Abstract
Actin filaments transiently associate with the endocytic machinery during clathrin-coated vesicle formation. Although several proteins that might mediate or regulate this association have been identified, in vivo demonstration of such an activity has not been achieved. Huntingtin interacting protein 1R (Hip1R) is a candidate cytoskeletal-endocytic linker or regulator because it binds to clathrin and actin. Here, Hip1R levels were lowered by RNA interference (RNAi). Surprisingly, rather than disrupting the transient association between endocytic and cytoskeletal proteins, clathrin-coated structures (CCSs) and their endocytic cargo became stably associated with dynamin, actin, the Arp2/3 complex, and its activator, cortactin. RNAi double-depletion experiments demonstrated that accumulation of the cortical actin-endocytic complexes depended on cortactin. Fluorescence recovery after photobleaching showed that dynamic actin filament assembly can occur at CCSs. Our results provide evidence that Hip1R helps to make the interaction between actin and the endocytic machinery functional and transient.
INTRODUCTION
Until recently, the actin cytoskeleton and the endocytic machinery were thought to operate independently. However, the actin cytoskeleton is an integral part of the cell cortex and there is growing evidence that F-actin plays a direct role during endocytic internalization (reviewed in Engqvist-Goldstein and Drubin, 2003). Genetic studies in yeast have demonstrated that Arp2/3-mediated F-actin assembly is required for the internalization step of endocytosis (reviewed in D'Hondt et al., 2000; Munn, 2001; Shaw et al., 2001; Engqvist-Goldstein and Drubin, 2003). Although actin may not play an obligatory role in clathrin-mediated internalization in all mammalian cells, there is increasing evidence that actin is involved. Merrifield et al. (2002) recently provided light-microscopy evidence that recruitment of actin to endocytic sites occurs at the moment that clathrin-coated pits (CCP) pinch off from the plasma membrane. It is currently not clear, however, which proteins are involved in this process, how it is regulated, and whether actin recruitment at CCPs involves filament recruitment or de novo filament assembly.
To learn more about the function of actin in clathrin-mediated endocytosis, a number of investigators have attempted to identify proteins that could function at the interface between the actin cytoskeleton and the endocytic machinery. In the past few years, several proteins including Hip1R, syndapin, cortactin, Abp1, intersectin and myosin VI have been identified as putative linker proteins (Qualmann and Kelly, 2000; Buss et al., 2001; Engqvist-Goldstein et al., 2001; Hussain et al., 2001; Kessels et al., 2001). Currently, in vivo evidence for such linker function is largely lacking. In this study, our aim was to determine the in vivo role of the putative linker protein Hip1R.
Hip1R is a component of CCPs and clathrin-coated vesicles (CCVs). It can simultaneously bind to both F-actin and to polymerized clathrin in vitro and can promote assembly of clathrin and F-actin into higher order structures (Engqvist-Goldstein et al., 1999, 2001). Furthermore, Hip1R contains an AP180-homology (ANTH) domain, which is also present in several other proteins involved in clathrin-mediated endocytosis. ANTH domains are known to interact with phosphatidylinositol-4, 5-biphosphate at the plasma membrane (Ford et al., 2001). Hip1R belongs to a conserved family of proteins that includes yeast Sla2p/End4p and mammalian Hip1. In yeast, Sla2p is absolutely required for endocytosis, for normal cell morphology and for a functional actin cytoskeleton (Holtzman et al., 1993; Wesp et al., 1997; Yang et al., 1999). Hip1 is closely related to Hip1R (∼50% overall amino acid sequence identity) and has been implicated in the pathology of Huntington disease (Kalchman et al., 1997; Wanker et al., 1997). Similar to Hip1R, Hip1 has been shown to be enriched in CCVs, to colocalize with markers for receptor-mediated endocytosis, and to bind to other endocytic proteins (clathrin, AP-2; Metzler et al., 2001; Mishra et al., 2001; Waelter et al., 2001; Legendre-Guillemin et al., 2002). Recently, phenotypic analysis of Hip1 knockout mice suggested that Hip1 functions in receptor-mediated endocytosis (Metzler et al., 2003), similar to its yeast homologue, Sla2p.
To test the hypothesis that Hip1R coordinates interactions between F-actin and clathrin at endocytic sites, we used RNA interference to reduce the expression of Hip1R and Hip1 in HeLa cells. Phenotypic analyses of cells expressing reduced levels of Hip1R provided evidence that F-actin polymerization occurs at coated-pits and that Hip1R plays a crucial role at the interface between the actin cytoskeleton and the endocytic machinery.
MATERIALS AND METHODS
RNA Preparation
Twenty-one–nucleotide RNAs were chemically synthesized, deprotected, and gel-purified (Dharmacon Research, Inc., Boulder, CO). The siRNA-A1 sequence designed to target both Hip1R and Hip1 corresponds to nucleotides 2629–2649 of human Hip1R (GenBank accession no. BAA31630) and nucleotides 2504–2524 of human Hip1 (GenBank accession no. NM005338). The siRNA-A2 sequence designed to target Hip1R, corresponds to nucleotides 2473–2493 of human or mouse Hip1R (GenBank accession nos. BAA31630 and AF221713, respectively). The siRNA-A3 designed to target Hip1R corresponds to nucleotides 184–204 of human Hip1R (GenBank accession no. BAA31630). The 2-nucleotide 3′ overhangs of 2′deoxythymidine were added to A1, A2 and A3 and are not part of the gene sequence. The siRNA-invA2 was designed as a control and does not target a gene. The sequence is the reverse sequence of the siRNA-A2.
For annealing of siRNA, 20 μl of each single-stranded RNA (50 μM) was incubated with 10 μl 5× annealing buffer (500 mM potassium acetate, 150 mM HEPES-KOH at pH 7.4, 10 mM magnesium acetate) for 1 min at 90°C and then 1 h at 37°C. The RNA duplexes were then stored at -20°C until used.
DNA Constructs
Construction of Hip1R (aa 1–1068)-6myc and Hip1R (aa 1–655)-6myc were described previously (Engqvist-Goldstein et al., 1999).
Treatment of HeLa Cells with siRNA
HeLa cells were plated 1 day before transfection in six-well plates (∼1.4 × 105 cells/well), 24-well plates (∼1.5 × 104 cells/well), or in T75 flasks. On the day of the transfection, cells were ∼30% confluent. For transfection of six-well plates, 3 μl of siRNA duplex (20 μM) was diluted into 200 μl OptiMem (Invitrogen Corp., Carlsbad, CA) in tube 1. In tube 2, 12 μl OligoFectamine (Invitrogen Corp.) was added to 48 μl Optimem. Tubes 1 and 2 were incubated for 7–10 min at room temperature before combining the solutions. The mixture was gently mixed and incubated for 20–25 min at room temperature 152 μl of OptiMem was then added to the mixture, bringing the total volume of siRNA/oligofectamine/OptiMem solution to 424 μl. This was added to cells grown in 2 ml of DMEM with 10% FBS and incubated for 2–4 days.
Western Blotting
For Western blotting, cells treated with siRNA were washed once with PBS and were subsequently put on ice. To lyse the cells, 150 μl of cold lysis buffer (20 mM HEPES, pH 7.4, 1 mM EDTA, 50 mM KCl, 1% NP-40) containing protease inhibitors (10 mg/ml leupeptin, 10 mg/ml aprotinin, 10 mg/ml soybean trypsin inhibitors, and 1 mM PMSF) was added directly to the six-well plate, and cells were scraped off and homogenized by passaging through a 27-gauge needle. The extracts were then spun at 4°C for 20 min at 15,000 rpm in a microcentrifuge. The resulting supernatant fractions were loaded on SDS-PAGE gels and analyzed by immunoblotting.
