Abstract
The correct asymmetric placement of inner organs is termed situs solitus and is determined early during development. Failure in symmetry breaking results in conditions ranging from randomized organ arrangement to a complete mirror image, often accompanied by severe congenital heart defects (CHD). We found that the zebrafish homolog of mammalian G protein-coupled receptor kinase 5 (GRK5) employs non-canonical, receptor-independent functions to secure symmetry breaking. Knockdown of GRK5’s closest homolog in zebrafish embryos, Grk5l, is sufficient to randomize cardiac looping and left-right asymmetry. Mechanistically, we found that loss of GRK5 increases mTORC1 activity. This causes elongation of motile cilia in the organ of laterality, a consequence known to be sufficient to trigger false organ arrangement. By fine-tuning mTORC1, GRK5 thus has an unanticipated function during early development, besides its well-characterized role in the adult heart. These findings thus could implicate GRK5 as a susceptibility allele for certain cases of CHD.
INTRODUCTION
Congenital heart disease (CHD) is a frequently occurring, developmental malformation and constitutes a high risk for infant mortality (Pierpont et al., 2007). It is characterized by abnormal development of the heart and its connected structures. Importantly, heart development is precisely controlled spatially as well as temporally. It starts long before the actual heart tube or its progenitor cells become evident. In particular the future heart’s shape and placement relative to the other internal organs is determined by early left-right (LR) symmetry breaking in vertebrates (Ramsdell, 2005). At the cellular level, the most important structure for asymmetry is the temporal organ of laterality. It is a cup-like structure at the distal end of the notochord. Motile cilia within the temporal organ of laterality induce a flow by coordinated movement that triggers the expression of genes only on the left side of the body and thus internal asymmetry along the body axis (Essner et al., 2002; Okada et al., 2005; Oteiza et al., 2008). Not surprisingly, disturbances in the formation or function of the temporal organ of laterality interfere with proper heart development. This is reflected by the fact, that CHD is very common in individuals with irregular asymmetry establishment (Sutherland and Ware, 2009). This condition, where internal organs appear to find their placement randomly is known as isomerism, situs ambiguous or heterotaxy. The seriousness of the condition varies widely. While some patients experience complete rearrangement of internal organs such as in left or right isomerism, there can be cases, in which the heart occurs to be the only affected structure (i.e. dextrocardia).
The molecular causes underlying ciliary diseases and as such situs anomalies have been the focus of many studies in the past decade. Many signaling cascades were found to be of importance including basic morphogens such as Sonic hedgehog (Shh) or Bone morphogenetic protein 4 (Bmp4) (Schilling et al., 1999). In addition to those the mTOR pathway has emerged as a key pathway in ciliary physiology. Interestingly, mTOR activity appears to be controlled through bending of cilia. In kidney cells, loss of cilia or the movement thereof resulted in unrestricted mTOR signaling and cell growth (Boehlke et al., 2010). It thus has been implicated in the development of polycystic kidney disease (PKD) (Zullo et al., 2010), which belongs to the family of cilia-based diseases. In zebrafish, mTOR signaling could be linked to symmetry breaking. Deregulated mTOR activity within the organ of laterality or during the stages, when this organ is functional, has been shown to randomize lateralization (DiBella et al., 2009; Yuan et al., 2012).
GRK5 is one of seven kinases in vertebrates, which were named after their ability to terminate GPCR signal transduction (Premont and Gainetdinov, 2007). It is highly abundant in the heart with the effect that GRK5 is a major regulator of cardiac GPCRs and thus has a vital influence on heart physiology (Chen et al., 2001; Eckhart et al., 2000; Liggett et al., 2008; Martini et al., 2008; Rockman et al., 1996). Recent studies revealed that GRK5 modulates signal transduction also from non-GPCR receptors and even non-receptor proteins (Huang et al., 2011b). Together with its action on adrenoceptors (Chen et al., 2001; Rockman et al., 1996), it is thus capable of significantly altering cardiac physiology by interacting with molecules such as NF κ B or histone deacetylases (Martini et al., 2008; Sorriento et al., 2010). These new functions are also promoted by GRK5’s ability to act not only close to the cell membrane, but also in other subcellular localizations, such as the nucleus (Gold et al., 2012; Michal et al., 2012).
Interestingly, GRK5 is already expressed in the developing heart (Premont et al., 1999). We thus hypothesized that GRK5 may play an additional, yet unreported role in heart formation during early development, besides controlling proper heart physiology in the adult organism.
