells. D, CDF inhibited the formation of second generation of spheres from mouse xenograft tumors derived from MiaPaCa-2 sphere cells. E, CDF decreased the expression of EZH2 mRNA in MiaPaCa-2 sphere cells.
Characterization of parental MiaPaCa-2 cells and tumors derived from either MiaPaCa-2 parental cells or MiaPaCa-2 sphere cells
We assessed the mRNA expressions of CSC markers such as EZH2, Lin28B, Nanog, and EpCAM in the parental MiaPaCa-2 cells, the MiaPaCa-2 sphere cells, and the MiaPaCa-2 tumor sphere cells. We found that the relative mRNA expressions of EZH2, Lin28B, Nanog, and EpCAM were significantly higher in the tumor sphere cells (Fig. 1B). We also assessed the protein expression of CSC markers such as EpCAM, Lin28B, Nanog, and EZH2 in the parental MiaPaCa-2 cells and tumors derived from either the MiaPaCa-2 parental cells or MiaPaCa-2 sphere cells (also called as tumor sphere cells). We found that the relative expressions of EpCAM, Lin28B, EZH2, and Nanog were significantly lower in the parental MiaPaCa-2 cells and tumors derived from MiaPaCa-2 parental cells (Fig. 1C), whereas the expression of all CSC makers was found to be highly expressed in the tumors derived from the MiaPaCa-2 sphere cells (Fig. 1C), suggesting CSC enrichment in the tumors derived from the pancreatospheres of the MiaPaCa-2 cells.
Effect of CDF on the sphere formation and EZH2 mRNA in MiaPaCa-2 tumor sphere cells
In our previous studies (18), we have shown that CDF was effective in the inhibition of pancreatospheres formation and as shown above that CSCs are highly enriched in the tumors derived from the MiaPaCa-2 pancreatospheres. Therefore, to examine the effect of CDF on self-renewal capacity of CSCs or CSC-like cells, we conducted sphere formation assay of the cells derived from the xenograft tumors established using pancreatospheres of MiaPaCa-2 cells (second generation of pancreatospheres or tumor sphere cells). We found significant inhibition of secondary pancreatospheres formation by CDF treatment in a dose-dependent manner (Fig. 1D). These results clearly documents that CDF can inhibit the formation of pancreatospheres, suggesting that CDF could indeed attenuate CSC function. We also examined the effect of CDF on EZH2 mRNA which is overexpressed in CSCs such as MiaPaCa-2 tumor sphere cells. The results showed that CDF could significantly inhibit the expression of EZH2 mRNA in tumor sphere cells (Fig. 1E).
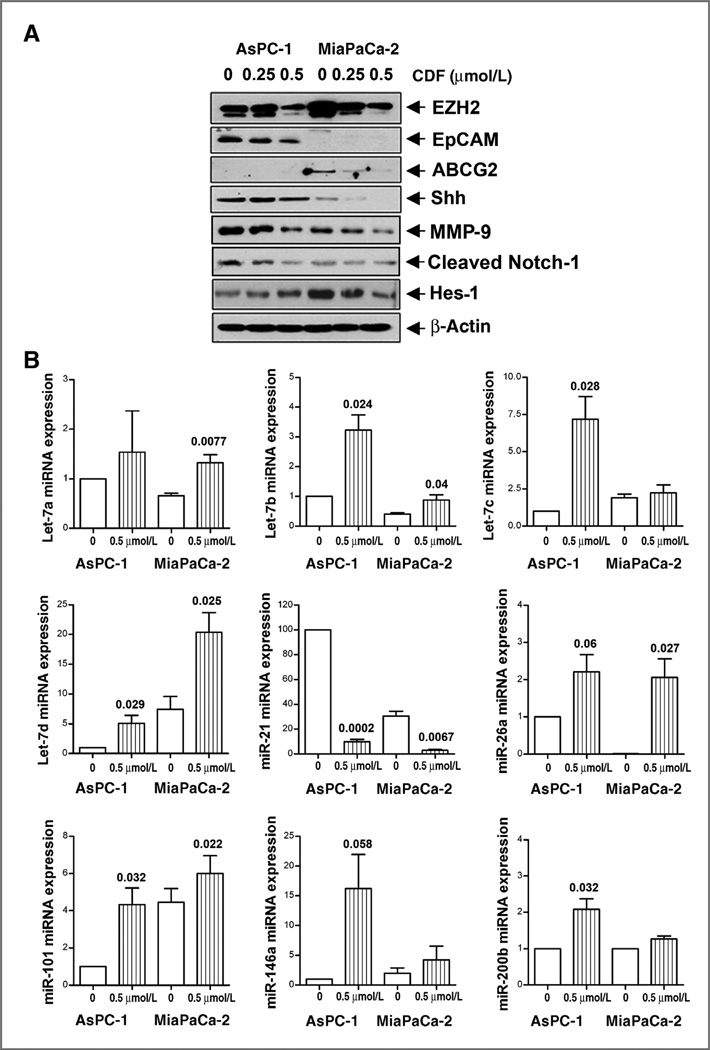
Effect of CDF on EZH2, EpCAM, ABCG2, Shh, Hes-1, and MMP-9 in AsPC-1 and MiaPaCa-2 cells
We assessed the expression of CSC markers (EZH2, EpCAM, Shh, and Hes-1) as well as a marker (ABCG2) for drug resistance. Figure 2A shows that CDF was effective in inhibiting the expression of EZH2, Shh, and cleaved Notch-1 in AsPC-1 and MiaPaCa-2 cells, and CDF remarkably decreased the expression of EpCAM in AsPC-1 cells. CDF treatment also remarkably decreased the expression of ABCG2 and Hes-1 in MiaPaCa-2 cells. We also found that CDF was very effective in inhibiting the expression of MMP-9, a biomarker for tumor metastasis, in AsPC-1 and MiaPaCa-2 cells (Fig. 2A).