Cell Culture and Indirect Immunofluorescence
Cell culture and immunofluorescence were performed as described previously (Engqvist-Goldstein et al., 1999). HeLa cells were grown in DMEM containing 10% FBS. For immunofluorescence analysis, cells were transfected with siRNA on glass coverslips in 24-well plates. Before processing for immunofluorescence, the cells were allowed to spread for 2–4 days in media containing siRNA. The following primary antibodies were used: anti-Hip1R antibody (Rbno.2855) was used at 1:50; anti-AP2 antibody (AP.6; Affinity Bioreagents, Golden, CO) was used at 1:100, anticlathrin antibody (X22; Affinity Bioreagents) was used at 1:500; anticlathrin LC antibody (CON.1; Covance Research Products, Madison, WI) was used at 1:200; antidynamin (Hudy 1, Upstate Biotechnology, Lake Placid, NY) was used at (1:100); antimyc antibody (A-14; Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:400; anticortactin antibody (4F11; Upstate Biotechnology) was used at 1:100; anti-Abp1 antibody (kindly provided by Michael M. Kessels) was used at 1:100; anti-Arp3 antibody (kindly provided by Matt Welch) was used at 1:50; anti-Hip1 antibody (kindly provided by Linton Traub) was used at 1:200; antipaxillin antibody (BD Transduction Laboratories, Lexington, KY) was used at 1:100, and antitubulin antibody (Accurate) was used at 1:400. Texas Red-X phalloidin (Molecular Probes, Eugene, OR) was used at 1:500 to stain F-actin.
For EGF labeling of CCVs, RNAi treated cells were incubated with 1 μg/ml EGF-rhodamine (Molecular Probes) for 1 h at 4°C. After several washes, cells were fixed and processed for immunofluorescence.
Transferrin Uptake Assays
For visual observation of transferrin uptake, HeLa cells were grown on glass coverslips as described above and were serum-starved for 45 min at 37°C in DMEM. They were then incubated with 25 μg/ml human transferrin conjugated to Texas Red (Molecular Probes) in serum-free medium for 30 min at 37°C. Cells were then fixed and processed for immunofluorescence.
For the biochemical transferrin assay, cells were grown in six-well plates and treated with siRNA as described above. Three days later, cells were washed one time with serum-free DMEM containing 20 mM HEPES, pH 7.4, and 1 mg/ml BSA (2 ml/well) and serum-starved in the same media for 1 h at 37°C. Cells were then put on ice, and the medium was replaced with the same medium (cold) containing human biotinylated-transferrin (2 μg/ml; Molecular Probes) and incubated for 1 h on ice. Cells were then washed twice with ice cold serum-free DMEM containing 20 mM HEPES, pH 7.4, containing 5 mg/ml BSA (5 ml/well) and moved to a 31°C water bath for various times (0, 4, 8, and 16 min) to allow transferrin internalization. To measure internalized transferrin, surface-bound transferrin was stripped by adding 2 ml ice-cold 10 mM HCl, 150 mM NaCl, pH 2.0, for 2 min to the cells followed by a wash with 10 ml of ice-cold PBS. This process was repeated once more. To measure total transferrin bound, cells were not subjected to the acid wash, but were put in 10 ml of ice-cold PBS. Cells were then lysed in PBS containing 1% Triton X-100 (250 μl) containing 1 mM PMSF and 10 μg/ml PIC and processed as described above. An ELISA assay was used to quantify the amount of biotinylated-transferrin in the lysate, as described in Buss et al. (2001). Percent transferrin uptake was calculated using data from three independent experiments.
Propidium Iodide Staining of Nonviable Cells
HeLa cells were treated with siRNA for 3 days as described above. The cells (∼106) were then trypsinized, washed with PBS, and resuspended in 1 ml PBS. Propidium iodide was added at a final concentration of 2 μg/ml and the cells were incubated on ice in the dark for 5 min. The cells were then analyzed by flow cytometry (Beckman-Coulter EPICS XL, Fullerton, CA; 15,000 events/run) with excitation at 488 nm and emission at 575 nm. Percent viability was calculated as the percentage of cells excluding propidium iodide, using data from three independent experiments.
Unroofing of HeLa cells for “Deep-etch” Electron Microscopy
HeLa cells grown on 3 × 3-mm pieces of glass (no. 1 glass coverslips) were transfected with siRNA (A3 or invA2) as described above (see treatment of HeLa cells with siRNA). Three days after starting the siRNA treatment, cells were “unroofed” (Heuser, 2000). Immediately after unroofing, the cells were fixed for 30 min in 2% glutaraldehyde in KHMgE medium, briefly washed in water, and then quick-frozen, freeze-dried, and rotary-replicated with platinum by standard procedures (Heuser, 2000).
3-D Reconstruction of the F-actin Structures Using Deconvolution Microscopy
HeLa cells treated with invA2 and A3 were processed for immunofluorescence as described above. Fluorescence data was collected using an API Deltavision DV4 Restoration microscope. Voxels were collected at 67-nm lateral and 100-nm axial intervals. Deconvolution was carried out using the API SoftWoRx software (API, San Francisco, CA) on a Linux 2.8 GHz workstation. Rendering of the three-dimensional data was carried using Imaris3.2 (Bitplane) on a Windows XP 2.8GHz workstation.
Imaging of GFP-actin in Living Cells
HeLa cells grown on 20-mm coverslips in six-well plates were transfected with siRNA (A1, A3, and invA2) as described above. One day after siRNA treatment, cells were transfected with GFP-actin using fugene 6 according to the manufacturer (Roche Diagnostics, Indianapolis, IN). Thirty-six to 48 h later, the coverslips were transferred to F-12 media (Invitrogen Corp.) containing 5% fetal calf serum and 10 mM HEPES, pH 7.4. Cells expressing low levels of GFP-actin were imaged using an inverted Nikon Eclipse TE300 fluorescence microscope (Nikon, Garden City, NY) with a Nikon Plan Apo (1.4NA) 100× objective and a Hamamatsu ORCA cooled CCD camera (Hamamatsu, Malvern, PA). GFP fluorescence was visualized with 1–5-s exposures using excitation filters at 465–495 nm and emission filters at 515–555 nm with a 505LP dichroic filter. During the experiments, the coverslips were maintained at 37°C using an aluminum chamber connected to a circulating water bath.
Fluorescence Recovery after Photobleaching
HeLa cells grown on 20-mm coverslips in six-well plates were prepared as described above (imaging of GFP-actin in live cells). Cells expressing low levels of actin-GFP were imaged using a 510 confocal laser scanning microscope (Zeiss) with a Zeiss 63×/0.90 W objective. The krypton/argon laser was used to excite GFP-actin at 488 nm and emission above 505 nm was collected. During the experiments, the coverslips were maintained at 35°C using a heated chamber connected to a dual automatic temperature controller (TC-344B, Warner Instrument Corp., Hamden, CT). A region of the cells containing the F-actin structures formed in A1 cells was selected and photobleached using the krypton/argon laser at 488 nm (20 iterations at ∼50% power). After photobleaching, fluorescence of the entire field was imaged every 3 s for at least a minute. The fluorescence intensity in the photobleached region was normalized to the fluorescence intensity measured in a nonbleached region at each time point to adjust for the decreased fluorescence due to overall bleaching of the entire field.
Rescue Experiments
Stable cell lines expressing murine Hip1R-6myc was generated using standard procedures. Several silent mutations were generated in these constructs (C2478G, C2481T, and A2484C) in the region targeted by siRNA-A2 (nucleotide 2473–2493). These were created to prevent siRNA-A2 from targeting mRNA derived from the stably expressed exogenous Hip1R. A stable cell line expressing 6myc alone was also generated as a control. Several clones for each construct were analyzed by Western blotting and immunofluorescence to confirm expression of exogenous Hip1R.
For rescue experiments, these stable cell lines expressing Hip1R were treated with siRNA (invA2, A1, and A3) as described above. Cells were then processed for immunofluorescence and stained with Texas Red-X phalloidin and antibodies against Hip1R, myc, clathrin, and cortactin as described above. For scoring the A1 phenotype, cells were stained for both F-actin and cortactin. The cells were scored as positive if they had at least three ring-like or tail-like structures containing both F-actin and cortactin. At least three independent experiments were performed scoring 200 cells per experiment.