RESULTS and DISCUSSION
GRK5 facilitates cardiac looping
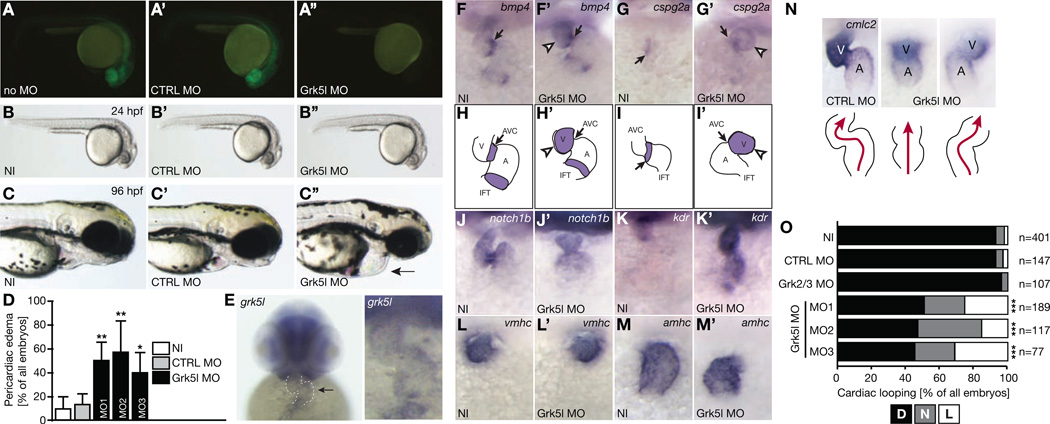
To investigate a potential role of GRK5 in the developing heart, we have used zebrafish as a model due to its advantages for developmental studies. We have applied a commonly used knockdown (KD) approach to generate GRK5 loss of function zebrafish embryos. Zebrafish have two genes located on two different chromosomes with homology to human GRK5. These homologs share little nucleotide sequence homology between one another. Multiple protein alignment analyses demonstrate that zebrafish Grk5l, encoded on chromosome 8, is the closer homolog to human GRK5 compared to zebrafish Grk5 on chromosome 10 (Figure S1A, B). Thus, a translation blocking antisense morpholino (MO) targeting the 5’-untranslated region (UTR) of Grk5l (Grk5l MO) was designed as well as a 5 bases mismatch control MO (CTRL MO). To confirm that the translation blocking MO targeting grk5l sufficiently depletes Grk5l, we co-injected capped mRNA encoding for a Grk5l-GFP fusion protein, which was preceded by parts of grk5l’s 5’-UTR. Embryos injected with Grk5l MO failed to produce GFP fluorescence (Figure 1A, A’, A’’). Moreover, KD efficiency was verified by Western blot (Figure S1C) and a MO binding assay (Figure S1D). Importantly, the 5’-UTRs of both grk5 variants in zebrafish are not conserved and MOs targeting grk5l are not predicted to bind to the UTR of grk5. We also did not find a feedback loop altering mRNA levels of grk5 upon KD of Grk5l (Figure S1E) suggesting that any phenotype observed was likely due to specific KD of Grk5l.
Figure 1. Grk5l KD in zebrafish affects early heart development.
A, A’, A’’, Grk5l MO prevents translation of co-injected RNA coding for Grk5l-GFP fused to parts of its 5’-UTR and results in fish showing no glowing (A’’), while RNA injection alone (A) or in combination with the CTRL MO (A’) produces strong GFP expression.
B, B’, B’’, 24 hpf zebrafish embryos injected with Grk5l MO (B’’) compared to non-injected embryos (NI) (B) or those injected with CTRL MO (B’).
C, C’, C’’, Live embryos (96 hpf). Arrow indicates pericardial edema in Grk5l morphants.
D, Bar graph displaying the percentage of embryos with pericardiac edema. n=3–10 experiments, 76 to 340 embryos. CTRL MO vs. Grk5l MO: p=0.0023; CTRL MO vs Grk5l MO2: p=0.0016, CTRL MO vs. Grk5l MO3: p=0.0213.
E, grk5l transcripts in the heart of 48 hpf zebrafish embryos (arrow and higher magnification).
F, F’, Bmp4 transcripts are upregulated and dispersed throughout the heart of Grk5l morphants (F’). n=10–35.
G, G’, Upregulation of cspg2a upon injection of Grk5l MO (G’). n=16–18.