Figure 2.
CDF decreased the expression of EZH2, EpCAM, ABCG2, Shh, MMP-9, cleaved Notch-1, and Hes-1 (A) and increased the miRNA expressions of let-7 family, miR-26a miR-101, miR-146a, and miR-200 (B) in pancreatic cancer cells. Western blot analysis and real-time RT-PCR of miRNAs were carried out as described under the Materials and Methods section.
Effect of CDF on the expression of miRNAs in AsPC-1 and MiaPaCa-2 cells by real-time RT-PCR
We examined the effect of CDF on the expression of miRNAs (let-7 family, miR-21, miR-26a, miR-101, miR-146a, and miR-200b) in pancreatic cancer cells. The results revealed that CDF treatment caused reexpression of the miRNAs such as let-7a,b, c,d, miR-26a, miR-101, miR-146a, and miR-200b that are lost in AsPC-1 and MiaPaCa-2 cells (Fig. 2B). Interestingly, miR-21 expression was very high in the AsPC-1 and MiaPaCa-2 cells and CDF was able to downregulate its expression. These data suggest that CDF differentially regulate the expression of these tumor-associated miRNAs in pancreatic cancer cells.
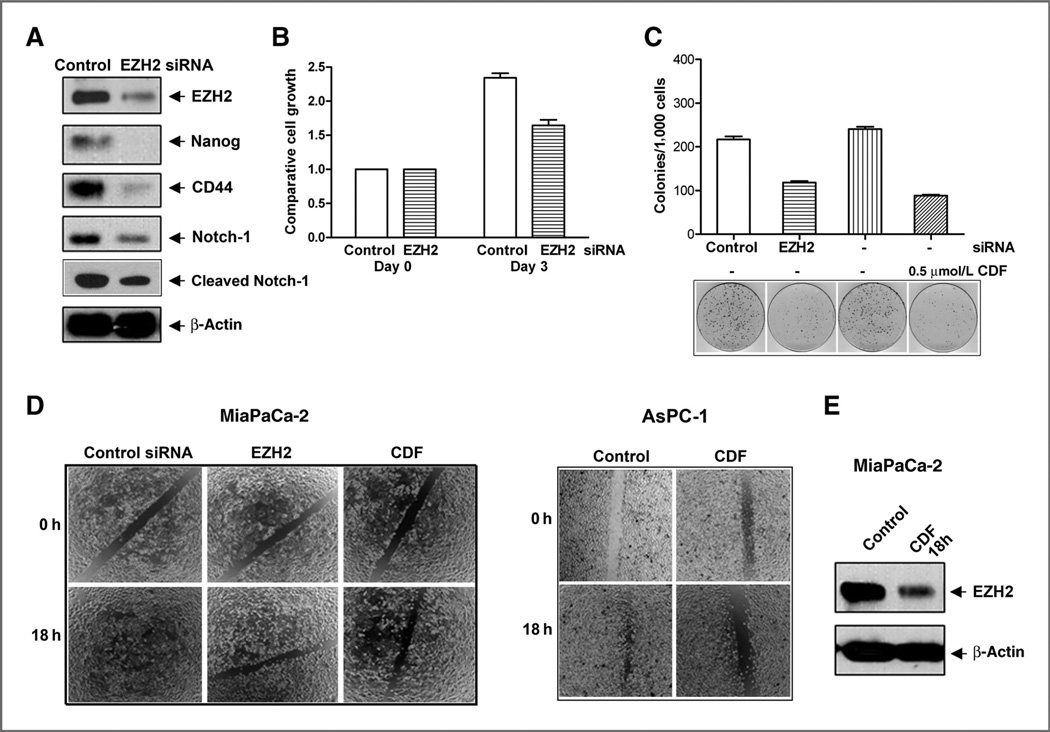
Effect of EZH2 deficiency on cell growth, clonogenicity, and cell migration of pancreatic cancer cells
To investigate the role of EZH2 in the regulation of cell growth, clonogenicity, and cell migration of pancreatic cancer cells, we carried out the EZH2 siRNA transfection experiments using MiaPaCa-2 cells. We found that EZH2 siRNA transfection resulted in decreased expression of EZH2, but most interestingly, the knockdown of EZH2 led to decreased expression of Nanog, CD44, Notch-1, and cleaved Notch-1 expression (Fig. 3A). The downregulation of EZH2 resulted in decreased cell growth (Fig. 3B), decreased clonogenicity (Fig. 3C), and decreased cell