Rescue experiments were also performed using transient expression of full-length murine Hip1R-6myc. HeLa cells were treated with siRNA (invA2, A1, and A3) as described above. After 1 day of siRNA treatment, cells were transfected with the above constructs using fugene 6 according to the manufacturer (Roche Diagnostics). Forty-eight hours later, cells were fixed and processed for immunofluorescence as described above. Scoring of the A1 and A3 phenotype in transiently transfected cells was done as described above for stable cell lines, except in these experiments only cells expressing the rescue construct, as judged by myc staining, were scored.
RESULTS
Hip1R Depletion by RNAi
Hip1R can bind directly to actin and to clathrin in vitro, and it is intimately associated with clathrin lattices and pits at the cell cortex. Nevertheless, its in vivo roles have not been established. As a test of Hip1R's biological function, its expression was silenced using RNA interference (Elbashir et al., 2001). If Hip1R is required to bring together actin and the endocytic machinery, lowering its levels might disrupt this linkage. Alternatively, if Hip1R is required to regulate the association between actin and the endocytic machinery or to coordinate cytoskeletal and endocytic activities, lowering Hip1R levels might alter the dynamics, the functionality, or both, of this cytoskeleton-endocytic association.
Three RNA duplexes (siRNA-A1, A2, and A3) directed against Hip1R were synthesized (see MATERIALS AND METHODS). As shown in Figure 1A, extracts prepared from cells transfected with either the A1, A2, or A3 duplex showed reduced Hip1R levels. The effect was strongest using A2, wherein Hip1R expression is <10% of the control (Figure 1B). A1 and A3 reduced Hip1R expression to 20–40% of the control (Figure 1, A and B). By design, A1 also affected expression of Hip1 (Figure 1A), which is a Hip1R isoform. Despite this added feature, we never observed phenotypic differences between A1 and A3 cells, and all phenotypes observed were rescued by reintroducing Hip1R (see below). The silencing of Hip1R and Hip1 appears specific because expression levels of other proteins such as clathrin, alpha-adaptin (AP-2 complex), and actin (Figure 1A and unpublished data) were not affected by A1, A2, or A3 duplexes. Furthermore, the negative control, invA2, did not affect expression of any of the proteins examined (Figure 1A).
Figure 1.
Hip1R siRNA duplexes reduce Hip1R expression in HeLa cells. (A) Western blots of siRNA-transfected cells using antibodies against Hip1R, Hip1, clathrin heavy chain (CL HC). (B) Western blot to quantify the reduction in Hip1R expression in A1 and A2 cells. In lanes 4–7 different amounts of extracts were loaded (10–100%) as indicated in the figure. (C) Hip1R immunofluoresence of cells treated with invA2, A1, or A2 (top panels). Bottom: DAPI staining of nuclei. Bars, 10 μm.
To determine the transfection efficiency of the siRNA, we performed anti-Hip1R immunofluorescence on cells treated with A1, A2, A3, and invA2 duplexes. As shown in Figure 1C, Hip1R staining was dramatically reduced by A2 compared with invA2. Most cells showed a similar reduction. There were only a few cells (<5%) that escaped silencing. Similar results were obtained for A1 and A3 duplexes, although the reduction of Hip1R expression was less pronounced (Figure 1C and unpublished data), which is consistent with the Western blotting data.
Deletion of SLA2, the budding yeast gene that encodes a Hip1R homologue, results in severe defects in actin organization, cell morphology, and endocytosis (Holtzman et al., 1993; Wesp et al., 1997; Yang et al., 1999). We therefore expected similar effects for A2 cells, which expressed Hip1R at the lowest levels. Indeed, the majority of cells treated with the A2 duplex (∼85%, n = 600) showed abnormal actin cytoskeleton organization and cell morphology (Figure 2D). A2 cells appeared rounder and smaller than control cells, displaying a disorganized actin cytoskeleton, wherein most cells lacked a normal cortical actin network and lacked proper stress fibers. Furthermore, many cells showed membrane blebbing.
Figure 2.
Hip1R “knock-down” cells have endocytic and actin defects. (A) Visual endocytosis assay. HeLa cells treated with siRNA (A1, A2, or invA2 as indicated) were labeled with Texas Red tranferrin for 30 min to visualize endocytic compartments. Top: DAPI staining of nuclei; bottom: transferrin labeling. (B) Biochemical endocytosis assay. The graph shows percent transferrin uptake as a function of total surface bound transferrin over time in A2-treated and mock-treated cells (control). Three independent experiments were performed for each data point. (C) The bar graph shows percent inhibition in biochemical transferrin uptake assay in A2 cells compared with control cells at different time points as indicated. (D) HeLa cells treated with siRNA (A1, A2, A3, or invA2 as indicated) were stained with DAPI (top) and Texas Red-X phalloidin (middle and bottom) to visualize F-actin. Bottom panels are enlargements of boxes in the middle panels. Bars, 10 μm.
It has previously been reported that cells lacking Hip1 are, like yeast sla2 mutants, defective for endocytosis (Wesp et al., 1997; Metzler et al., 2003). We confirmed that A2 cells, expressing Hip1R at <10% of normal levels, also show a defect in transferrin internalization (Figure 2, A–C). In a visual assay, A2 cells showed reduced transferrin uptake compared with invA2 cells (Figure 2A). Approximately 89% of control cells (n = 300) showed a clear perinuclear labeling, whereas only ∼37% of A2 cells (n = 300) showed a clear perinuclear labeling. For A1 cells, which showed less effective Hip1R silencing, ∼82% of cells (n = 300) showed clear perinuclear staining. To make sure that the observed reduction in transferrin uptake in the A2 cells was due to defects in the internalization step, we performed a biochemical endocytosis assay that measures uptake of internalized transferrin as a function of total surface bound transferrin. As shown in Figure 2, B and C, the rate of internalization was slower in A2 cells than in control cells. These effects are not due to general necrosis because the nuclei of these cells appeared normal as visualized by DAPI staining (Figure 2D), and cell viability as assessed by propidium iodide labeling was similar to that of control cells (Supplementary Figure 1). It should be noted that A2 cells also bind less transferrin (∼40%) than control cells (unpublished data), possibly reflecting a defect in other trafficking events. The reduction in transferrin binding also explains why the endocytic defect in A2 cells appears more severe using the visual assay for transferrin uptake than using the biochemical assay. In contrast to A2 cells, A1 and A3 cells showed normal cell morphologies and no defect in transferrin endocytosis could be detected (Figure 2). The lack of more pronounced endocytic defects in A2 cells or of a detectable endocytosis defect in A1 and A3 cells, was not unexpected. A1 and A3 cells still express Hip1R at 20–40% of normal levels. In A2 cells, the remaining Hip1R (10%) may be sufficient for endocytosis, albeit at reduced rates. Alternatively, Hip1R's importance for endocytosis may vary in different mammalian cell types, just as actin's importance varies (reviewed in Engqvist-Goldstein and Drubin, 2003).
