Abstract
Mutation of the template region in the RNA component of telomerase can cause incorporation of mutant DNA sequences at telomeres. We made all 63 mutant sequence combinations at template positions 474–476 of the yeast telomerase RNA, TLC1. Mutants contained faithfully incorporated template mutations, as well as misincorporated sequences in telomeres, a phenotype not previously reported for Saccharomyces cerevisiae telomerase template mutants. Although growth rates and telomere profiles varied widely among the tlc1 mutants, chromosome separation and segregation were always aberrant. The mutants showed defects in sister chromatid separation at centromeres as well as telomeres, suggesting activation of a cell cycle checkpoint. Deletion of the DNA damage response genes DDC1, MEC3, or DDC2/SML1 failed to restore chromosome separation in the tlc1 template mutants. These results suggest that mutant telomere sequences elicit a checkpoint that is genetically distinct from those activated by deletion of telomerase or DNA damage.
INTRODUCTION
Telomeres, the ends of linear chromosomes, are DNA–protein complexes required for the complete replication of DNA and for chromosome stability (Blackburn, 2000c, 2001). The ribonucleoprotein enzyme telomerase adds DNA repeat sequences to telomeres (Greider and Blackburn, 1985; Greider and Blackburn, 1989). Deletion of telomerase causes progressive shortening of telomeres in dividing cells and eventual cellular senescence (Blackburn, 2000b).
Telomerase contains an enzymatically catalytic protein subunit (Est2p in Saccharomyces cerevisiae, TERT in other organisms) and an RNA molecule that contains a short template sequence (TLC1, TER) (Counter et al., 1997; Nakamura et al., 1997; Weinrich et al., 1997; Bryan et al., 1998). Like other reverse transcriptases, a triad of aspartates in the conserved reverse transcriptase domain directly participates in catalysis and is essential for telomerase activity (Counter et al., 1997). The templating sequence within the telomerase RNA component not only provides the sequence information used by telomerase to direct synthesis of new telomeric DNA but also contributes to other enzymatic properties. In Tetrahymena, single-base mutations in the template cause primer slippage, loss of fidelity, and premature dissociation of product (Gilley et al., 1995; Gilley and Blackburn, 1996). More dramatically, a three-base change in the template region of TLC1 in S. cerevisiae telomerase, tlc1-476gug, completely abolishes enzyme activity in vitro and in vivo (Prescott and Blackburn, 1997). Single or double point mutations to the same three bases mutated in 476gug still retained in vitro core telomerase activity, suggesting that the ablation of activity in the triplet gug mutant results from the combined effect of all three substitutions (Prescott and Blackburn, 2000).
In addition to its templating and enzymatic properties, the telomerase RNA template also affects telomere length regulation. Telomere length is maintained within a tight range characteristic of a given organism (Greider, 1996). The TLC1 template sequence normally directs the synthesis of telomeric TG1–3 repeats, which contain specific DNA binding sites for proteins involved in telomere length regulation and protection. Thus, changes within the templating sequence can have a direct influence on the binding of these proteins and consequently, can influence telomere length and integrity. In S. cerevisiae, sequence-specific binding of Rap1p to telomeric DNA nucleates a higher order DNA–protein complex that controls the accessibility of nucleases, telomerase, and proteins involved in recombination and DNA repair. This structure protects the telomeres from degradation and maintains a tight species- and strain-specific length distribution (Hardy et al., 1992; Kyrion et al., 1992; Marcand et al., 1997; Wotton and Shore, 1997; Krauskopf and Blackburn, 1998).
Telomerase RNA template mutants have been expressed and characterized in budding yeasts, mammalian cells, and Tetrahymena (Blackburn, 2000a). They cause incorporation of mutant telomeric DNA sequences, in some cases, leading to uncontrolled elongation, degradation, and increased single strandedness at telomeres (Blackburn, 2000a). In the budding yeast Kluyveromyces lactis, certain template mutant cells caused “monster cell” phenotypes, characterized by variable and often increased DNA content in enlarged and misshapen cells (Smith and Blackburn, 1999). In Tetrahymena, template mutations cause chromosome fusion, failed chromosomal separation, and accumulation of cells in late anaphase (Kirk et al., 1997). However, it is not known if mutant telomere sequences are seen as DNA damage or how template mutations affect cell cycle progression.
Here, we systematically examine the effects of mutating a core 3-base region of the template sequence of S. cerevisiae RNA. Our collection of 63 mutants, together with wild type, correspond to every possible sequence of template positions 474, 475, and 476 of TLC1. We examined telomere profile and growth phenotype for all mutants and classified them into six categories. We chose three representative mutants in which telomeres were respectively long, very short, or extensively degraded; in each mutant, we examined cell morphology, budding kinetics, chromosome dynamics and activation of DNA damage checkpoints. Hence, our results indicate that mutant telomeric sequences elicit a checkpoint response that is distinct from the DNA damage or telomerase loss checkpoints.
MATERIALS AND METHODS
Yeast Strain Construction
All yeast strains used in this study (except for intermediate strains yEHB5012, yEHB5013, yEHB5025, and yEHB5026 described below) are listed in Table 1 and were constructed using standard genetic techniques. Plasmid and oligo sequences are available upon request. Diploids were isolated on selective media at 23°C and subsequently sporulated at 23°C. Strain yEHB4003 was made in the S288C genetic background (Brachmann et al., 1998) and was constructed by disrupting the TLC1 gene with TRP1 and the RAD52 gene with LEU2. The strain carries pRS316TLC1, a CEN/ARS, URA3 plasmid containing the wild-type TLC1 with its endogenous promoter and terminator.