migration (Fig. 3D). CDF decreased cell migration in MiaPaCa-2 and AsPC-1 cells after 18 hours of treatment (Fig. 3D). The Western blot analysis shows that CDF also decreased the expression of EZH2 in MiaPaCa-2 cells after 18 hours of treatment (Fig. 3E). These results suggest that EZH2 mediates cell growth, clonogenicity, and cell migration of pancreatic cancer cells and that the inhibition of cell growth/survival, clonogenicity, cell migration by CDF treatment is indeed mediated through the downregulation of EZH2 in pancreatic cancer cells.
Figure 3.
EZH2 siRNA decreased the relative protein levels of EZH2, Nanog, CD44, and Notch-1 expression (A), cell growth (B), clonogenicity (C), in MIAPaCa-2 cells and the wound-healing capacity (D) of MiaPaCa-2 and AsPC-1 cells. The transfection of EZH2 siRNA was conducted in MiaPaCa-2 cells. The transfected cells were then collected for cell growth assay, Western blot analysis, clonogenic assay, and wound-healing assay as described under the Materials and Methods section. CDF decreased the wound-healing capacity (D) and EZH2 expression (E) in pancreatic cancer cells after 18 hours of treatment.
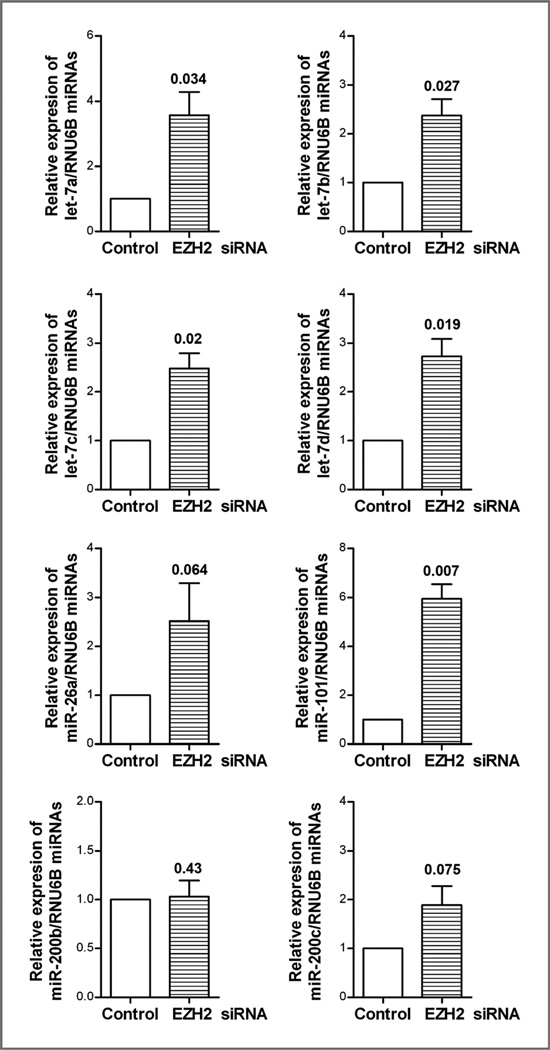
Effect of EZH2 siRNA on the expression of miRNAs (let-7 family, miR-26a, miR-101, and miR-200b) in MiaPaCa-2 cells
We examined the effect of EZH2 inactivation by EZH2 siRNA on the expression of miRNAs (let-7 family, miR-26a, miR-101, and miR-200b,c) in MiaPaCa-2 cells by real-time RT-PCR. We found that the inactivation of EZH2 led to reexpression of let-7 family, miR-26a, miR-101, and miR-200c which are typically lost in pancreatic cancer cells (Fig. 4), and these results are consistent with CDF treatment results as presented above in Fig. 2B.