Accumulation of Cortical Actin Tails and Other Unusual Cortical Actin Structures in Response to Lowered Hip1R Levels
Because A1 and A3 cells lacked obvious global morphological abnormalities, we reasoned that any unusual features that we could detect in these cells would most likely have resulted specifically from the lowered Hip1R levels. We therefore chose to investigate in A1 and A3 cells the association between actin cytoskeletal proteins and the endocytic machinery. This association is normally very transient (Merrifield et al., 2002). Strikingly, both A1 and A3 cells accumulated unusual actin structures near the cell cortex. Approximately 60% of A1 cells (n = 600) and ∼75% of A3 cells (n = 300) accumulated unusual actin structures near the cell cortex (Figure 2D). These structures took a variety of forms including tails, rings, and more complex structures (Figure 3C). Control cells and untreated cells also had some cortical actin structures. However, these were smaller and much less abundant (Figure 2D). Approximately 1% (n = 600) of invA2 cells have actin structures that are similar to those in A1/A3 cells. As mentioned above, most A2 cells have a completely disorganized actin cytoskeleton with a rounded morphology. However, those few A2 cells that displayed a relatively normal actin cytoskeleton and cell morphology also accumulated the unusual cortical actin structures (unpublished data). We speculate that the global defects in cortical integrity or structure that are observed in most A2 cells may interfere with the formation or stability of the unusual cortical actin structures. Because these structures are more abundant in A1/A3 cells than in A2 cells, for the remainder of this study we characterize these cortical actin structures in A1/A3 cells exclusively. In the interest of space and simplicity, because we observed no phenotypic differences between cells treated with A1 or A3 duplexes, we typically show data from cells treated with only one of these two siRNA duplexes.
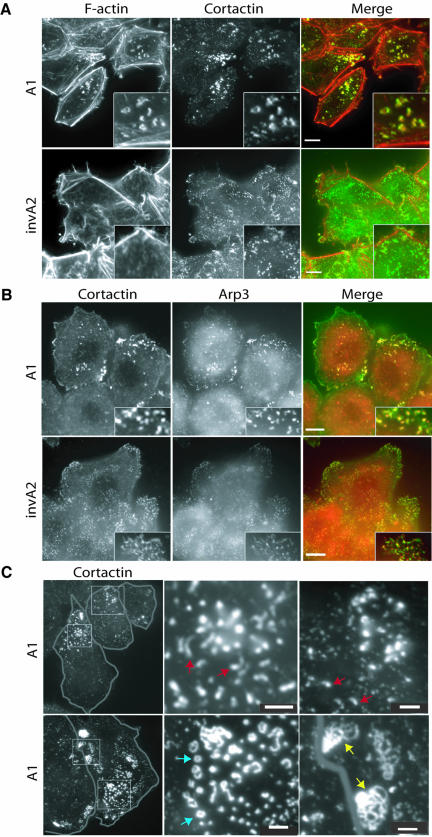
Figure 3.
Cortactin and the Arp2/3 complex are specifically recruited to the cortical F-actin structures. HeLa cells treated with siRNA (A1 or invA2) for 3 days were stained with fluorescent phalloidin or different antibodies as indicated. (A) Texas Red-X phalloidin (left panels), cortactin (middle panels), merge (right panels). Boxes in the bottom right corners are enlarged regions of the micrographs. Bars, 10 μm. (B) Cortactin (left), Arp3 (middle), merge (right). Bars, 10 μm. (C) This figure shows the different types of F-actin/cortactin structures observed in A1-treated cells. Left panels show low magnification views of cells wherein the periphery of each cell is marked. Red arrows indicate individual tail-like F-actin/cortactin structures (top). Blue arrows indicate ring-like F-actin/cortactin structures (bottom). Yellow arrows indicate more complex F-actin/cortactin structures (bottom). Bars, 2.5 μm.
Different actin-based cellular structures contain different combinations of actin-associated proteins that dictate their properties and regulation. Because the composition of the cortical actin structures formed in A1/A3 cells can provide information about their origins and properties, we next asked which other actin-associated proteins are present in these structures. Because Arp2/3-mediated F-actin assembly is required for the internalization step of endocytosis in yeast, we next attempted to stain the cortical actin structures observed in A1/A3 cells with antibodies against different actin-binding proteins involved in Arp2/3-mediated actin polymerization. We found that cortactin, the Arp2/3 complex and Abp1 are enriched in these structures (Figure 3 and unpublished data). Similar to Hip1R, cortactin, and Abp1 are actin-binding proteins implicated in both endocytic and actin functions, and they may function at the interface between the actin cytoskeleton and the endocytic machinery (Kessels et al., 2000, 2001; McNiven et al., 2000; Cao et al., 2003; Mise-Omata et al., 2003). Cortactin and Abp1 both colocalize with the Arp2/3 complex in cells (Kessels et al., 2000; McNiven et al., 2000). Additionally, cortactin and yeast Abp1 have recently been shown to activate the Arp2/3 complex (Goode et al., 2001; Uruno et al., 2001; Weaver et al., 2001). In control cells, cortactin shows a fine punctate staining pattern at the cell cortex, with some enrichment in F-actin ruffles (Figure 3A). Note that in A1/A3 cells cortactin does not get recruited to actin stress fibers. Instead, it appears to almost exclusively localize to the cortical F-actin structures that accumulate in response to reduction in Hip1R expression (Figure 3A). Other cytoskeletal proteins such as paxillin, which is a focal adhesion protein, and tubulin, which is part of the microtubule cytoskeleton, did not colocalize with these F-actin structures (Supplementary Figure 2 and unpublished data). These observations demonstrate that only a subset of cytoskeletal proteins associate with the unusual cortical actin structures that accumulate in cells with lowered Hip1R levels.
Because cortactin colocalized with the unusual cortical actin structures that formed in cells with reduced Hip1R levels, but not with other F-actin structures in these cells, we were able to use this protein as a specific marker for the cortical actin structures. The cortactin/F-actin cortical structures are quite heterogeneous in shape and size (Figure 3C). Some appear as spots, whereas others are tail-like and ring-like. In A1/A3 cells ∼20% (n = 200) of the structures are spots, ∼30% (n = 200) are tail-like structures, and 50% (n = 200) are more complex structures (interconnected rings and lines). Each of these types of structures contains the Arp2/3 complex in addition to F-actin and cortactin (Figures 3B and 4, B and C).
Figure 4.
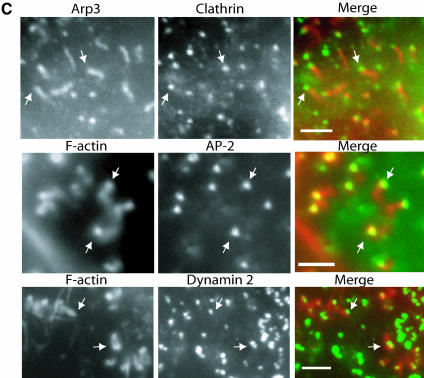
The F-actin structures that accumulate in the absence of Hip1R are associated with endocytic components. HeLa cells treated with siRNA (A1 or invA2) for 3 days were stained with different antibodies as indicated. (A) Clathrin (green) colocalizes with the F-actin (red) structures in A1 cells. The middle panel shows A1 cells containing several F-actin spots. Arrows indicate colocalization between clathrin and F-actin. Arrowheads indicate spots that only stain with clathrin. Bars, 10 μm. (B) Coincidence of clathrin and actin in A1 cells containing complex F-actin structures (see arrows). Top two panels show Texas Red-X phalloidin staining (red) and clathrin staining (green). Bars, 10 μm. Bottom panel shows Arp3 staining (green) and clathrin staining (red). Asterisk indicates a stress fiber. Bar, 10 μm. (C) Clathrin, AP-2, and dynamin (green) localize to one end of the tail-like F-actin structures (red) in A1 cells (see arrows). Bars, 2 μm.
Clathrin, AP-2, Dynamin, and an Endocytic Cargo Protein Associate with the Cortical F-actin Structures
Multicolor fluorescence microscopy has revealed that actin and endocytic proteins associate transiently during endocytosis (Merrifield et al., 2002). However, little is known about how productive interactions between the actin cytoskeleton and endocytic machinery are formed and regulated. Because Hip1R can bind to actin and is a component of CCPs and CCVs, we next asked whether the actin tails and the unusual actin structures at the cell cortex in A1/A3 cells are associated with any endocytic components. To address this question, we stained A1/A3 cells with antibodies against several endocytic components. In control cells, clathrin localizes to spots at the cell cortex that represent assembled clathrin (clathrin lattices, CCPs, and/or CCVs; Figure 4A, top panel). We will refer to these as clathrin-coated structures (CCSs). As shown in Figure 4A, F-actin does not colocalize with CCSs in control cells, although in some cells coated-pits appear to align along stress fibers as previously reported (Bennett et al., 2001). It should be noted that because HeLa cells are not polarized, alignment of coated-pits with stress fibers is only found in a small percentage of untreated HeLa cells (unpublished data).