Table 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| yEHB4003 | MATαade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 tlc1Δ::TRP1 rad52Δ::LEU2 {pRS316TLC1} |
| yEHB5001 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δlys2Δ |
| yEHB5004 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1 ade2-1 can1-100 bar1Δlys2ΔtelIV::LacO::LEU2 |
| yEHB5031 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δlys2Δ tlc1Δ::KAN {pRS313-tlc1-aCA-(D)} |
| yEHB5032 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1 ade2-1 can1-100 bar1Δ lys2Δ telIV::LacO::LEU2 tlc1Δ::KAN {pRS313-tlc1-aCA-(D)} |
| yEHB5033 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δlys2Δ tlc1Δ::KAN {pRS313-tlc1-Cuc-(E)} |
| yEHB5034 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1 ade2-1 can1-100 bar1Δ lys2Δ telIV::LacO::LEU2 tlc1Δ::KAN {pRS313-tlc1-Cuc-(E)} |
| yEHB5035 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δlys2Δtlc1Δ::KAN {pRS313-tlc-1Cgg-(SS)} |
| yEHB5036 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1 ade2-1 can1-100 bar1Δ lys2Δ telIV::LacO::LEU2 tlc1Δ::KAN {pRS313-tlc-1Cgg-(SS)} |
| yEHB5029 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ cdc13-1 |
| yEHB5076 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ top2-4 |
| yEHB5077 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1 ade2-1 can1-100 bar1Δlys2Δ telIV::LacO::LEU2 top2-4 |
| yEHB5092 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δcdc13-5 |
| yEHB5094 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1 ade2-1 can1-100 bar1Δlys2ΔtelIV::LacO::LEU2 cdc13-5 |
| yEHB5056 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN TLC1::tlc1-aCA-(D)::HIS3 |
| yEHB5115 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δlys2Δ ddc1Δ::KAN |
| yEHB5121 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ LYS2 mec3Δ::TRP |
| yEHB5122 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ LYS2 ddc1Δ::KAN TLC1::tlc1-aCA-(D)::HIS3 |
| yEHB5097 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::URA3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ mec3Δ::TRP TLC1::tlc1-aCA-(D)::HIS3 |
| yEHB5150 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN ddc2Δ::NAT sml1Δ::ADE |
| yEHB5158 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN ddc2Δ::NAT sml1Δ::ADE {pRS313-tlc1-aCA-(D)} |
| yEHB5161 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN ddc2Δ::NAT sml1Δ::ADE {pRS313-tlc1-Cuc-(E)} |
| yEHB5164 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN ddc2Δ::NAT sml1Δ::ADE {pRS313-tlc-1Cgg-(SS)} |
| yEHB5144 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC1-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc1-Cuc-(E)} |
| yEHB5145 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC1-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc1-aCA-(D)} |
| yEHB5146 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC1-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc-1Cgg-(SS)} |
| yEHB5147 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC2-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc1-Cuc-(E)} |
| yEHB5148 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC2-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc1-aCA-(D)} |
| yEHB5149 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ddc1::DDC2-GFP-KanMX6 tlc1Δ::TRP1 {pRS313-tlc-1Cgg-(SS)} |
| yEHB5141 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ ddc2Δ::NAT sml11Δ::ADE |
| yEHB5203 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ ddc2Δ::NAT sml11Δ::ADE ddc1Δ::KAN |
| yEHB5204 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ cdc13-1 |
| yEHB5201 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ cdc13-1 ddc1Δ::KAN |
| yEHB5194 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ ddc2Δ::NAT sml11Δ::ADE cdc13-1 |
| yEHB5195 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ ddc2Δ::NAT sml11Δ::ADE cdc13-1 ddc1Δ::KAN |
| yEHB5185 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ mad2Δ::URA |
| yEHB5222 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN mad2Δ::URA {pRS313-tlc1-aCA-(D)} |
| yEHB5223 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN mad2Δ::URA {pRS313-tlc1-Cuc-(E)} |
| yEHB5224 | MATa ura3-1 leu2-3,112 his3-11::pCUP1-GFP12-LacI12::HIS3 trp1-1::LacO::TRP1 ade2-1 can1-100 bar1Δ lys2Δ tlc1Δ::KAN mad2Δ::URA {pRS313-tlc-1Cgg-(SS)} |
Plasmids are indicated in brackets.
For cytological assays, the W303 genetic background was used and template mutants were derived from yEHB5001 or yEHB5004 in which chromosome IV was marked as described previously (Straight et al., 1996), either 12 kb from the centromere (yEHB5001) or 100 kb from the telomere (yEHB5004) with 256 tandem repeats of the lactose repressor operator sequence. Both strains contain copper-inducible pCUP1-GFP12-lacI12::HIS3. The strains were modified for these experiments in three steps. First, the HIS3 marker was converted to URA3 by using pDS317 to create strains yEHB5012 from yEHB5001 (centromere marked) and yEHB5013 from yEHB5004 (telomere marked). Second, TLC1 was expressed using pRS317(LYS2), whereas the endogenous TLC1 was deleted and marked with KAN in yEHB5012(cen) and yEHB5013(tel) through polymerase chain reaction (PCR) integration to make yEHB5025(cen) and yEHB5026(tel). The integration product was made using primers oEHB4075 and oEHB4076 to PCR amplify pFA6a-kanMX6 (Longtine et al., 1998). And third, the tlc1 template mutations, tlc1aCA(D), tlc1Cuc(E), and tlc1Cgg(SS), were introduced on pRS313. Template mutants were passaged six times after counterselection of TLC1. Cells from the sixth passage were used for subsequent analyses or further genetic manipulation.
yEHB5029 (cdc13-1) was made by crossing yEHB5025 with yEHB5023. Strains yEHB5076 and yEHB5077 (top2-4) were a gift from N. Bhalla (University of California, San Francisco, San Francisco, CA). Centromere-marked (yEHB5092) or telomere-marked (yEHB5094) strains of cdc13-5 were made by disruption of CDC13 in yEHB5025(cen) and yEHB5026(tel), by using pVL1215 (pEHB5005), a gift of V. Lundblad (Baylor College of Medicine, Houston, TX). In yEHB5056 (tlc1(D)) TLC1 was deleted and marked with KAN through PCR integration, by using primers oEHB4075 and oEHB4076 to PCR amplify pFA6a-kanMX6 (as described above). TLC1 was expressed using pRS317(LYS2), and the tlc1(D) was introduced in pRS303. yEHB5115(Δddc1) and yEHB5121(Δmec3) were made by crossing a yEHB5025(cen) with yEHB5072 or yEHB5070, respectively (aka YJB4567 and YJB4527, both gifts from J. Berman, University of Minnesota, St. Paul, MN). yEHB5122(Δddc1, tlc1(D)) was made by crossing yEHB5056 with yEHB5072, and yEHB5097(Δmec3, tlc1(D)) was made by crossing yEHB5056 with yEHB5070. yEHB5150 (Δsml1,Δddc2,Δtlc1) was made in three steps: First, the deletion of SML1 was made by transformation by using PCR integration. Primers oEHB1100 and oEHB1101 (a gift from Simon Chan, University of California, San Francisco, CA) were used to amplify pRS402(ADE2). Product was integrated into yEHB5025(cen) to make yEHB5137. The deletion of DDC2 was made by transformation and PCR integration into yEHB5137 by using primers oEHB5023 and oEHB5019 for amplification of pAG25-NAT1MX4 to make yEHB5141. Deletion of TLC1 was carried out as described above for yEHB5025, and subsequent introduction of template mutations, (D), (E), and (SS), was done with pRS313 to create yEHB5158, yEHB5161, and yEHB5164, respectively.
yEHB10007(Ddc1-GFP) and yEHB10008(Ddc2-GFP) (a gift from Shang Li, University of California, San Francisco, CA), made in the S288C genetic background, were used as the parent strains for the construction of yEHB5144-5149. In these strains, TLC1 was deleted, and tlc1-template mutations were introduced as for yEHB4003 described above.
Construction of Template Mutants
Plasmid pR313TLC1 contains TLC1 with 614 base pair 5′- and 222 base pair 3′-flanking sequences inserted at BamHI-XhoI of pRS313 as reported previously (Prescott and Blackburn, 1997). Plasmid pRS313TLC1tempcassette was made by changing nucleotides 456G to C and 458A to T in TLC1 to create a SphI site, and by changing nucleotide 490T to C and inserting G at nucleotide 490 to create a SalI site. Primers oEHB4031 and oEHB4032, which have randomized nucleotides corresponding to positions 474–476 of TLC1, were annealed and cloned into the SphI and SalI sites of pRS313TLC1tempcassette. Transformants were sequenced and each mutant was identified to create the whole collection of template mutants.
Southern Blot Analysis of Telomeres
Strain yEHB4003, carrying pRS316TLC1, was transformed with various mutant TLC1 plasmids. Cells were grown in -Ura-His medium to keep both the wild-type and mutant plasmids (streak 0). They were streaked on 5-Fluoroorotic acid-His to select against the wild-type TLC1 plasmid (streak 1). Cells were then streaked on -His plates continuously. Genomic DNA was prepared from cells after certain numbers of streaks as indicated, digested with XhoI, and run on 0.8% agarose gels. DNA was transferred from gels to Hybond N+ membranes and probed with a γ32P end-labeled wild-type telomeric repeat oligonucleotide as described previously (Prescott and Blackburn, 1997). A similar protocol was used to confirm the telomere profiles of template mutants used for cell cycle analysis.