Figure 4.
Effect of EZH2 siRNA on the expression of miRNAs in MiaPaCa-2 cells. After 72 hours of the transfection with EZH2 siRNA, the transfected cells were collected for real-time RT-PCR for assessing the expression of miRNAs as described under the Materials and Methods section.
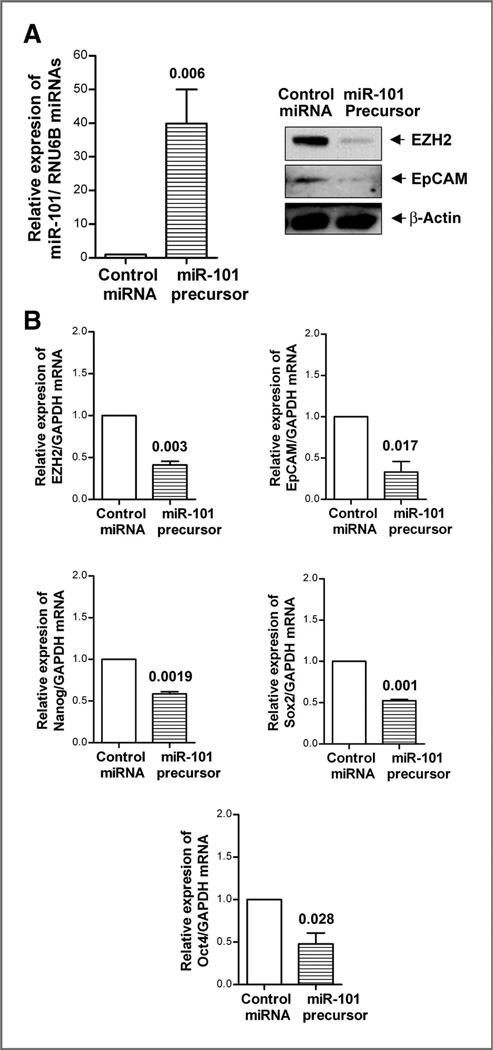
The role of miR-101 in the regulation of EZH2, EpCAM, Sox2, Oct4, and Notch-1 in MiaPaCa-2 cells
To investigate the role of miR-101 in the regulation of CSC marker genes, we carried out transfection experiment using miR-101 precursor. We found that the transfection of miR-101 precursor led to increased expression of miR-101 as expected, which led to decreased expression of EZH2 and EpCAM proteins in MiaPaCa-2 cells (Fig. 5A). Moreover, we found that the reexpression of miR-101 also led to decreased mRNA expression of EZH2, EpCAM, Nanog, Sox2, and Oct4 mRNAs in MiaPaCa-2 cells (Fig. 5B), which suggests that miR-101 plays a key role in the regulation of CSC marker genes such as EZH2, EpCAM, Nanog, Sox2, and Oct4.
Figure 5.
Reexpression of miR-101 decreased the expression of EZH2 and EpCAM proteins (A) and mRNAs of EZH2, EpCAM, Nanog, Sox2, and Oct4 (B) in MiaPaCa-2 cells. The transfection of miR-101 precursor was conducted in MiaPaCa-2 cells as described under the Materials and Methods section. Real-time RT-PCR and Western blot analysis were conducted to measure the relative mRNA and protein levels, respectively.