Strikingly, in A1/A3 cells F-actin colocalized with CCSs and appeared enriched in these structures (Figure 4A, see arrows in the enlargement, and unpublished data). Approximately 97% (n = 200) of the F-actin/cortactin structures in A1/A3 cells also contained clathrin. F-actin and the clathrin colocalized with F-actin spots (Figure 4A), F-actin rings (Figure 4B), and F-actin tails (Figure 4C, top panels). Interestingly, clathrin appeared as a spot at one end of each actin (Arp2/3) tail (Figure 4C, top panel).
Clathrin functions not only in endocytosis, but also in several other trafficking events (e.g., TGN budding). Therefore, to determine whether we were observing endocytic structures, we asked whether AP-2, an adaptor complex that is involved specifically in clathrin-mediated endocytosis, is present in the cortical actin structures of A1/A3 cells. Similar to clathrin, AP-2 is enriched in the F-actin/cortactin structures induced by loss of Hip1R expression (Figure 4C, middle panel, and unpublished data), suggesting that these structures were derived from the plasma membrane. AP-2, like clathrin, localizes to one end of the tail-like structures (Figure 4C, middle panel; see arrows). Approximately 98% (n = 200) of the F-actin/cortactin structures in A1/A3 cells also contained AP-2.
Dynamin is critically important for the fission event in clathrin-mediated endocytosis and associates with the endocytic complex at a late stage of the internalization process (Merrifield et al., 2002). It is thought that dynamin gets recruited to the necks of CCPs (reviewed in Hinshaw, 2000). We next asked whether dynamin is present in the actin/endocytic structures that accumulate in A1/A3 cells because such a colocalization would provide evidence that these structures represent mature coated pits. Similar to clathrin and AP-2, dynamin localized to one end of the tail-like structures in A1/A3 cells (Figure 4C, bottom panel). Approximately 95% (n = 200) of the F-actin/cortactin structures in A1/A3 cells also contained dynamin.
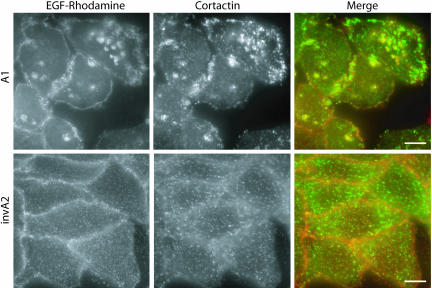
Finally, we asked whether the cortical actin/cortactin structures contained endocytic cargo. Because transferrin labeling of CCPs is very weak, we used fluorescently labeled epidermal growth factor (EGF), another cargo for clathrin-mediated endocytosis. Strikingly, we found that EGF accumulates at the cortical actin/cortactin structures (Figure 5 and unpublished data). Taken together, this strongly suggests that the actin structures in A1/A3 cells associates with the endocytic machinery at some stage during CCV formation.
Figure 5.
EGF colocalizes with the cortical actin structures. HeLa cells treated with invA2 or A1 were incubated with EGF-rhodamine at 4°C to label CCPs. Left: EGF staining (red); middle: cortactin staining (green); and right: merged images. Bar, 10 μm.
The Cortical F-actin Structures Appear to be Tethered to the Cell Cortex via the Endocytic Machinery and Are Sites of Rapid and Continuous Actin Polymerization
The data presented so far suggests that partial loss of Hip1R function causes accumulation of cargo-containing endocytic structures that are stably associated with filamentous actin and actin nucleating proteins. We next asked whether these various proteins are tethered to the cell cortex and whether the actin structures are oriented in a specific manner in relation to the endocytic machinery. To address this question, we generated 3-D reconstructions using deconvolution microscopy of intact cells. As shown for A3 cells in Figure 6, A and B, the actin structures are connected to AP-2 very close to the presumed plane of the plasma membrane. This is consistent with our data showing that these structures are detectable in intact cells using total internal reflection microscope (unpublished data). The tail-like F-actin structures extend into the cytoplasm and are oriented away from the presumed plane of the plasma membrane. Note that some tail-like F-actin structures are connected to one AP-2 spot (Figure 6B, arrow), whereas others appear to be connected to several spots (Figure 6B, arrowhead). Even in the more complex ring structures, F-actin appeared to localize away from the cell cortex, whereas AP-2 was cortically localized (Supplementary Figure 3).
Figure 6.
The F-actin/cortactin structures are tethered to the cell cortex via association with the endocytic machinery. (A and B) HeLa cells treated with A3 were stained Texas Red-X phallodin (red) and anti–AP-2 antibodies (green). (A) A gallery of images showing the first 15 deconvolved focal planes (0.1 μm/section) of actin/AP-2 structures. The top left image shows the first plane from the ventral surface. Bar, 1 μm. (B) Two orientations of a volume rendering of the actin/AP2 structures shown in A. Renderings are oriented with plasma membrane at bottom (bottom image) or in the plane of the viewer (top image). Arrow shows a tail-like F-actin structure connected to one AP-2 spot. Arrowhead shows a tail-like actin structure, wherein several AP-2 spots are connected to F-actin. Small grid boxes, 200 × 200 nm. (C) A gallery of “deep etch” electron micrographs of A3-treated HeLa cells. These micrographs show anaglyph stereo views of the inner surface of “unroofed” cells. Bar, 0.5 μm.
Formation of a stable complex between actin cytoskeletal proteins and the endocytic machinery provided a unique opportunity for an ultrastructural analysis of proteins that normally associate in only a very transient manner. Because an ultrastructural understanding actin's association with the endocytic machinery is required to understand how actin contributes to endocytosis, we determined the ultrastructure of CCSs and their association with actin in unroofed cells. “Deep etch” electron micrographs of A3 cells showed unusually close actin/clathrin associations that took a variety of forms from cup-shaped crescents of actin around the polygonal coats of clathrin (Figure 6C, top panels), to spiderlike tufts of actin that lay beside (or sometimes even directly on top of) the clathrin coats (Figure 6C, middle panels), and rarely, to tapered “tails” of actin that directly contacted certain tightly curved clathrin lattices (Figure 6C, bottom right panel).
These sorts of intimate and persistent actin/clathrin associations were not seen in control HeLa cells grown without exposure to the “A3” siRNA (unpublished data), nor have they been seen before in any of a wide variety of other cultured cells examined by unroofing over the years (Heuser, 1980; Heuser and Anderson, 1989; Fujimoto et al., 2000; Engqvist-Goldstein et al., 2001). In addition, the collections of F-actin filaments observed in A3 cells have an unusual configuration, not commonly observed on the ventral surfaces of these HeLa (or any other) cultured cells. Namely, they are made up of unusually short and unusually tightly intertwined actin filaments, and they tend to stand up from the membrane in “tufts,” much like the configuration of actin filaments at the very leading edges of actively motile cells (or the configuration of actin in the dynamic, tuft-like “podosomes” seen in osteoclasts and some transformed cells). This is quite unlike the usual configuration of F-actin filaments normally seen on the ventral surfaces of cultured cells, which are usually relatively long and either loosely associated in flat, open meshes or tightly associated laterally into stress fibers. Finally, it is worth mentioning that these unusual collections of F-actin filaments do not appear to be altering the structure of the clathrin lattices themselves, which appear in their usual (and full) range of curvature in these cells, regardless of whether they are ensconced in actin or not.
Two critical remaining questions are whether the actin/endocytic complexes are motile and whether the actin associated with the endocytic complexes is undergoing dynamic turnover characteristic of polymerization-driven processes or are composed of more static actin structures often associated with myosin-based processes. Having identified a situation in which actin is stably associated with endocytic proteins provided a unique opportunity to ask whether these actin filaments are dynamically turning over.