Telomere Cloning and Sequencing
Telomere cloning was done as described previously (Tzfati et al., 2000). Briefly, genomic DNA was ligated to a 3′ end amino-modified oligo RA20. The ligated genomic DNA was PCR-amplified with oligos RA23 and 1SUBT. The PCR product was gel purified, digested with EagI and PstI, and cloned into pBluescript KS-. Clones were then sequenced.
Cytological Techniques and Microscopy
Microscopy to analyze chromosome dynamics was performed using an Eclipse E600 microscope (Nikon, Tokyo, Japan) with a 100× PL APO 1.4 numerical aperture oil immersion objective. Data were visualized with a Coolsnap fx charge-coupled device camera and software (Roper Scientific, Tucson, AZ). CuSO4 was added to a final concentration of 0.25 mM to all experiments involving strains with marked chromosomes to induce expression of the green fluorescent protein (GFP)-LacI fusion. All chromosome analysis experiments were carried out by arresting cells in 1 μg/ml α-factor (Bio-Synthesis, Lewisville, TX) at 23°C for 4 h and then washing cells twice in α-factor–free media. Cells were then resuspended in fresh YPD at 23°C, and 1-ml samples were collected every 20 min and held on ice until a time course was complete. To fix cells, harvested samples were pelleted and resuspended in 100 ml of 4% paraformaldehyde, 3.4% sucrose at room temperature for 15 min. Cells were washed once in 1 ml of 0.1 M potassium phosphate, 1.2 M sorbitol buffer and resuspended in the same buffer. Cells were sonicated before microscopy. Only cells that responded to α-factor were scored. Indirect immunofluorescence was carried out as described previously (Rose et al., 1990). 4′6-Diamidino-2-phenylindole was obtained from Molecular Probes (Eugene, OR) and used at 1 mg/ml final concentration. Rat anti-α-tubulin antibodies were obtained from Accurate Chemical & Scientific (Westbury, NY) and used at a 1:1000 dilution. Goat anti-rat Texas Red antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA) and used at a 1:1000 dilution. For quantification of Ddc1-GFP and Ddc2-GFP foci, 1-ml samples were harvested, held on ice, and visualized live, without fixation. In all microscopy experiments, at least three sets of 100 cells for each time point were counted.
RESULTS
Growth and Telomere Phenotypes of 63 TLC1 Template Mutants
We systematically mutated each of the three nucleotides corresponding to TLC1 positions 474–476 to all possible sequences, to determine which of these template bases are required for yeast telomerase activity in vivo. This resulted in a complete collection of 63 mutants. To prevent generation of telomerase-independent, Rad52p-mediated survivors (Lundblad and Blackburn, 1993), which might complicate the interpretation of telomere length profiles, we deleted the RAD52 gene.
We analyzed the growth phenotypes of all 63 mutants. Only the tlc1-476gug led to complete loss of telomerase activity and senescence identical to that caused by tlc1 deletion described originally (Prescott and Blackburn, 2000). Cells that express tlc1-476gug and are Δrad52 stopped growth completely 50–75 generations after the loss of the wild-type TLC1, with no survivors generated (Prescott and Blackburn, 2000). In contrast, the other 62 mutants were still able to form colonies 20 streaks (∼400–500 generations) after removal of the wild-type TLC1. Mutants showed different degrees of compromised growth, based on colony size. We scored each mutant for growth and the results are summarized in Figure 1C. Figure 1D shows the growth of one representative mutant from each growth class at the sixth streak after loss of the wild-type TLC1.
Figure 1.
Summary of telomere profile and growth phenotypes of 63 mutants. (A) Sequence of the template cassette. The wild-type TLC1 sequence surrounding the template region is shown on top. The template region is in capitals and positions 476–474 are underlined. Mutations that create SphI and SalI sites are in capitals. The SphI and SalI sites are underlined with arrows pointing to the bases where the restriction enzymes cut. Sequence of the Rap1p consensus binding site is shown in 3′-5′ direction underneath. (B) Sequences of the oligos used to construct the template mutant library. (C) Summary of telomere profile and growth phenotypes of 63 mutants. Sequence is shown in 3′-5′ direction. Telomere length for each mutant is characterized as described in text. Cell growth for each mutant is scored as wild type or close to wild-type growth (+++), distinguishably sicker than wild type (++), very sick (+), or senescent (-). Stars indicate mutants that showed immediate slow growth but later recovered. Mutants that were recovered in the screen in Forstemann et al. (2003) are checked. Mutants that were further analyzed for their cellular phenotypes as described in RESULTS are marked by dots. (D) Growth of representative mutants was scored as described in C. Cells were scored ∼100 generations (five streaks) after the loss of the wild-type sequence. Cell were grown in liquid culture to OD600 = 1.0 and serially diluted by 1:3. Equal volumes of the diluted cultures were spotted on the plate. The sequence and the telomere profile class for each mutant are indicated.
The 63 tlc1 template mutants fell into six classes based on Southern blot analyses of their telomere length profiles: 1) wild-type length (WT); 2) progressively shortened telomeres, which led to senescence at the same rate as telomerase-null cells (S); 3) elongated telomeres (E); 4) mixed populations of telomeres (short, but tightly regulated plus elongated with a broad size distribution) (M); 5) elongated and degraded telomeres (D); and 6) short but stably maintained telomeres (SS). The Southern blot analysis results are summarized in Figure 1C, and representative blots are shown in Figure 2.
Figure 2.
Southern blots of one representative mutant from each class. Genomic DNA was prepared from cells after the indicated number of streaks, and then was digested with XhoI and probed with a wild-type telomeric repeat oligonucleotide. The sequences of positions 476–474 of TLC1 for each mutant are shown on the top. For pRS313 and 476gug, cells for last time point were ∼10 generations before senescence.
Only one template mutant, tlc-476aCc, had a wild-type telomere profile with ∼350-base pair-long telomeric repeat tracts and a normal growth phenotype. The template RNA directs the synthesis of the Rap1p binding site and Rap1p binding is strongly influenced by mutations in the 474–476 sequence (Prescott and Blackburn, 2000). This “wild-type” allele has two point mutations, 476C to A and 474A to C. It can potentially copy this template into TGTGTGTGGGTGG repeats, with a 10/13 nucleotide match to the Rap1p consensus binding site (Figure 1A). Apparently, sufficient Rap1p binding affinity is retained by the sequences incorporated into these mutant telomeres to support normal telomere length regulation.
Telomeres in the five elongated (E) mutants, 476auA, 476CuA, 476Cuc, 476Cug, and 476uuc were much longer than wild type. All five mutant sequences share a common C to U change at position 475. This is consistent with previous results that this position is critical for Rap1p binding (Krauskopf and Blackburn, 1998). The telomeres in two of these mutants, 476uuc(E), and 476Cuc(E), were >10 kb at the 10th streak, longer than any previously reported S. cerevisiae mutant.