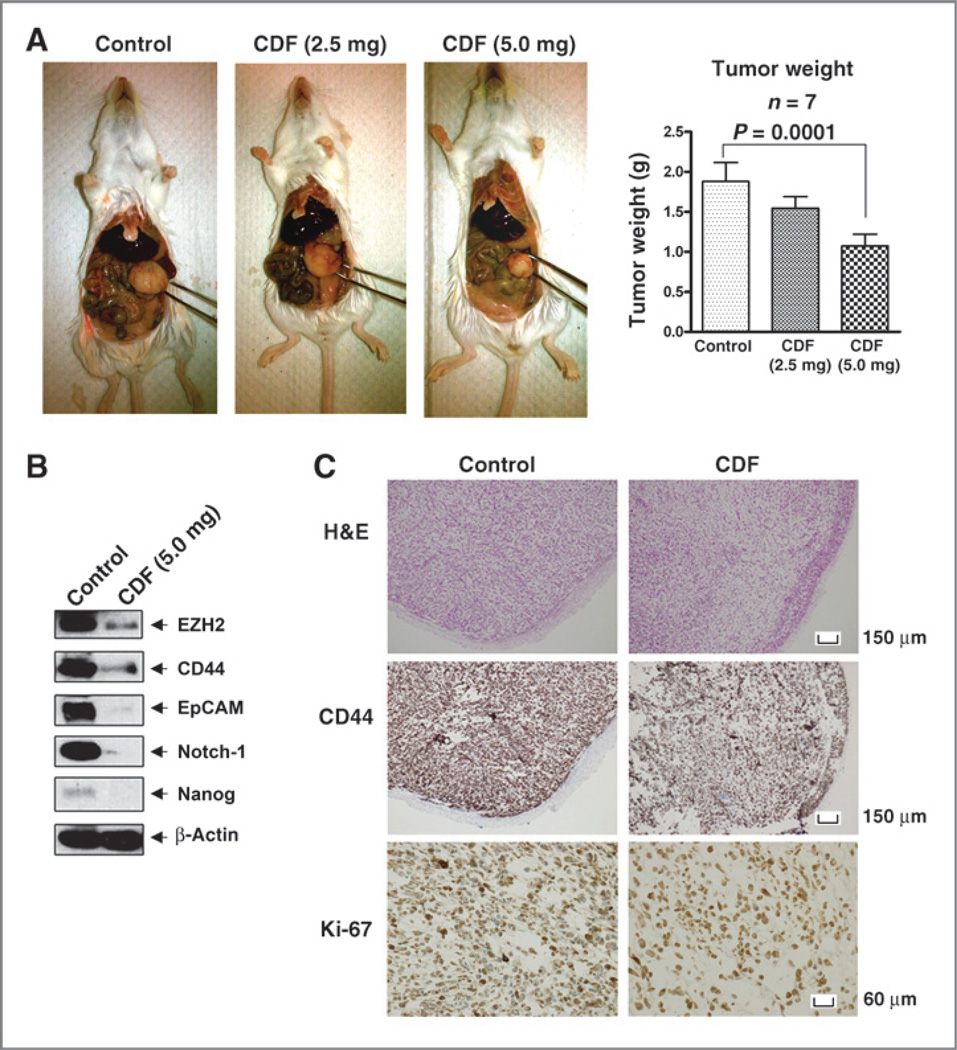
Effect of CDF on tumor growth and on the expression of EZH2, CD44, EpCAM, Notch-1, and Nanog in tumor remnants in vivo
We carried out animal experiment using orthotopic mouse model of pancreatic cancer in which the tumors were derived from MiaPaCa-2 cells injected into the pancreas. We found that group with 5 mg CDF per mouse could significantly decrease tumor size and weight, compared with the control group or group with 2.5 mg CDF per mouse (Fig. 6A), which is consistent with our previous findings (17). Interestingly, we found that CDF treatment could decrease the protein expression of EZH2, CD44, EpCAM, Notch-1, and Nanog in pancreatic tumor remnant (Fig. 6B). Immunohistochemical study of pancreatic tumors revealed that the intensity scores of Ki-67 and CD44 from the control and CDF groups were 3 and 2, respectively, indicating that CDF treatment reduced the expression of Ki-67 and CD44, in pancreatic tumor tissue remnants (Fig. 6C).
Figure 6.
CDF decreased tumor growth (A), the relative protein levels of EZH2, CD44, EpCAM, Notch-1, and Nanog by Western blot analysis (B), the cell proliferation, Ki-67, and CD44 (C) immunohistochemistry using pancreatic tumor tissues orthotopically derived from human pancreatic cancer MiaPaCa-2 cells showing reduced expression of Ki-67 and CD44. H&E, hematoxylin and eosin.