First, to probe the dynamic properties of these F-actin structures in live cells, we performed time-lapse video microscopy of cells expressing GFP-actin. Living A1/A3 cells contained F-actin structures similar to those seen in fixed cells shown above (Figure 7A and unpublished data). The structures also labeled with clathrin (unpublished data). Strikingly, the tail-like F-actin structures appeared to be tethered at one end, whereas the other end appeared to wave back and forth (Figure 7A and arrows in movie 1). The tethered ends of the tail-like structures never moved during several minutes of observation (100%, n = 30). The more complex ring-like F-actin structures showed very little lateral mobility and appeared to be tethered to the cell cortex along their entire lengths (Figure 7A and arrow in movie 2).
Figure 7.
The F-actin structures that accumulate in the absence of Hip1R contain rapidly polymerizing actin. (A) Time lapse videomicroscopy of invA2- or A1-treated HeLa cells expressing GFP-actin. The F-actin structures in A1 cells were observed at 3-s intervals. Top panel shows low magnification views of the cells. Bars, 10 μm. Bottom panel is enlargements of boxes in the top panel showing the first frame of movie 1 and movie 2. Bars, 2.5 μm. (B) FRAP analysis of A1-treated HeLa cells expressing GFP-actin. Top (FRAP 1): analysis of small spots/tail-like F-actin structures; bottom (FRAP 2): analysis of a larger ring-like structures. We observed the F-actin structures in A1 cells at 3-s intervals >40 frames. The cells were photobleached in a defined area after two frames as indicated in the figure (see asterisk). The graph shows fluorescence intensity (arbitrary units) as a function of time (s), where the data have been normalized against a reference region that was not photobleached. Bars, 10 μm.
To determine whether the F-actin structures are actively polymerizing F-actin and turning over, we performed fluorescence recovery after photobleaching (FRAP) on A1 cells expressing GFP-actin. As shown in Figure 7B, these structures are dynamically assembling actin filaments with a half-time for recovery of 23 ± 15 s (n = 8). Stress fibers in the same cells recovered from photobleaching much more slowly, with a half-time for recovery of >2 min (unpublished data). The recovery time for the unusual F-actin structures in A1 cells is similar to that reported for GFP-actin in podosomes (Ochoa et al., 2000), dynamic membrane structures involved in adhesion and degradation of the extracellular matrix.
Cortactin Is Required for the Formation of the Cortical Actin Structures in A1/A3 Cells
Cortactin is a good candidate for a protein involved in actin assembly at endocytic sites because it is an Arp2/3 activator that binds to dynamin and because cortactin is involved in clathrin-mediated endocytosis (Uruno et al., 2001; Weaver et al., 2001; Cao et al., 2003). We used RNAi to ask whether cortactin is required for the formation of cortical actin/endocytic structures. Cortactin expression was lowered by RNAi in HeLa cells that were also treated with A1 or A3 duplexes. It should be noted that when using two siRNA duplexes, we have found that the efficacy of reducing expression of each individual protein is slightly decreased compared with using only one siRNA. Therefore, the penetrance of the phenotype even in cells treated with a control duplex is somewhat reduced. The expression of cortactin in the double RNAi-treated cells is <5% (C. Barroso and D.G. Drubin, unpublished results) and the expression of Hip1R is ∼50%. Nevertheless, in cells treated with control siRNA + A3, 15.3 ± 1.3% (n = 400) of the cells clearly contained the unusual cortical actin structures. However, when cells are treated with siRNA against cortactin + A3, only 2.4 ± 0.2% (n = 400) of the cells display this phenotype, which is similar to what is observed with control siRNA (1.1 ± 0.2%, n = 600). Similar results were obtained with A1 (unpublished data). This result suggests that cortactin is required for the assembly of actin structures in association with endocytic proteins.
Specificity of Hip1R Phenotypes
Observing that similar cortical actin/endocytic complexes formed in response to treatment with any of three siRNA duplexes strongly suggested that this phenotype resulted specifically from Hip1R depletion. Two further tests of the specificity of our siRNA results were designed. First, we showed that overexpression of a Hip1R truncation mutant that lacks the C-terminal actin-binding domain (Hip1R aa1–655) also induced formation of similar cortical actin/endocytic complexes (Supplementary Figure 4). This Hip1R mutant competes with endogenous Hip1R for localization to cortical clathrin structures (Engqvist-Goldstein et al., 2001 and unpublished data). We therefore speculate that the truncated Hip1R associates nonproductively with CCSs.
Additionally, we constructed stable cell lines that express mouse Hip1R-6myc. A1 and A3 should not target this construct (see MATERIALS AND METHODS). We also constructed as a control a stable cell-line expressing only the 6myc-tag. Hip1R-6myc localizes like endogenous Hip1R (Engqvist-Goldstein et al., 2001). Neither A1 nor A3 target mouse Hip1R-6myc, whereas endogenous Hip1R was still targeted (Figure 8B and unpublished data). Expression of mouse Hip1R-6myc completely suppressed the formation of the cortical actin/endocytic structures in A1 and A3 cells (Figure 8A and unpublished data). The A1 treatment in control cells (6myc) causes 61% (n = 300) of the cells to have the cortical actin/endocytic structure phenotype, whereas only 0.5% (n = 300) of A1-treated Hip1R-6myc–expressing cells have the F-actin/cortactin phenotype. The occurrence of this phenotype is similar to what is observed in control invA2 cells (0.8%, n = 300). The actin phenotype in both A1 and A3 cells was also completely rescued when Hip1R-6myc was expressed transiently (unpublished data). Taken together, these experiments demonstrate that the formation of actin/endocytic complexes is caused by loss of Hip1R expression.
Figure 8.
Phenotypes associated with A1 and A3 are due to loss of Hip1R function. (A and B) Stable HeLa cell lines expressing mouse Hip1R-6myc or 6myc (control) were treated with A1 or invA2 for 3 days as indicated. (A) Cells were fixed and stained with DAPI (top) and anticortactin antibodies (middle and bottom). (B) Western blotting of cell extracts from these cells. Black arrow (top) indicates exogenous Hip1R-6myc, and gray arrow (bottom) indicates endogenous Hip1R.
DISCUSSION
Although it is now generally accepted that the actin cytoskeleton participates in endocytosis, little is known about the mechanisms that mediate this association. In this study, by analyzing loss of function phenotypes in cells expressing Hip1R at reduced levels, we demonstrated that this protein is critically important for coordinating activities of the actin cytoskeleton and the endocytic machinery.
A3-treated cells, which express Hip1R at 20–40% of normal levels, and A1-treated cells, which express Hip1R and Hip1 at 20–40% of normal levels, accumulated actin tails and other unusual cortical actin structures at the cell cortex. A2 cells, which showed the most pronounced reduction in Hip1R expression, also exhibited this phenotype in the subset of cells with unperturbed morphologies and adhesion. Interestingly, the cortical structures formed in response to RNAi-mediated Hip1R silencing contained both actin polymerization (e.g., the Arp2/3 complex and cortactin) and endocytic machinery. These two machineries normally are expected to associate only transiently. Because the F-actin structures are connected to spots labeled with clathrin, AP-2, and dynamin, it is likely that these endocytic proteins represent CCPs and/or CCVs derived from the plasma membrane. This conclusion is further supported by our light and electron microscopy studies showing in unroofed cells that these structures are tethered to the cell cortex and that they are <200 nm from the plasma membrane because they were visible in intact cells using TIRF microscopy. Finally, the observation that EGF colocalized with the cortical actin structures showed that loss of Hip1R function causes stable association of cargo-containing CCPs and/or CCVs with actin. It is currently not clear whether Hip1R loss of function causes a complete block, or a delay, in CCV internalization because we were unable to follow internalization of individual CCVs in HeLa cells. Because A1 and A3 cells express Hip1R at 20–40% of normal levels and because only a fraction of the total number of coated-pits are affected in these cells, it was not surprising that these cells did not have a detectable endocytic defect.