Initial shortening of telomeres followed by rapid elongation was a feature common to three classes of mutants: E, M, and D. In these mutants, telomeres shortened during the first 50–100 generations after loss of the wild-type TLC1 (Figure 2; see 0 and 1 streak lanes). This was followed by rapid deregulation of telomere length within the next ∼50 generations. For the E mutants, the initially shortened telomere population disappeared and the entire population became lengthened. In contrast, in the M mutants, the shortened telomere subpopulation became stabilized for at least another 300 generations. A comparable population of shortened telomeres was also maintained in a subset of the D mutants with mild degradation, but it was not evident in D mutants with severely degraded telomeric DNA (see Figure 2 and below for more discussion of D mutants).
Similar mixed telomere phenotypes were previously reported for telomerase RNA template mutants in the yeast K. lactis (Krauskopf and Blackburn, 1996; Krauskopf and Blackburn, 1998). In these mutants, which retained the Rap1p consensus binding site and normal in vitro Rap1p binding, telomeres were initially well-regulated at shorter-than-wild-type length for many generations, but they subsequently underwent rapid lengthening. It was proposed that this rapid elongation occurred upon the eventual replacement of the bulk of the wild-type telomeric tract by mutant sequence (Krauskopf and Blackburn, 1998). Our mutants may reflect a similar situation.
Three tlc1 Template Mutants Caused High Misincorporation Rates
The fact that 62 mutants continued to grow for hundreds more generations than Δtlc1 or tlc1-476gug mutants, in the absence of Rad52p, indicated that these mutant telomerases are active in vivo. To confirm telomerase activity, we analyzed the telomeres of three mutants for the incorporation of the predicted mutant nucleotides, by using a previously developed PCR-based technique (Tzfati et al., 2000). As expected, mutant sequences were found in the telomeres, sometimes as multiple repeats (see Figure 3 for representative clones). A striking finding was that, in addition to the expected mutant sequences, all three mutants contained significant numbers of misincorporated bases in their telomeres (indicated as bold and italicized bases in Figure 3). Figure 3D lists all the misincorporated repeats found. 476agc(SS) and 476Cuc(E) have four and three misincorporated repeats of 42 total repeats synthesized by the mutant telomerase respectively (10 and 7%). 476uug(SS) has four misincorporated repeats of 31 total repeats (13%). As controls, telomeres were cloned from cells expressing only the wild-type TLC1. These contained no misincorporated bases of the ∼2200 bases sequenced (Lin and Blackburn, unpublished data). Thus, like Tetrahymena (Gilley et al., 1995), mutations in the template region of yeast telomerase also cause reduced fidelity. This is the first report of this type of base misincorporation by mutant-template telomerases in vivo for S. cerevisiae.
Figure 3.
Three mutants have high misincorporation rates. (A–C) Representative telomeric sequences cloned from three mutants. (D) Complete list of misincorporated sequences seen in three mutants. The TG-rich strand is shown. Correctly matched nucleotides that are synthesized from the mutant template are in lowercase. Misincorporated nucleotides are underlined and in bold.
The telomeres in tlc1-476agc(SS) and tlc1-476uug(SS) were both shorter than wild type, but stably maintained. However, by the sixth streak after loss of the wild-type TLC1, tlc1-476agc(SS) grew like wild type, whereas tlc1-476uug(SS) was very sick (Figure 1). Telomeres were cloned from these two mutants at the sixth streak. In both cases, the telomeres contained tandem repeats of mutant sequence, indicating that the enzyme was able to copy the template completely (see Figure 3, A–C, for representative clones). Therefore, the contrasting growth properties of these two mutants did not reflect any obvious difference in their efficiency of mutant repeat incorporation or length maintenance.
Telomeres of tlc1-476Cuc(E) became much longer than wild-type 120 generations after removal of wild-type TLC1 (Figure 2). To clone full-length telomeres from tlc-476Cuc(E), genomic DNA was extracted from the first streak after removal of the wild-type TLC1, when the bulk telomere length was still similar to wild type. Of the eight cloned tlc1-476Cuc(E) telomeres, two contained only wild-type sequences, but the other six contained up to 14 repeats of the expected mutant sequence (our unpublished data). Interestingly, long tracts of wild-type telomeric sequence were interspersed with mutant sequences. Because these telomeres were cloned from rad52Δ cells that contained only mutant tlc1 for ∼30 cell generations, we speculate that these wild-type sequences resulted from copying the parts of the template that remained wild type in 476Cuc(E). The enzyme may have dissociated before it reached the mutant sequence, or the mutant DNA was cleaved off.
Degraded Telomeres Are Associated with an Immediate Slow Growth Phenotype
Eleven of the 63 mutants had telomeres that looked both elongated and degraded (D). As reported previously for tlc1–476A [here referred to as tlc1-aCA(D)] (Chan et al., 2001), telomeric DNA hybridization signal in Southern blots from these cells was extremely broad, extending from the wells of the gel to the bottom, with no discrete bands (Figure 2). No common pattern of base substitution was discernible for this telomere profile class: although 476CaA, 476Cac, and 476Cag share a common C-to-A mutation in position 475, in 476aCA, 476gCA, and 476uCA, positions 475 and 474 are still wild type. The severity of the degradation phenotype varied from very severe (476aCA, 476gCA, 476uCA, 476Cac, and 476CCg) or intermediate (476Cgc, 476CaA, 476aCu, 476uac, and 476Cag) to least severe, in which isolated bands become distinguishable (476uCA; Figure 1C).
The four (D) mutants with the most severe telomeric DNA degradation phenotype showed slower growth within ∼20 generations after the loss of the wild-type TLC1 (Figure 1C). These mutants had heterogeneous colony sizes and extended population-doubling times (Figure S1; our unpublished data). This immediate slow growth phenotype, with high percentages of enlarged, misshapen monster cells, was reported previously for tlc1476aCA(D) (Chan et al., 2001). This mutant telomerase was active and the predicted mutant sequence was incorporated into telomeres in vivo, likely causing these phenotypes (Chan et al., 2001). Such behavior contrasts with the delayed phenotype characteristic of senescence, which only becomes apparent at 50–75 generations after the loss of functional TLC1. Furthermore, senescent rad52Δ cells stop growing completely 50–75 generations after the loss of the wild-type TLC1. In contrast, the (D) mutant cells apparently adapted, regaining relatively healthy growth after 50 generations and continuing to grow throughout the rest of the experiment (>400 generations). Thus, we conclude that the immediate slow growth in (D) mutants is the consequence of incorporation of mutant telomeric sequence rather than lack of telomerase activity.
The tlc1 Template Mutations Cause Aberrant Chromosome Separation
To further analyze the cellular consequences of mutant-sequence telomeres, we examined budding kinetics and chromosome dynamics in representatives of three distinct tlc1 mutant telomere length classes: tlc1-476aC(D), tlc-4761Cuc(E), and tlc1–476Cgg(SS). These cause degraded, elongated, and short and stable telomeres, respectively. Although some cellular phenotypes of telomerase RNA template mutants have been described previously in other eukaryotes (Kirk et al., 1997; Smith and Blackburn, 1999; Kim et al., 2001), direct analysis of their chromosome behavior has not been reported. To visualize chromosome movement in individual cells, we used a method that marks a single chromosome with a green spot at a specified location (Straight et al., 1996, 1997) (diagrammed in Figure S2A), by using a tandem array of lac operators inserted either 12 kb from the centromere (centromere marked) or 100 kb from one telomere (telomere marked) of chromosome IV, the largest chromosome in S. cerevisiae (Figure S2A). Lactose repressor fused to GFP (GFP-LacI) expressed in these cells allows visualization of chromosome IV, which during the course of a normal cell cycle, is seen as one green spot (unreplicated or replicated but unseparated) or two spots (replicated and separated). The three tlc1 template mutations were transformed into haploid strains containing either the centromere or telomere-marked chromosome IV. After release from an α-factor arrest, we compared the timing of sister chromatid separation in the centromere and telomere marked strains. Budding kinetics, sister chromatid separation, and chromosome segregation were measured for all strains during the course of a single cell cycle.