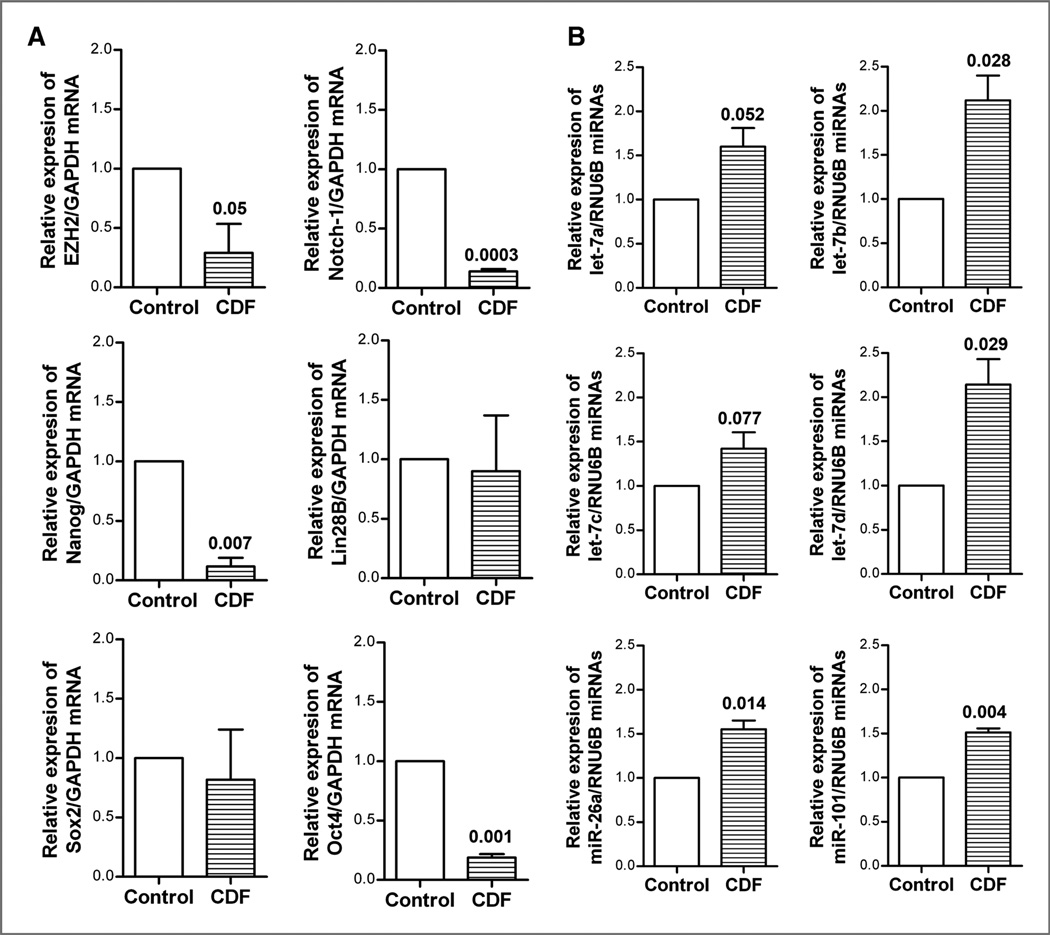
Effect of CDF on gene expression and miRNA expression in vivo
We examined the effect of CDF treatment of orthotopic tumors on both the mRNA and miRNA expression in tumor remnants. We found that CDF treatment significantly decreased the relative mRNA levels of EZH2, Notch-1, Nanog, and Oct4 in pancreatic tumor tissues (Fig. 7A), suggesting that CDF could inhibit the mRNA expression of these CSC marker genes in vivo. Similarly, we also found that CDF treatment led to increased expression of let-7 family, miR-26a, and miR-101 in tumor tissue remnants (Fig. 7B) and these in vivo results are consistent with our in vitro findings.
Figure 7.
CDF decreased the mRNA expression of EZH2, Notch-1, Nanog, and Oct4 in pancreatic tumor tissues (A), and increased miRNA expression of let-7a,b,c, miR-26a, and miR-10 in pancreatic tumors (B) derived from human pancreatic cancer MiaPaCa-2 cell-induced tumor implanted orthotopically in the pancreas of severe combined immunodeficient (SCID) mice, and real-time RT-PCR was carried out as described under the Materials and Methods section.
Discussion
Emerging evidence suggests that EZH2 plays a key role in tumorigenesis by maintaining stem cell function including CSCs through epigenetic repression of cell fate–regulating genes and tumor suppressor genes (8, 14, 16). Increased expression of EZH2 has been observed in variety of tumors. For example, in prostate cancer, the overexpression of EZH2 was associated with aggressive and metastatic disease (11, 24). Similarly, in breast cancer, the expression of EZH2 was found to be elevated in invasive and metastatic tumor, and its increased expression was positively associated with poor prognosis (25, 26). The increased expression of EZH2 has also been found in other tumors including pancreatic cancer (26, 27). Recently, EZH2 has been identified to be associated with tumor angiogenesis, the self-renewal capacity, and the gene expression of CSC markers associated with tumor aggressiveness (12, 26, 28–31). In our current study, we have shown that the decreased EZH2 expression using EZH2 siRNA transfection led to decreased cell proliferation, migration, and the protein expressions of CSC markers such as Nanog, CD44, and Notch-1 in pancreatic cancer cells. Moreover, EZH2 deficiency led to increased expression of miRNAs such as let-7a,b,c,d, miR-26a, miR-101, and miR-200 in pancreatic cancer cells, suggesting that these miRNAs are involved in the regulation of EZH2 and that inactivation of EZH2 could contribute to inhibit tumor aggressiveness. Our results on the effects of CDF on pancreatospheres formation of tumor cells derived from the tumors induced by the pancreatospheres of MiaPaCa-2 cells, clearly suggest that CSCs could be eliminated by CDF by attenuating EZH2 and other CSC-specific genes through reexpression of miRNAs.