H, H’, Illustration of normal distribution of bmp4 in controls (H) and widespread expression in Grk5l morphants (H’).
I, I’, Cartoon showing the regular as well as aberrant distribution of cspg2a in controls (I) and Grk5l MO (I’) injected fish.
J, J’, Notch1b fails to accumulate in the future valve region upon Grk5l KD (J’). n=16–25.
K, K’, Kdr is strongly upregulated upon KD of Grk5l (K’) compared to control fish (K). n=5–22.
L, L’, Ventricular fate as shown by WMISH for vmhc is properly established when Grk5l is lost. n=27–32.
M, M’, Amhc expression in the atrium. n=18–23.
N, WMISH for cmlc2 at 50 hpf revealing altered cardiac looping in Grk5l morphants (arrow). Cartoon depicts heart morphology and blood flow. A, atrium, V, ventricle.
O, Summary of heart looping after injection with MOs targeting either Grk2/3 or Grk5l. For Grk5l, three different MOs were tested. D, D-loop, N, no loop, L, L-loop. CTRL MO vs. Grk5l MO: p<0.0001; CTRL MO vs Grk5l MO2: p<0.0001; CTRL MO vs Grk5l MO3: p<0.0001.
F to M: All images: 48 to 52 hpf.
MO-mediated KD of Grk5l results initially in viable embryos which are morphologically very similar to non-injected WT (NI) embryos or embryos injected with the CTRL MO (Figure 1B, B’, B’’). After KD of Grk5l, we allowed embryos to develop further and observed that Grk5l KD induces pericardiac edema and cardiac dilatation (Figure 1C to D). Two additional MOs targeting Grk5l either upstream of the first MO in the 5’-UTR or downstream around the start codon could validate this cardiac phenotype (Figure 1D and S1F). We thus examined Grk5l expression in the heart and observed grk5l transcripts in cardiac tissue at 48 hours post fertilization (hpf) (Figure 1E). Furthermore, depletion of Grk5l affected also expression of genes in the heart, which were shown to be important for valve seeding (Figure 1F–K’). Chamber formation, however, occurred regularly (Figure 1L to M’). But importantly, hearts of Grk5l KD embryos injected with any of the three MOs either failed to loop or showed an inverse loop in roughly half of all embryos (Figure 1N, O), which might be attributed to failure in establishing LR asymmetry. KD of the single zebrafish homolog of GRK2 and GRK3, Grk2/3, displayed regular D-loops (Figure 1O). Interestingly, loss of the other Grk5 in zebrafish resulted also in abnormal heart looping, although to a lesser extent. Combined KD of both, Grk5 and Grk5l, did not worsen the phenotype (Figure S1G, H). This implies that this function in early heart looping may be executed by both grk5 genes. From these data we conclude that Grk5l influences heart development during its early formation.
Grk5l governs symmetry breaking
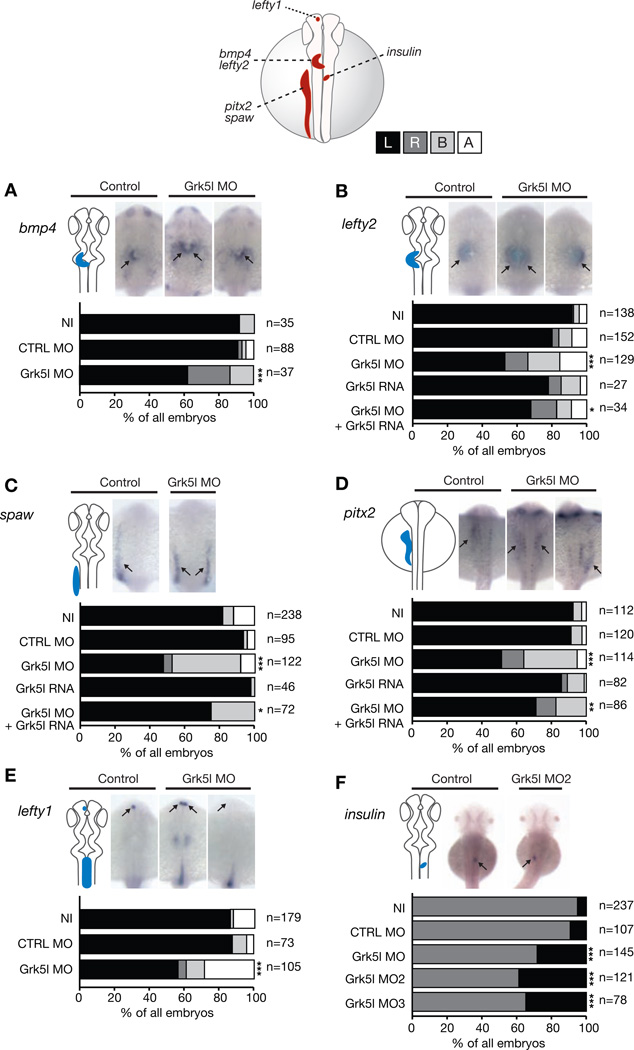