Several factors suggest that the stable association between endocytic and actin cytoskeleton machinery in cells depleted for Hip1R is a specific phenotype. First, three different siRNA duplexes, each of which targets Hip1R expression, each caused this phenotype. Second, restoring Hip1R expression rescued the phenotype. Third, overexpression in HeLa cells of a Hip1R mutant that lacks the C-terminal domain and that competes with endogenous Hip1R for association with CCSs induced formation of the actin-endocytic protein complexes.
Several lines of evidence suggest that the Hip1R loss of function phenotype is a reflection of its normal biological role. First, when Hip1R is depleted, actin polymerization occurs specifically at sites (CCPs and CCVs) where Hip1R normally localizes, and not at other sites in the cell. Second, Hip1R can physically connect actin and clathrin in vitro (Engqvist-Goldstein et al., 2001). Third, yeast cells lacking the Hip1R-related protein Sla2p were recently shown to accumulate similar actin tails connected to endocytic complexes (Kaksonen et al., 2003).
It is particularly noteworthy that very similar loss-of-function phenotypes have now been observed for Hip1R in mammalian cells and Sla2p in yeast. The fact that endocytosis in mammalian cells but not in yeast shows a clear dependence on dynamin and clathrin, whereas endocytosis in yeast but not mammals shows an absolute dependence on actin, has raised questions about the mechanistic relatedness of these processes. The common Hip1R/Sla2p loss-of-function phenotype argues strongly for mechanistic conservation.
Actin tails or plumes resembling actin comet tails that propel some intracellular pathogens through the cytoplasm have been observed to associate with a variety of endocytic structures (CCSs, endosomes, lysosomes, caveolae, and macropinosomes), and other trafficking vesicles (Heuser and Morisaki, 1992; Frischknecht et al., 1999; Merrifield et al., 1999; Kaksonen et al., 2000; Rozelle et al., 2000; Schafer et al., 2000; Taunton et al., 2000; Kanzaki et al., 2001; Lee and De Camilli, 2002; Merrifield et al., 2002; Orth et al., 2002; Pelkmans et al., 2002). During clathrin-mediated endocysosis, it is now known that actin appears transiently at CCPs just as they begin to move into the cytoplasm (Merrifield et al., 2002). Currently, very little is known about how actin gets recruited to endocytic sites and how this interaction is regulated during CCV formation. Our data show that Hip1R is critical for proper association between the endocytic machinery and the actin polymerization machinery. We hypothesize that when Hip1R expression is reduced, actin polymerization at coated pits is no longer properly regulated, resulting in accumulation of actin filaments that fail to facilitate CCP release. The F-actin structures that form in A1/A3 cells are quite heterogeneous in appearance (spots, tails, rings, and more complex structures). We speculate, however, that F-actin spots give rise to tails, which then give rise to the more complicated ring-like structures as more and more actin accumulates at CCPs. This conclusion is based on the observation that more rings appear in cells treated for longer times with A1/A3 duplexes (unpublished data), and the fact that we observe many structures that appear to be intermediates between tails and rings. It is interesting that the actin tail structures that we observe are stationary in relation to the cell cortex, which is different from actin comet tails that are known to propel intracellular pathogens and vesicles through the cytoplasm. We speculate that when Hip1R levels are reduced, actin polymerization occurs at CCPs, which are anchored at the plasma membrane and therefore cannot be propelled into and through the cytoplasm. Although we originally speculated that Hip1R might be required for association of the actin cytoskeleton with the endocytic machinery, Hip1R appears instead to be required to coordinate the activities of the actin cytoskeleton and the endocytic machinery.
Several lines of evidence suggest that the Arp2/3 complex is involved in endocytic internalization, wherein it nucleates assembly of new actin filaments at endocytic sites. Mutants of the Arp2/3 complex and mutants of Arp2/3 activators (e.g., Pan1 and yeast WASP) are defective in endocytic internalization in budding yeast (reviewed in Engqvist-Goldstein and Drubin, 2003). Furthermore, the Arp2/3 complex and Scar (WASP-related proteins) localize to sites of macropinocytic and phagocytic internalization and are important for phagocytosis and macropinocytosis in Dictyostelium (reviewed in Maniak, 2002). The Arp2/3 complex, however, has not yet been directly implicated in clathrin-mediated endocytosis. Two activators of the Arp2/3 complex, N-WASP and cortactin, however, have recently been implicated in clathrin-mediated endocytosis (Zhang et al., 1999; Kessels and Qualmann, 2002; Cao et al., 2003). Cortactin distribution is severely affected in cells expressing Hip1R at reduced levels and we showed that cortactin is required for the formation of the unusual cortical actin structures. Cortactin directly activates the Arp2/3 complex and inhibits debranching of the resulting filament networks in vitro (Uruno et al., 2001; Weaver et al., 2001). Because cortactin localizes to actin-rich membrane structures such as lamellipodia, podosomes, invadopodia, rocketing endosomes, and CCPs, it has been speculated that cortactin functions to promote F-actin assembly on membranes. In support of this conclusion, Schafer et al. (2002) recently showed that cortactin causes association of F-actin bundles with PtdIns(4,5)P2-containing liposomes in vitro. Additionally, Cao et al. (2003) showed that microinjection of anticortactin antibodies inhibits transferrin uptake, suggesting that cortactin functions in clathrin-mediated endocytosis. Cortactin has been shown to localize to CCPs, where it frequently localizes to the edges of the forming coated pits (Cao et al., 2003). We have previously demonstrated that Hip1R is intimately associated with clathrin during CCV formation (Engqvist-Goldstein et al., 1999, 2001). Hip1R frequently localizes to the edges of the forming coated pits, similar to cortactin, and it appears to connect clathrin to filamentous actin at the cortex (Engqvist-Goldstein et al., 2001). We recently discovered that Hip1R binds to cortactin (Engqvist-Goldstein, Å.E.Y., and D.G. Drubin, unpublished results). Because Hip1R is required for proper localization of cortactin in vivo and because cells expressing reduced levels of Hip1R show constitutive polymerization of actin at coated pits, we speculate that Hip1R might negatively regulate F-actin assembly at CCPs, perhaps by affecting cortactin. The absence of such a regulatory mechanism may hinder productive coupling of actin polymerization to the endocytic machinery.
Supplementary Material
Acknowledgments
We thank Ann Fischer for her advice on cell culture, Holly Aaron for advice on FRAP, and Steve Ruzin for advice on deconvolution microscopy. We also thank Kimberly Cunningham and Robyn Roth for technical help. Finally, we thank Michael M. Kessels, Matt Welch, and Linton Traub for reagents used in this study. This work was supported by grants from the National Institutes of Health to D.G.D. (GM65462) and to J.E.H. (GM29647).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–09–0639. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-09-0639.
Online version of this article contains supplementary figures and videos. Online version is available at www.molbiolcell.org.