As documented earlier that miRNAs function as endogenous posttranscriptional gene regulators by specific binding to the 3′ untranslated region (3′-UTR) of target mRNAs to mediate protein synthesis or mRNA stability (32, 33). The miRNAs are currently recognized as important and key regulators of the expression of most genes, and consequently, they play critical roles in wide variety of biologic processes, including cell differentiation, proliferation, death, metabolism, and energy homeostasis (33, 34). A large number of miRNAs have been reported to be associated with tumorigenesis by regulation of the expression/transcription of many tumor-related genes (35). Emerging evidence suggests that miRNAs may play an important role in the regulation of stem cell function in tumors including pancreatic cancer (36). However, their exact roles in the pathogenesis of pancreatic cancer are not fully understood. Moreover, it has been reported that the let- 7 family function as a potential tumor suppressor and has been found to be down-regulated in tumors, resulting in enhanced expression of Ras and c-Myc in tumor cells (35, 37, 38). In addition, the downregulation of let-7 has been observed in pancreatic cancer (35, 38). A recent study has shown that the expression of CSC marker genes could be regulated bidirectionally and associated with the downregulation of let-7 expression (39), suggesting that let- 7 may play a role in the regulation of CSC function. Our findings clearly show that inactivation of EZH2 led to increased let-7 expression, and thus CDF-mediated reexpression of let-7 family of miRNAs could be responsible for the biologic effects of CDF in pancreatic cancer cells and tumors.
Emerging evidence also suggests that miR-200 serve as a potential tumor suppressor, primarily by repressing the acquisition of epithelial-to-mesenchymal transition (EMT) phenotype during the development and progression of tumors (17, 22, 35, 40, 41). Decreased expression of miR-200 has been observed in many tumors including pancreatic cancer, which is associated with tumor invasion and metastasis (35, 42, 43). The in vitro studies revealed that the downregulation of miR-200 enhances the acquisition of EMT phenotype in the cancer cells and that reexpression of miR-200 results in the reversal of EMT phenotype (44), which is consistent with our previous findings in pancreatic cancer cells as summarized in our recent review article (36). A recent study has shown that loss of miR-200b increases breast CSC growth and invasive capacity by targeting CSC gene Suz12, suggesting a key role of miR-200 in the deregulation of CSC function (45). Our results on the EZH2 siRNA transfection study showed that the inhibition of EZH2 led to increased expression of let-7 and miR-200 in pancreatic cancer cells, suggesting that EZH2 could be involved in the regulation of let-7 and miR-200 in cancer cells and that CDF could lead to the reexpression of let-7 and miR-200, which would be highly valuable in reverting tumor cell aggressiveness or in the killing of CSCs.
Furthermore, miR-26a and miR-101 are known potential tumor suppressors and have been reported to modulate the cancer epigenome by repressing the polycomb group protein, EZH2 (9, 10, 30, 46–48). Decreased expression of miR-26a and miR-101 has been found in a wide variety of cancers such as gastric cancer, breast cancer, prostate cancer, nasopharyngeal carcinoma, and pancreatic cancer (49). Reexpression of miR-26a or miR-101 in cancer cells leads to the downregulation of EZH2, resulting in the inhibition of tumor metastasis and invasion (9, 30, 47, 50). Our findings showed that reexpression of miR-101 results in the downregulation in the expression of EZH2 and EpCAM and that inactivation of EZH2 results in the upregulation of miR-101 in pancreatic cancer cells, showing a negative mutual feedback loop of EZH2 and miR-101. These findings are also consistent with treatment of cells with CDF, suggesting that miRNAs that are lost in pancreatic cancer cells could be reexpressed using our novel agent CDF (18, 19). In our previous reports, we showed that CDF inhibited tumor growth by attenuating multiple targets such as NF-κB, Akt, COX-2 signaling pathways, EMT phenotype and CSC function, PTEN, miR-21, and miR-200 in vitro and in vivo (17, 18), which is consistent with our current findings showing that CDF could inhibit the expression of EZH2, Notch-1, and Oct4 through reexpression of let-7a,b,c,d, miR-26a, and miR-101 in the pancreatic cancer cells as well as in tumor remnants. Collectively, our results together with our published reports clearly suggest that CDF could be a useful agent for the prevention of tumor progression and/or treatment of pancreatic cancer which would be accomplished by attenuating CSC function and overcoming therapeutic resistance.
Acknowledgments
The authors thank Ms. Amanda C. Sehmer for editing the manuscript.