Based on the behavior of template mutants in Tetrahymena (Kirk et al., 1997), our initial expectation was that these assays might reveal mutant chromosomes that were able to separate at their centromeres but that had delayed separation, or remained attached, at their ends. Such behavior is also exhibited by the previously characterized yeast topoisomerase II mutant top2-4. Top2p functions to resolve the concatenations between sister chromatids so they can fully separate from each other at anaphase (DiNardo et al., 1984; Uemura et al., 1987; Holm et al., 1989; Shamu and Murray, 1992). Using chromosome IV GFP-marked at the centromere, the telomere, or midway along a chromatid arm (Bhalla et al., 2002), it was shown that in the top2-4 mutant, centromere separation precedes telomere separation. Therefore, as a control, we first confirmed that our assay revealed such differential centromere and telomere separation kinetics in top2-4 mutant cells. Cells were synchronized in G1 by growth in α-factor for 4 h, followed by release into fresh medium without α-factor. Samples were collected every 20 min and fixed for later analysis. Only “shmooed” cells, capable of growing and responding to α-factor, were considered for this analysis. Cells were counted and classified as unbudded, or small, large, or rebudded. We confirmed that the telomerically located GFP spots in top2-4 cells separated more slowly than centromeric GFP spots in the first cell cycle after α-factor release, reassuring us that this assay could, in our hands, reveal defects that lead to telomere fusion, but still allow centromere separation (Figure 4B; our unpublished data).
Figure 4.
Template mutations lead to impaired chromosome separation and segregation. Strains bearing tlc1 template mutations contained Lac operator repeats integrated at the TRP1 locus near the centromere (yEHB5025-derived) or near the telomere (yEHB5026-derived). Template mutations were well-established at telomeres before cytological analysis (six passages, ∼120 generations). (A) Classes 1–7 show all spot-conformations that were observed during microscopic analysis in either cen- or telmarked strains. Sister chromatid separation and sister chromatid segregation were scored as indicated. (B) Distribution of marked chromosomes at representative time point, t = 100, for Wt cen; top2-4cen and top2-4tel; tlc1cen mutants (D) (E), and (SS); cdc13-1cen at 30°C and 23°C; cdc13-5cen; cdc13-1cen in combination with Δddc1, Δddc2, or both at 23°C and Δmad2 alone or in combination with tlc1cen (D).
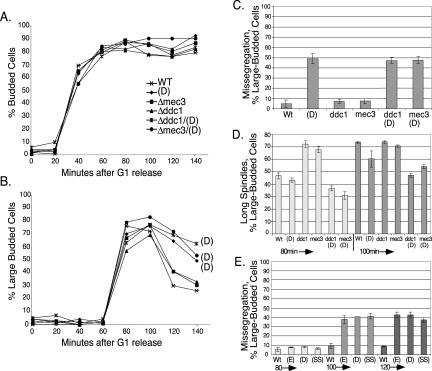
We then carried out the same analysis for the tlc1(D), (E), and (SS) mutants. After release from α-factor, budding kinetics for all three mutants were the same as wild type, suggesting that they proceed with normal kinetics through the cell cycle. Fluorescence-activated cell sorting analysis showed that the tlc1 cells accumulate 2N DNA content at rates similar to wild type for the (E) and (SS) mutants, and with slightly delayed kinetics for the (D) mutant tlc1-476aCA (Figure S2, B and C). However, in all three mutants, chromosome segregation was defective. Chromosome dynamics after release from α-factor were assayed by counting additional cells every 20 min and sorting them into seven categories based on the number and position of GFP-marked chromosome spots (Figure 4). In all GFP-chromosome assays, only shmooed cells, those responding to α-factor and still capable of entering the cell cycle, were scored. In shmooed, large-budded cells, categories 1–3, the most common, are found during the course of a normal WT cell cycle. Categories 4–7 are rare in WT cells but were found in significant numbers in the tlc1 mutants. Sister chromatid separation was scored as the percentage of cells in categories 2, 3, 4, 5, and 7. Chromosome segregation was made up of category 3 only, and consists of cells with sister chromatids properly separated and segregated into mother and daughter cells. Chromosomes unsegregated was measured as the percentage of cells in all categories except 1, 3, and 6, until the 100-min time point and beyond, when categories 1 and 6 were included. The (D) mutant, in particular, had 2–10% monster cells, with more in early time points, and the (SS) mutant accumulated ∼5% monster cells in some later passages. Total cell viability (colony-forming ability) was quantified and was ∼90% of wild type for all three mutants (our unpublished data). Monster cells were not included in the GFP-chromosome assays. Figure 4A summarizes how cells were scored and Figure 4B shows the distribution of cell categories at the 100 min time point for several strains.
In all three tlc1 mutants, quantification of the above categories uncovered aberrant separation of replicated GFP-marked chromosomes and improper segregation of the marked chromosomes to daughter cells. Figure 5A shows that in wild-type cells, sister chromatid separation began 80 min after release from G1 and was complete by 120 min. In tlc1 mutant cells, sister separation also began at 80 min, but the percentage of cells in which chromatids separated, or went on to segregate, was never as high. This low percentage of separation and high degree of unsegregated chromosomes was a consistent property of all three tlc1 template mutants (Figure 5B). Strikingly, for all tlc1 template mutants, there was no difference in segregation kinetics between the centromere and telomere-marked strains. This suggested that the lack of separation was occurring along the entire length of the chromosome, not just at the telomeres. Furthermore, for cells with a single spot at late time points (classes 1 and 6 in Figure 4A), the spot occurred in daughter cells almost as often as mother cells. As noted above, the high viability of the template mutant strains (our unpublished data) indicated that the 20–30% of shmooed cells with chromosome abnormalities were not simply dead.
Figure 5.
Analysis of chromosome dynamics. (A) Sister chromatid separation is delayed in the tlc1 template mutants, but there is no difference between centromere and telomere marked strains. (B) The number of chromosomes that fail to separate and segregate in the template mutants is high for all three classes, (D), (E), and (SS), but there is no difference between centromere- and telomere-marked strains.