Morphological defects of cardiac structures in the context of looping irregularities are indicators of disrupted LR asymmetry development. Similarly, human heterotaxy patients, who suffer from random placement of thoracic and/or abdominal organs, often develop dextrocardia as well as valvular or septal defects (i.e. common single ventricle). Thus, frequent complications in heterotaxy patients include CHD. To test if Grk5l might govern proper cardiogenesis by ensuring asymmetry development, we analyzed genes which are expressed unilaterally at specific developmental stages (Sutherland and Ware, 2009). Using whole mount in situ hybridization (WMISH) at late somite stages (ss), we found that genes expressed in the left heart region, bmp4 and the nodal target gene lefty2, are misexpressed upon Grk5l KD (Figure 2A, B). We next wondered, if Grk5l’s impact on lateralization could be seen earlier on the level of the lateral plate mesoderm (LPM), which also gives rise to the heart. The LPM-specific genes southpaw (spaw) and pitx2 are ambiguously expressed in Grk5l morphants (Figure 2C, D). However, this false pattern can be partially rescued by co-injection of MO-insensitive synthetic grk5l RNA (Figure 2C, D). Similarly, the correct distribution of lefty2 could be rescued by grk5l RNA (Figure 2B). Analysis of lefty1 expression in the left diencephalon as well as pancreas placement in 2 days-post-fertilization (dpf) embryos demonstrated further that the lateralization defect upon loss of Grk5l is general (Figure 2E, F). Grk5l may therefore be a so far unanticipated gene involved in the determination of LR asymmetry.
Figure 2. Grk5l governs LR asymmetry development.
WMISH for leftward marker genes at 20–22 ss with the respective expression domain outlined next to the images. Bar graphs display percentages of expression on the left side (black), on the right (dark grey), on both sides of the midline (light grey) or no expression at all (white).
A, bmp4 expression in the heart region is randomized upon Grk5l KD. CTRL MO vs Grk5l MO: p=0.0056.
B, Random distribution of lefty2 can be partially rescued by grk5l RNA. CTRL MO vs Grk5l MO: p<0.0001; Grk5l MO vs Grk5l MO+grk5l RNA: p=0.0425.
C, spaw in the LPM is affected upon loss of Grk5l, but can be rescued by co-injection of grk5l mRNA. CTRL MO vs Grk5l MO: p<0.0001; Grk5l MO vs Grk5l MO+grk5l RNA: p=0.0002.
D, Ambiguous pitx2 reversed by mRNA encoding for grk5l. CTRL MO vs Grk5l MO: p<0.0001; Grk5l MO vs Grk5l MO+grk5l RNA: p=0.0068.
E, Lefty1 transcripts in the dorsal diencephalon are ambiguously distributed, when Grk5l is lost. CTRL MO vs Grk5l MO: p<0.0001.
F, Loss of Grk5l interferes with pancreas placement, as can be seen by the expression of insulin. CTRL MO vs Grk5l MO: p=0.0009; CTRL MO vs Grk5l MO2: p<0.0001; CTRL MO vs Grk5l MO3: p<0.0001.