Grant Support
This work was financially supported by Puschelberg and Guido Foundations; National Cancer Institute, NIH grants 5R01CA131151, 3R01CA131151-02S1, 5R01CA132794, and 1R01CA154321-01A1 (F.H. Sarkar).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Arslan AA, Helzlsouer KJ, Kooperberg C, Shu XO, Steplowski E, Buenode-Mesquita HB, et al. Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Arch Intern Med. 2010;170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, et al. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- 4.Gaviraghi M, Tunici P, Valensin S, Rossi M, Giordano C, Magnoni L, et al. Pancreatic cancer spheres are more than just aggregates of stem marker positive cells. Biosci Rep. 2010;31:45–55. doi: 10.1042/BSR20100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CJ, Li C, Simeone DM. Human pancreatic cancer stem cells: implications for how we treat pancreatic cancer. Transl Oncol. 2008;1:14–18. doi: 10.1593/tlo.08013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller MT, Hermann PC, Heeschen C. Cancer stem cells as new therapeutic target to prevent tumour progression and metastasis. Front Biosci (Elite Ed) 2010;2:602–613. doi: 10.2741/e117. [DOI] [PubMed] [Google Scholar]
- 7.Pasini D, Bracken AP, Helin K. Polycomb group proteins in cell cycle progression and cancer. Cell Cycle. 2004;3:396–400. [PubMed] [Google Scholar]
- 8.Sparmann A, van LM. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee R, Mani RS, Russo N, Scanlon CS, Tsodikov A, Jing X, et al. The tumor suppressor gene raplGAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 2011;30:4339–4349. doi: 10.1038/onc.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 12.Yu YL, Chou RH, Chen LT, Shyu WC, Hsieh SC, Wu CS, et al. EZH2 regulates neuronal differentiation of mesenchymal stem cells through PIP5K1 C-dependent calcium signaling. J Biol Chem. 2011;286:9657–9667. doi: 10.1074/jbc.M110.185124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 15.Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin Cancer Res. 2005;11:8570–8576. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, et al. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Bao B, Ali S, Kong D, Sarkar SH, Wang Z, Banerjee S, et al. Anti-tumor activity of a novel compound-CDF is mediated by regulating miR-21, miR-200, and PTEN in pancreatic cancer. PLoS One. 2011;6:e17850. doi: 10.1371/journal.pone.0017850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Padhye S, Yang H, Jamadar A, Cui QC, Chavan D, Dominiak K, et al. New difluoro Knoevenagel condensates of curcumin, their Schiff bases and copper complexes as proteasome inhibitors and apoptosis inducers in cancer cells. Pharm Res. 2009;26:1874–1880. doi: 10.1007/s11095-009-9900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, et al. Notch-1 nduces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307:26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, et al. Overexpression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–2306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, et al. Upregulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Kong D, Banerjee S, Li Y, Adsay NV, Abbruzzese J, et al. Down-regulation of platelet-derived growth factor-D inhibits cell growth and angiogenesis through inactivation of Notch-1 and nuclear factor-kappaB signaling. Cancer Res. 2007;67:11377–11385. doi: 10.1158/0008-5472.CAN-07-2803. [DOI] [PubMed] [Google Scholar]
- 24.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47:701–706. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toll AD, Dasgupta A, Potoczek M, Yeo CJ, Kleer CG, Brady JR, et al. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41:1205–1209. doi: 10.1016/j.humpath.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, et al. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, Han HD, Mangala LS, li-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 34.DeSano JT, Xu L. MicroRNA regulation of cancer stem cells and therapeutic implications. AAPS J. 2009;11:682–692. doi: 10.1208/s12248-009-9147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, et al. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13:109–118. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catto JW, Alcaraz A, Bjartell AS, De Vere WR, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 38.Kent OA, Mullendore M, Wentzel EA, Lopez-Romero P, Tan AC, Alvarez H, et al. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol Ther. 2009;8:2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunaratne PH. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cel Res Ther. 2009;4:168–177. doi: 10.2174/157488809789057400. [DOI] [PubMed] [Google Scholar]
- 40.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong D, Banerjee S, Ahmad A, Li Y, Wang Z, Sethi S, et al. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 2010;5:e12445. doi: 10.1371/journal.pone.0012445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moriyama T, Ohuchida K, Mizumoto K, Yu J, Sato N, Nabae T, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067–1074. doi: 10.1158/1535-7163.MCT-08-0592. [DOI] [PubMed] [Google Scholar]
- 43.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, et al. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar FH, Li Y, Wang Z, Kong D. Pancreatic cancer stem cells and EMT in drug resistance and metastasis. Minerva Chir. 2009;64:489–500. [PMC free article] [PubMed] [Google Scholar]
- 45.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Xie D, Yin LW, Man CC, Yao H, Chan CY, et al. RNAi targeting EZH2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo . Cancer Lett. 2010;297:109–116. doi: 10.1016/j.canlet.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 48.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai) 2010;42:363–369. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 50.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]