References
- Bennett, E.M., Chen, C.Y., Engqvist-Goldstein, A.E., Drubin, D.G., and Brodsky, F.M. (2001). Clathrin hub expression dissociates the actin-binding protein Hip1R from coated pits and disrupts their alignment with the actin cytoskeleton. Traffic 2, 851-858. [DOI] [PubMed] [Google Scholar]
- Buss, F., Arden, S.D., Lindsay, M., Luzio, J.P., and Kendrick-Jones, J. (2001). Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 20, 3676-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Orth, J.D., Chen, J., Weller, S.G., Heuser, J.E., and McNiven, M.A. (2003). Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol. Cell. Biol. 23, 2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hondt, K., Heese-Peck, A., and Riezman, H. (2000). Protein and lipid requirements for endocytosis. Annu. Rev. Genet. 34, 255-295. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., and Drubin, D.G. (2003). Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287-332. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., Kessels, M.M., Chopra, V.S., Hayden, M.R., and Drubin, D.G. (1999). An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J. Cell Biol. 147, 1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., Warren, R.A., Kessels, M.M., Keen, J.H., Heuser, J., and Drubin, D.G. (2001). The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J. Cell Biol. 154, 1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, M.G., Pearse, B.M., Higgins, M.K., Vallis, Y., Owen, D.J., Gibson, A., Hopkins, C.R., Evans, P.R., and McMahon, H.T. (2001). Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291, 1051-1055. [DOI] [PubMed] [Google Scholar]
- Frischknecht, F., Moreau, V., Rottger, S., Gonfloni, S., Reckmann, I., Superti-Furga, G., and Way, M. (1999). Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401, 926-929. [DOI] [PubMed] [Google Scholar]
- Fujimoto, L.M., Roth, R., Heuser, J.E., and Schmid, S.L. (2000). Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic 1, 161-171. [DOI] [PubMed] [Google Scholar]
- Goode, B.L., Rodal, A.A., Barnes, G., and Drubin, D.G. (2001). Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J. Cell Biol. 153, 627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J. (1980). Three-dimensional visualization of coated vesicle formation in fibroblasts. J. Cell Biol. 84, 560-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E. (2000). The production of `cell cortices'for light and electron microscopy:follow the middle road. Traffic 1, 545-552. [DOI] [PubMed] [Google Scholar]
- Heuser, J.E., and Anderson, R.G. (1989). Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 108, 389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E., and Morisaki, J.H. (1992). Time-lapse video microscopy of endosomal “rocketing” of La/Zn-treated cells. Mol. Biol. Cell Abstr. 3, 172. [Google Scholar]
- Hinshaw, J.E. (2000). Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 16, 483-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman, D.A., Yang, S., and Drubin, D.G. (1993). Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122, 635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, N.K. et al. (2001). Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927-932. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Peng, H.B., and Rauvala, H. (2000). Association of cortactin with dynamic actin in lamellipodia and on endosomal vesicles. J. Cell Sci. 113(Pt 24), 4421-4426. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475-487. [DOI] [PubMed] [Google Scholar]
- Kalchman, M.A. et al. (1997). HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet. 16, 44-53. [DOI] [PubMed] [Google Scholar]
- Kanzaki, M., Watson, R.T., Khan, A.H., and Pessin, J.E. (2001). Insulin stimulates actin comet tails on intracellular GLUT4-containing compartments in differentiated 3T3L1 adipocytes. J. Biol. Chem. 276, 49331-49336. [DOI] [PubMed] [Google Scholar]
- Kessels, M.M., Engqvist-Goldstein, A.E., and Drubin, D.G. (2000). Association of mouse actin-binding protein 1 (mAbp1/SH3P7), an Src kinase target, with dynamic regions of the cortical actin cytoskeleton in response to Rac1 activation. Mol. Biol. Cell 11, 393-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels, M.M., Engqvist-Goldstein, A.E., Drubin, D.G., and Qualmann, B. (2001). Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J. Cell Biol. 153, 351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels, M.M., and Qualmann, B. (2002). Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J. 21, 6083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., and De Camilli, P. (2002). Dynamin at actin tails. Proc. Natl. Acad. Sci. USA 99, 161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin, V., Metzler, M., Charbonneau, M., Gan, L., Chopra, V., Philie, J., Hayden, M.R., and McPherson, P.S. (2002). HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin. Identification of a novel interaction with clathrin light chain. J. Biol. Chem. 277, 19897-19904. [DOI] [PubMed] [Google Scholar]
- Maniak, M. (2002). Conserved features of endocytosis in Dictyostelium. Int. Rev. Cytol. 221, 257-287. [DOI] [PubMed] [Google Scholar]
- McNiven, M.A., Kim, L., Krueger, E.W., Orth, J.D., Cao, H., and Wong, T.W. (2000). Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 151, 187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield, C.J., Feldman, M.E., Wan, L., and Almers, W. (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4, 691-698. [DOI] [PubMed] [Google Scholar]
- Merrifield, C.J., Moss, S.E., Ballestrem, C., Imhof, B.A., Giese, G., Wunderlich, I., and Almers, W. (1999). Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat. Cell Biol. 1, 72-74. [DOI] [PubMed] [Google Scholar]
- Metzler, M., Legendre-Guillemin, V., Gan, L., Chopra, V., Kwok, A., McPherson, P.S., and Hayden, M.R. (2001). HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J. Biol. Chem. 276, 39271-39276. [DOI] [PubMed] [Google Scholar]
- Metzler, M. et al. (2003). Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 22, 3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise-Omata, S., Montagne, B., Deckert, M., Wienands, J., and Acuto, O. (2003). Mammalian actin binding protein 1 is essential for endocytosis but not lamellipodia formation: functional analysis by RNA interference. Biochem. Biophys. Res. Commun. 301, 704-710. [DOI] [PubMed] [Google Scholar]
- Mishra, S.K., Agostinelli, N.R., Brett, T.J., Mizukami, I., Ross, T.S., and Traub, L.M. (2001). Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J. Biol. Chem. 276, 46230-46236. [DOI] [PubMed] [Google Scholar]
- Munn, A.L. (2001). Molecular requirements for the internalisation step of endocytosis: insights from yeast. Biochim. Biophys. Acta 1535, 236-257. [DOI] [PubMed] [Google Scholar]
- Ochoa, G.C. et al. (2000). A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150, 377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, J.D., Krueger, E.W., Cao, H., and McNiven, M.A. (2002). The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc. Natl. Acad. Sci. USA 99, 167-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans, L., Puntener, D., and Helenius, A. (2002). Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science 296, 535-539. [DOI] [PubMed] [Google Scholar]
- Qualmann, B., and Kelly, R.B. (2000). Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J. Cell Biol. 148, 1047-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozelle, A.L. et al. (2000). Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311-320. [DOI] [PubMed] [Google Scholar]
- Schafer, D.A., D'Souza-Schorey, C., and Cooper, J.A. (2000). Actin assembly at membranes controlled by ARF6. Traffic 1, 896-907. [DOI] [PubMed] [Google Scholar]
- Schafer, D.A., Weed, S.A., Binns, D., Karginov, A.V., Parsons, J.T., and Cooper, J.A. (2002). Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 12, 1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, J.D., Cummings, K.B., Huyer, G., Michaelis, S., and Wendland, B. (2001). Yeast as a model system for studying endocytosis. Exp. Cell Res. 271, 1-9. [DOI] [PubMed] [Google Scholar]
- Taunton, J., Rowning, B.A., Coughlin, M.L., Wu, M., Moon, R.T., Mitchison, T.J., and Larabell, C.A. (2000). Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno, T., Liu, J., Zhang, P., Fan, Y., Egile, C., Li, R., Mueller, S.C., and Zhan, X. (2001). Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3, 259-266. [DOI] [PubMed] [Google Scholar]
- Waelter, S. et al. (2001). The huntingtin interacting protein HIP1 is a clathrin and alpha-adaptin-binding protein involved in receptor-mediated endocytosis. Hum. Mol. Genet. 10, 1807-1817. [DOI] [PubMed] [Google Scholar]
- Wanker, E.E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J., and Lehrach, H. (1997). HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6, 487-495. [DOI] [PubMed] [Google Scholar]
- Weaver, A.M., Karginov, A.V., Kinley, A.W., Weed, S.A., Li, Y., Parsons, J.T., and Cooper, J.A. (2001). Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370-374. [DOI] [PubMed] [Google Scholar]
- Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A.L., and Riezman, H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Cope, M.J., and Drubin, D.G. (1999). Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol. Biol. Cell 10, 2265-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. et al. (1999). Antigen receptor-induced activation and cytoskeletal rearrangement are impaired in Wiskott-Aldrich syndrome protein-deficient lymphocytes. J. Exp. Med. 190, 1329-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.