The Mutant-Telomere Response Is Distinct from Known DNA Damage Checkpoints
One explanation for a whole-chromosome delay in chromatid separation and segregation could be activation of a cell cycle checkpoint. Therefore, we used our assay to examine chromosome dynamics by using two mutant alleles of CDC13 that cause telomere perturbations: cdc13-1, which activates a cell-cycle arrest, and cdc13-5, which does not. When grown at the nonpermissive temperature (30°C), the cdc13-1 mutant contains extensive single-stranded DNA at telomeres and arrests in G2/M with a large bud and a single nucleus, via the RAD9-dependent checkpoint pathway (Burke and Church, 1991; Weinert and Hartwell, 1993; Garvik et al., 1995). Figure 4B shows the breakdown of marked chromosome segregation categories at the 100-min time point for wild-type cells, the tlc1 template mutants, cdc13-5 and cdc13-1 mutants at different temperatures. When grown at 30°C, cdc13-1 accumulated large-budded cells with single chromosome spots, as expected for cells that arrest at G2/M with a single nucleus. Interestingly, when grown at the permissive temperature (23°C), cdc13-1 behaved like the tlc1 template mutants: the percentage of unsegregated chromatids was high, with no significant difference in kinetics between telomere and centromere-marked strains (Figure 4B; our unpublished data). We combined cdc13-1 with deletions of DDC1 and/or DDC2, components of the DNA damage checkpoint and found that the missegregation phenotype of cdc13-1 at 23°C is completely relieved by combining cdc13-1 with Δddc1, Δddc2, or Δddc1Δddc2 (Figure 4B and Figure S4). This suggests that the DNA damage checkpoint is activated in response to the telomere damage created by the cdc13-1, even at 23°C.
The mutant carrying the cdc13-5 allele neither activates the DNA damage checkpoint nor causes a temperature-sensitive phenotype, even though it has long, G-rich, single-stranded telomeric overhangs during S phase (Chandra et al., 2001). cdc13-5 also contained unsegregated chromatids, with equal fractions seen in both the telomere- and the centromere-marked strains (Figure 4B; our unpublished data). Hence, mutations of a telomeric component, Cdc13p, other than in the TLC1 template also lead to a chromosome segregation defect. Furthermore, in the case of the cdc13-5 mutant, this segregation defect occurs independently of activation of any previously described known checkpoint.
Complete deletion of TLC1 causes cell cycle arrest, activates a checkpoint (Enomoto et al., 2002; IJpma and Greider, 2003), and causes chromosomal fusions when telomeres become short (Hackett et al., 2001). Consistent with this, we examined chromosome dynamics in an asynchronous Δtlc1 culture and found that 60% of cells were large budded (compared with ∼30% in an asynchronous wild-type culture). Among the large-budded cells, only 17% had properly separated and segregated chromosomes (class 3). The majority, 75%, had a single spot (classes 1 and 6) and the rest fell into the aberrant segregation classes (4, 5, and 7). Because most of these cells were arrested, they did not respond to α-factor and thus we could not do synchronized time course analysis.
If the tlc1 template mutations activate a DNA damage checkpoint that blocks chromosome separation, then disruption of the checkpoint-sensing complexes might restore normal chromosome dynamics. In our assay, the restoration would manifest as a higher percentage of large-budded cells with two fully separated and segregated GFP spots (class 3 cells). DDC1, MEC3, and DDC2 were shown to be important for activating the checkpoint in response to loss of telomerase in a Δtlc1 background (Enomoto et al., 2002; IJpma and Greider, 2003). Ddc1p is a member of the PCNA-like trimer that loads directly at sites of DNA damage, and Ddc2p associates with Mec1p (an ATR kinase) in a complex that is also recruited to sites of damage (Melo and Toczyski, 2002). First, we combined tlc1(D) with deletion of DDC1, DDC2, or MEC3 and examined budding and chromosome dynamics for each single and double mutant (Figure 6). Overall, budding kinetics was the same for all strains (Figure 6A). However, in both tlc1(D) and the double mutants tlc1(D)/mec3 and tlc1(D)/ddc1, the number of cells with large buds was higher than wild type, beginning at 100 min, and remained high for the rest of the time course (Figure 6B). Furthermore, like tlc1(D), the double mutants were slow to separate sister chromatids and showed a high degree of unsegregated chromosomes (Figure 6C; our unpublished data).
Figure 6.
(A) Overall, budding is normal for tlc1(D), Δddc1, Δmec3, and tlc1(D) double mutants, but (B) there is an enrichment of large-budded cells in tlc1(D); tlc1(D)Δddc1; and tlc1(D)Δmec3. (C) The number of unsegregated chromosomes is as high in tlc1(D)Δddc1 and tlc1(D) Δmec3, as it is in tlc1-(D). Data for the 100-min time point is shown. (D) The number of cells with long spindles rapidly increases in DNA damage checkpoint mutants Δddc1 and Δmec3 but remains low in tlc1(D), tlc1(D)Δddc1, and tlc1(D) Δmec3 at the 80- and 100-min time points (E) The percentage of unsgegregated chromosomes remains high for all three template mutants, (D), (E), or (SS), when combined with Δddc2/Δsml1.
Spindle staining with anti-tubulin antibody provided further evidence that neither DDC1 nor MEC3 is required for the checkpoint, resulting in the arrest of chromosome separation in tlc1(D) mutant cells. After release from G1 arrest, strains deleted for DDC1 or MEC3 alone accumulated cells with long spindles faster than wild type, as expected for cells that can no longer pause and respond to DNA damage (Figure 6D, 80-min time point bars). However, tlc1(D)/ddc1 and tlc1(D)/mec3 double mutants, like tlc1(D), all grew long spindles more slowly than wild-type cells or ddc1 or mec3 single mutants. The timing of this delay in spindle formation (Figure 6D) coincided with the slower chromosome segregation and the persistence of large-budded cells described above.
Next, we combined Δddc2/Δsml1 with each of the three tlc1 template mutations (E, D, or SS). Like Δmec1 lethality, Δddc2 lethality is suppressed by Δsml1. Again, the fraction of unseparated chromatids was comparable with that seen for the each template mutation alone (Figure 6E). Thus, in contrast to the telomere defect caused by the cdc13-1 mutation, mutant-template telomerase RNAs cause a DNA damage checkpoint-independent response.
Finally, because DDC1 and DDC2 were not required for the checkpoint, we tested whether the delayed spindle elongation in tlc1 template mutants is due to activation of the spindle assembly checkpoint pathway. To address this possibility, we combined each of the three template mutants (E, D, or SS) with Δmad2 and looked for restoration of proper chromosome segregation. However, chromosome missegregation remained high for all three mutants. This result suggests that the spindle assembly checkpoint is not solely responsible for detecting or responding to aberrations at mutant telomeres (Figure 4B and Figure S5).
In summary, our results show that aberrant-sequence telomeres, whether very short, elongated, or highly degraded, all arrest chromosome separation and lead to impaired chromosome segregation and failure to progress into anaphase. Furthermore, disruption of the DNA damage checkpoint or the spindle assembly checkpoint in cells with these telomere defects, via deletion of MEC3, DDC1, DDC2, or MAD2 does not restore cell cycle progression. These findings imply that neither of the major DNA damage-sensing complexes, nor the spindle checkpoint, is solely responsible for activating a cell cycle checkpoint in response to the type of telomere aberration caused by mutant telomeric DNA sequences.
DISCUSSION
Roles of Template Bases in Telomerase Enzymatic Activity
Here, we have analyzed all possible mutations within an essential three-base sequence at the core of the yeast telomerase RNA template sequence. We have shown that among the 63 substitution mutations of positions 474–476, only one, tlc1-476gug, completely abolishes enzymatic activity, implying that telomerase enzyme activity will tolerate almost any sequence at TLC1 positions 474–476 except 476gug. However, although the remaining mutants do not senesce, many of them (34/63, or 54%) remain compromised for telomerase function in vivo, as judged by their shortened telomeres.