Cilia length depends on Grk5l
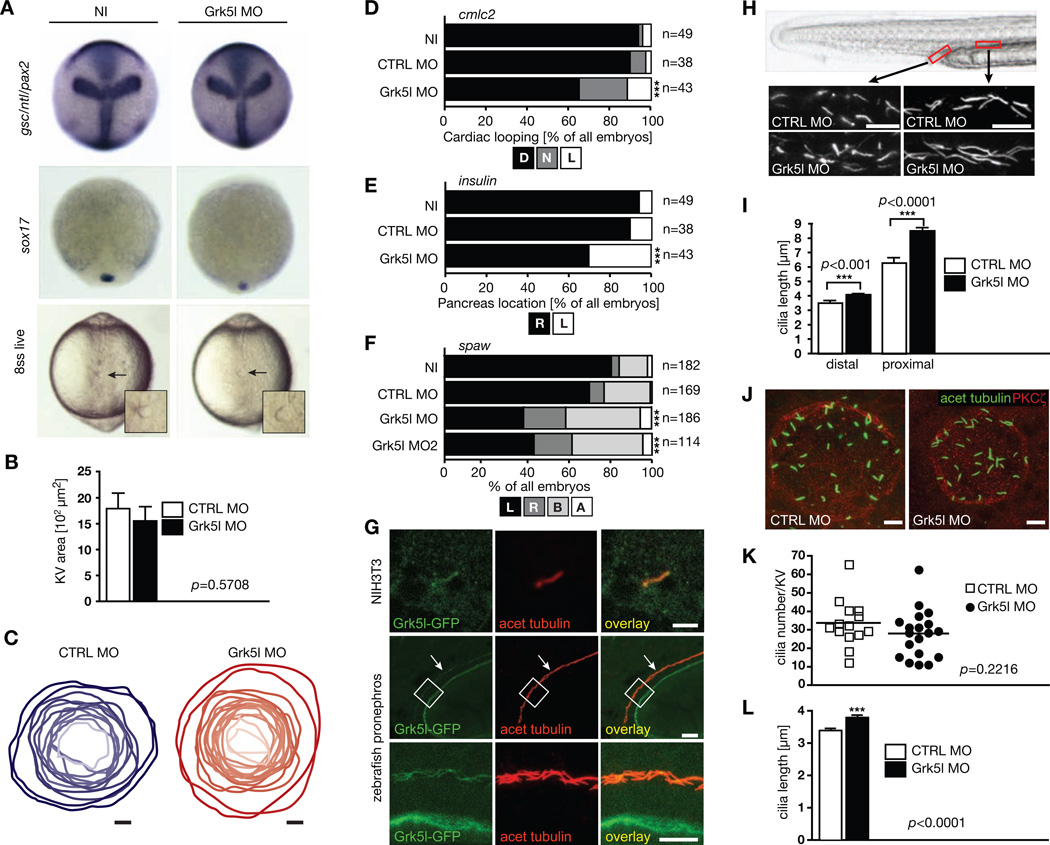
A prominent model in LR asymmetry development depends on the formation and function of cilia in the temporary organ of laterality (Kupffer’s vesicle, KV, in zebrafish or the node in mice) (Sutherland and Ware, 2009). In line with this, zebrafish mutants for ciliary proteins often develop asymmetry defects, hydrocephalus and a distinct body curvature (Sun et al., 2004). Such curved body axis could also be observed upon depletion of Grk5l and was most prominent in embryos injected with MO3 (Figure S1F). We thus questioned whether Grk5l might act on or within the KV, especially since we found transcripts of grk5l, which is expressed throughout development (Figure S2A–K), enriched in the tailbud region of embryos (Figure S2D–G).
First we tested, if the asymmetry defect was due to improper KV establishment. We analyzed midline formation as well as dorsal forerunner cell (DFC) clustering, both essential processes preceding KV differentiation (Bisgrove et al., 1999; Oteiza et al., 2008). Neither of those was altered upon Grk5l KD (Figure 3A). Additionally, the KV area was not changed (Figure 3B, C) indicating that Grk5l may potentially exert its function within the KV itself.
Figure 3. Grk5l acts in the organ of laterality.
A, Combined WMISH for gsc, ntl and pax2 (10hpf). DFCs cluster normally at 90% epiboly as shown by sox17 (n=32–35). Live imaging at 8 ss shows that the KV forms. Arrows indicate KV, higher magnification in inset.
B, KV-specific KD did not alter KV area as assessed by PKCζ staining at 8 ss. n=14–19 embryos. CTRL MO: 1793±299 µm2, Grk5l MO: 1555±277 µm2.
C, Outline of KVs upon injection of CTRL MO (blue) or of Grk5l MO (red). Dependent on the respective size, the KV is shown in darker or brighter shading. n=12–17 embryos.
D, Impaired heart looping by Grk5l depletion in KV cells detected by cmlc2 ISH at 50 hpf. CTRL MO vs Grk5l MO: p<0.0001.
E, Insulin-positive cells of the pancreas are more often on the left side of the midline upon KV-specific KD of Grk5l. CTRL MO vs Grk5l MO: p=0.0007.