Further evidence for altered enzymatic activity came from sequencing the telomeric DNA cloned from several of our mutants. Telomerase normally makes a faithful copy of its RNA template sequence. Specific mutations of the Tetrahymena telomerase template cause primer slippage, loss of fidelity, and premature dissociation of products (Gilley et al., 1995; Gilley and Blackburn, 1996). Here, we report, for the first time in S. cerevisiae that three tlc1 template mutations lead to high levels of base misincorporation. Hence, the ability of template bases to affect properties of the polymerization reaction may be general among telomerases.
Cellular Consequences of Mutating the Telomerase RNA
Thorough mutagenesis of a small essential template region showed that mutant telomeric sequences led to great variation in telomere integrity, with telomere profiles ranging from severe shortening to extensive lengthening. Many mutants had highly degraded telomeres, with no consistently maintained length. This wide variation of bulk telomere sizes confirms and extends previous work in other systems showing that template mutations can greatly influence both positive and negative regulation of telomere length (Krauskopf and Blackburn, 1996; Prescott and Blackburn, 1997; Krauskopf and Blackburn, 1998; Prescott and Blackburn, 2000; Chan et al., 2001).
The effects of our set of template mutants on cell growth also varied widely in severity and time of onset. Although the senescent phenotype of 476gug was caused by total loss of telomerase activity, several mutants that retained enzymatic activity grew slowly. Two types of slow growth were observed: an initial defect that improved with passaging, or normal growth followed by increased sickness in later passages. Mutants with severely degraded telomeres fell into the first group, with growth becoming faster ∼50 generations after the loss of wild-type TLC1. Growth continued for as long as cells were observed (>400 generations), despite the accumulation of progressively more degraded and single-stranded telomeric DNA. It seems that these cells incorporated mutations but then adapted, in a Δrad52 background, by unknown mechanism(s). The second group of slow growers contained short and stable telomeres, and the onset of slowing of growth was delayed to ∼100 generations after loss of the wild-type TLC1. We speculate that a critical number of mutant repeats had to be added to telomeres, possibly in combination with a critical degree of shortening, to affect cell growth.
Our mutant collection was assembled without stringent growth requirements. Recently, Forstemann et al. (2003) screened libraries of randomly mutagenized tlc1 template sequences for complementation of the Δtlc1 senescence phenotype. Thirty-two clones were recovered from a library containing random substitutions throughout positions 477–473. Most of them contained C/A-rich sequences, like the wild-type S. cerevisiae sequence. All 32 isolates had wild-type growth, and among them, six template sequences were the same as in the best growers of our collection (Figure 1C). The difference in our results can be explained by the more stringent growth requirements of the screen by Forstemann et al. (2003).
We found no simple correlation between telomere length and cell growth. Mutants with identical telomere length profiles in Southern blots grew well or poorly depending on individual sequence changes, indicating that the specific telomeric sequences, rather than bulk telomere length, determined their growth properties.
Mutant-Sequence Telomeres Elicit a Unique Checkpoint Response
Chromosome dynamics had not previously been analyzed in any telomerase template mutant in any organism. Interestingly, our direct analysis of chromosomes in three diverse template mutants revealed a common cell cycle arrest response: specifically, whether telomeres were short, long, or extremely degraded, tlc1 mutations consistently led to a high level of delayed sister chromatid separation and an increase in cells with unsegregated chromosomes. Furthermore, deletion of the known DNA damage checkpoint genes MEC3, DDC1 or DDC2 did not restore cell cycle progression or proper chromosome dynamics to template mutant cells. These findings imply that neither of the established DNA damage sensing complexes is solely responsible for activating a cell cycle checkpoint in response to telomere damage caused by mutant telomeric DNA sequences.
This cellular response to tlc1 mutations, even those that lead to shortened telomeres, is distinct from the response to telomeric shortening and senescence caused by TLC1 deletion. Enomoto et al. (2002) and IJpma and Greider (2003) have independently reported that when telomeres become short after deletion of TLC1, a G2/M arrest occurs that is dependent on MEC1, MEC3, DDC2, and RAD24. Despite the short telomeres of the (SS) mutants, there was no activation of the senescence phenotype, and the cellular response was different, because the components of the ATR complex Mec1p and Ddc2p were not required to activate a checkpoint in response to short, long, or degraded telomeres in the tlc1(SS), (E), and (D) template mutants. The Enomoto et al. (2002) study specifically ruled out involvement of Tel1p the ATM-kinase in S. cerevisiae, in the cell cycle arrest response to telomerase deficiency. These cell cycle responses imply that yeast has more than one mechanism for responding to mutant telomeres. Furthermore, response to telomerase deletion the yeast contrasts with results seen in human cells. Specifically, overexpression of a dominant-negative form of the human telomere-protective protein TRF2 causes a pronounced ATM-dependent cellular response (de Lange, 2002). Together, these results suggest that ATM and ATR have varied responses to different types of telomeric lesions, both within and between organisms.
Our results suggest that mutant telomeric sequences may not be seen by the cell in the same way as general genomic DNA damage, because deletion of DDC1 or DDC2 did not relieve the chromosome defect. However, all three mutations chosen for cellular analysis accumulated Ddc1-GFP and Ddc2-GFP foci in mutant cells, a characteristic associated with activation of a DNA damage checkpoint (Figure S3). Very early after introduction of mutant template tlc1 alleles (<20 generations), both Ddc1-GFP and Ddc2-GFP manifested as bright nuclear foci in a subpopulation of cells (Figure S3). The Ddc2-GFP foci persisted throughout six serial passages (∼120 generations). On further passaging, the number of Ddc1-GFP foci gradually increased in (SS) mutant cells but gradually decreased in the (D) mutant. These findings echoed the slow onset of growth defects in the (SS) mutants and the immediate growth defect seen for the (D) mutants, as described above; they may reflect DNA damage foci formed at sites of secondary DNA damage, such as chromosome breaks after end-to-end chromosome fusion, rather than foci on the telomeres.
Surveillance of mutant telomere sequences may require a combination of DNA damage checkpoint proteins and/or participation of more than one checkpoint pathway. This is the case with deletion of Taz1p, the Schizosaccharomyces pombe orthologue of hTRF2. Taz1p promotes proper chromosome segregation, DNA repair, and chromosome end protection: both DNA damage and spindle assembly checkpoint proteins are required for Δtaz1 cells to survive (Miller and Cooper, 2003). A similar response is seen in cells with deletions of the nonhomologous end-joining and telomere-protection proteins yKu70 and yKu80. ΔKu70 and ΔKu80 cells are temperature sensitive and have short telomeres and single-stranded Y′ subtelomeric repeats (Gravel et al., 1998; Polotnianka et al., 1998; Smith and Jackson, 1999). Maringele and Lydall (2002) recently showed that subsets of both the DNA damage checkpoint (CHK1, MEC1, and RAD9) and the spindle assembly checkpoint (MAD2) pathways are required for efficient cell cycle arrest of yKu70Δ mutants grown at the nonpermissive temperature. Other alterations of chromosome structure can activate multiple checkpoints, and many conditions that activate the DNA damage or DNA replication checkpoints also activate the spindle checkpoint (Garber and Rine, 2002). Notably, the arrest of cdc13-1 mutants is an exception; arrest in these cells is dependent on a large group of DNA damage checkpoint genes: CHK1, MEC1, and RAD9 as well as RAD17, RAD24, MEC3, DDC1, and DUN1, but it does not require the spindle assembly checkpoint pathway (Maringele and Lydall, 2002). In contrast, the tlc1 template mutant response shown here seems to work independently of the spindle assembly checkpoint. We have shown that mutant telomeric repeats elicit a response, directly or indirectly, that is distinct in its genetic dependence, from that induced by cdc13-1 telomerase deficiency or other DNA-damaging agents. Together, these findings suggest that defects at telomeres activate various checkpoint responses depending on the molecular nature of disruption to telomere integrity. A future challenge will be to link specific types of telomere damage to precise patterns of checkpoint activation.
Supplementary Material
Acknowledgments
We thank J. Berman for Δmec3 and Δddc1 strains; V. Lundblad for pVL1215, used to construct cdc13-5; Needhi Bhalla for top2-4 strains; Justine Melo and Genevieve Vidanes for technical advice with Ddc1-GFP and Ddc2-GFP protein imaging; and Shivani Nautiyal, Dan Levy, Carol Anderson, Sveta Makovets, Dave Toczyski, and Dave Morgan for critical reading of the manuscript.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–10–0740. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–10–0740.
Online version of this article contains supplementary material. Online version is available at www.molbiolcell.org.
References
- Bhalla, N., Biggins, S., and Murray, A.W. (2002). Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13, 632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, E.H. (2000a). The end of the (DNA) line. Nat. Struct. Biol. 7, 847-850. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2000b). Telomere states and cell fates. Nature 408, 53-56. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2000c). Telomeres and telomerase. Keio J. Med. 49, 59-65. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661-673. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Bryan, T.M., Sperger, J.M., Chapman, K.B., and Cech, T.R. (1998). Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. USA 95, 8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D.J., and Church, D. (1991). Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 3691-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.W., Chang, J., Prescott, J., and Blackburn, E.H. (2001). Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11, 1240-1250. [DOI] [PubMed] [Google Scholar]
- Chandra, A., Hughes, T.R., Nugent, C.I., and Lundblad, V. (2001). Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15, 404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter, C.M., Meyerson, M., Eaton, E.N., and Weinberg, R.A. (1997). The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. USA 94, 9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange, T. (2002). Protection of mammalian telomeres. Oncogene 21, 532-540. [DOI] [PubMed] [Google Scholar]
- DiNardo, S., Voelkel, K., and Sternglanz, R. (1984). DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. USA 81, 2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, S., Glowczewski, L., and Berman, J. (2002). MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann, K., Zaug, A.J., Cech, T.R., and Lingner, J. (2003). Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 31, 1646-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, P.M., and Rine, J. (2002). Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161, 521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik, B., Carson, M., and Hartwell, L. (1995). Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15, 6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley, D., and Blackburn, E.H. (1996). Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol. Cell. Biol. 16, 66-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley, D., Lee, M.S., and Blackburn, E.H. (1995). Altering specific telomerase RNA template residues affects active site function. Genes Dev. 9, 2214-2226. [DOI] [PubMed] [Google Scholar]
- Gravel, S., Larrivee, M., Labrecque, P., and Wellinger, R.J. (1998). Yeast Ku as a regulator of chromosomal DNA end structure. Science 280, 741-744. [DOI] [PubMed] [Google Scholar]
- Greider, C.W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65, 337-365. [DOI] [PubMed] [Google Scholar]
- Greider, C.W., and Blackburn, E.H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405-413. [DOI] [PubMed] [Google Scholar]
- Greider, C.W., and Blackburn, E.H. (1989). A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331-337. [DOI] [PubMed] [Google Scholar]
- Hackett, J.A., Feldser, D.M., and Greider, C.W. (2001). Telomere dysfunction increases mutation rate and genomic instability. Cell 106, 275-286. [DOI] [PubMed] [Google Scholar]
- Hardy, C.F., Sussel, L., and Shore, D. (1992). A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6, 801-814. [DOI] [PubMed] [Google Scholar]
- Holm, C., Stearns, T., and Botstein, D. (1989). DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 9, 159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJpma, A.S., and Greider, C.W. (2003). Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.M., Rivera, M.A., Botchkina, I.L., Shalaby, R., Thor, A.D., and Blackburn, E.H. (2001). A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl. Acad. Sci. USA 98, 7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K.E., Harmon, B.P., Reichardt, I.K., Sedat, J.W., and Blackburn, E.H. (1997). Block in anaphase chromosome separation caused by a telomerase template mutation, Science 275, 1478-1481. [DOI] [PubMed] [Google Scholar]
- Krauskopf, A., and Blackburn, E.H. (1996). Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383, 354-357. [DOI] [PubMed] [Google Scholar]
- Krauskopf, A., and Blackburn, E.H. (1998). Rap1 protein regulates telomere turnover in yeast. Proc. Natl. Acad. Sci. USA 95, 12486-12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion, G., Boakye, K.A., and Lustig, A.J. (1992). C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 5159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and Blackburn, E.H. (1993). An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73, 347-360. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Wotton, D., Gilson, E., and Shore, D. (1997). Rap1p and telomere length regulation in yeast. Ciba Found Symp. 211, 76-93; discussion, 93–103. [DOI] [PubMed] [Google Scholar]
- Maringele, L., and Lydall, D. (2002). EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16, 1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, D. (2002). A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14, 237-245. [DOI] [PubMed] [Google Scholar]
- Miller, K.M., and Cooper, J.P. (2003). The telomere protein Taz1 is required to prevent and repair genomic DNA breaks. Mol. Cell 11, 303-313. [DOI] [PubMed] [Google Scholar]
- Nakamura, T.M., Morin, G.B., Chapman, K.B., Weinrich, S.L., Andrews, W.H., Lingner, J., Harley, C.B., and Cech, T.R. (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955-959. [DOI] [PubMed] [Google Scholar]
- Polotnianka, R.M., Li, J., and Lustig, A.J. (1998). The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8, 831-834. [DOI] [PubMed] [Google Scholar]
- Prescott, J., and Blackburn, E.H. (1997). Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11, 528-540. [DOI] [PubMed] [Google Scholar]
- Prescott, J.C., and Blackburn, E.H. (2000). Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol. Cell. Biol. 20, 2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Heiter, P. (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shamu, C.E., and Murray, A.W. (1992). Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 117, 921-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.D., and Blackburn, E.H. (1999). Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J. Cell Biol. 145, 203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G.C., and Jackson, S.P. (1999). The DNA-dependent protein kinase. Genes Dev. 13, 916-934. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Belmont, A.S., Robinett, C.C., and Murray, A.W. (1996). GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6, 1599-1608. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., Marshall, W.F., Sedat, J.W., and Murray, A.W. (1997). Mitosis in living budding yeast: anaphase A but no metaphase plate, Science 277, 574-8. [DOI] [PubMed] [Google Scholar]
- Tzfati, Y., Fulton, T.B., Roy, J., and Blackburn, E.H. (2000). Template boundary in a yeast telomerase specified by RNA structure. Science 288, 863-867. [DOI] [PubMed] [Google Scholar]
- Uemura, T., Ohkura, H., Adachi, Y., Morino, K., Shiozaki, K., and Yanagida, M. (1987). DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50, 917-925. [DOI] [PubMed] [Google Scholar]
- Weinert, T.A., and Hartwell, L.H. (1993). Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134, 63-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich, S.L., et al. (1997). Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17, 498-502. [DOI] [PubMed] [Google Scholar]
- Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748-760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.