F, KV-directed ablation of Grk5l randomizes spaw. CTRL MO vs Grk5l MO: p<0.0001; CTRL MO vs Grk5l MO2: p=0.0002.
G, Grk5l localizes to primary cilia in NIH3T3 cells and motile cilia in the developing zebrafish kidney. Images show Grk5l-GFP after transfection in cells or injection of capped RNA into 1 cell stage embryos. Scale bars: 3 µm (NIH3T3), 20 µm (low magnification of pronephros), 10 µm (higher magnification).
H, Elongation of motile cilia of the zebrafish pronephros (2 dpf). Cilia in the distal (CTRL MO: 3.571±0.116 µm, Grk5l MO: 4.090±0.0852 µm) and proximal part (CTRL MO: 6.347±0.318 µm, Grk5l MO: 8.526±0.220 µm) were analyzed. Approximate areas of analysis are indicated in red in the live image of a 2 dpf zebrafish tail. Scale bar: 10 µm.
I, Bar graph summarizing pronephric cilia measurements (n=32–101 cilia).
J, Motile cilia at 8 ss with MOs targeted to KV cells. Images were selected for cilia length. Scale bar: 10 µm.
K, Cilia number was not altered in the KV by MOs targeted to the KV (8 ss). n=14–19 embryos. CTRL MO: 34.14±3.372 cilia; Grk5l MO: 28.53±2.960 cilia.
L, Grk5l depletion in the KV increases cilia length. n=460–513 cilia at 8 ss. CTRL MO: 3.344±0.049 µm; Grk5l MO: 3.748±0.054 µm.; p<0.0001, two-tailed, unpaired t-test.
See also Figure S3.
To test this, we targeted the MO to DFCs, which are the cells giving rise to the KV. This can be achieved by injection at the 1k cell stage. MO or RNA is then specifically delivered to DFCs and thus the KV (Amack and Yost, 2004). This chimeric KD of Grk5l was sufficient to interfere with proper heart looping as well as pancreas placement (Figure 3D, E). In addition we observed randomization of spaw expression (Figure 3F). We next wondered if Grk5l would localize to cilia. We detected Grk5l in primary cilia when expressed in NIH3T3 cells and in motile cilia in the pronephric duct (Figure 3G). We therefore investigated if Grk5l KD would affect cilia formation in zebrafish. Motile cilia of the developing pronephros were longer in Grk5l morphants compared to control embryos (Figure 3H, I). Those results suggested that a ciliary defect in the KV may possibly be the basis for the Grk5l phenotype. While we did not observe a significant change in cilia number (Figure 3J, K), we found cilia of Grk5l-depleted KVs were significantly longer than in control injected embryos (Figure 3J, L). These findings are again in line with a common observation that zebrafish with ciliary deficiencies display a pronounced body curvature, which we had also observed (Figure S1F). These results suggest that Grk5l helps to regulate cilia formation in the developing embryo and by that ensures symmetry breaking.
Grk5l fine-tunes mTORC1 activity during symmetry breaking
We next wanted to identify the molecular mechanism causing the Grk5l KD phenotype. Overstimulation of α1b-adrenoceptors (α1b-AR) can cause lateralization defects (Fujinaga et al., 1994). As GRK5 attenuates elevated α1b-AR signaling (Eckhart et al., 2000), we hypothesized that the observed phenotype could be caused by loss of Grk5l-mediated α1b-AR desensitization. However, WD-4101, an α1b-AR specific inhibitor did not rescue situs inversus frequency in Grk5l KD embryos (Figure S3A). We thus wondered if the observed phenotype would potentially be due to Grk5l modulating signaling not at the GPCR level. However, no differences were found when we studied target gene expression of the Shh or retinoic acid signaling in Grk5l morphants (Figure S3B–D). Both pathways had been implicated in symmetry breaking before (Huang et al., 2011a; Schilling et al., 1999).
Previously, we have reported that Grk5 facilitates canonical Wnt signaling (Chen et al., 2009). As Grk5l might share this function, we tested its effect on axin2 (Figure S3E). Grk5l appears to be a positive modulator of canonical Wnt signaling. Canonical Wnt signaling has also been reported to steer LR asymmetry development (Caron et al., 2012). However, impaired Wnt signaling is associated with shorter cilia, while we see elongated cilia. Grk5l’s action on Wnt signaling may therefore not necessarily be related to its role in asymmetry development.
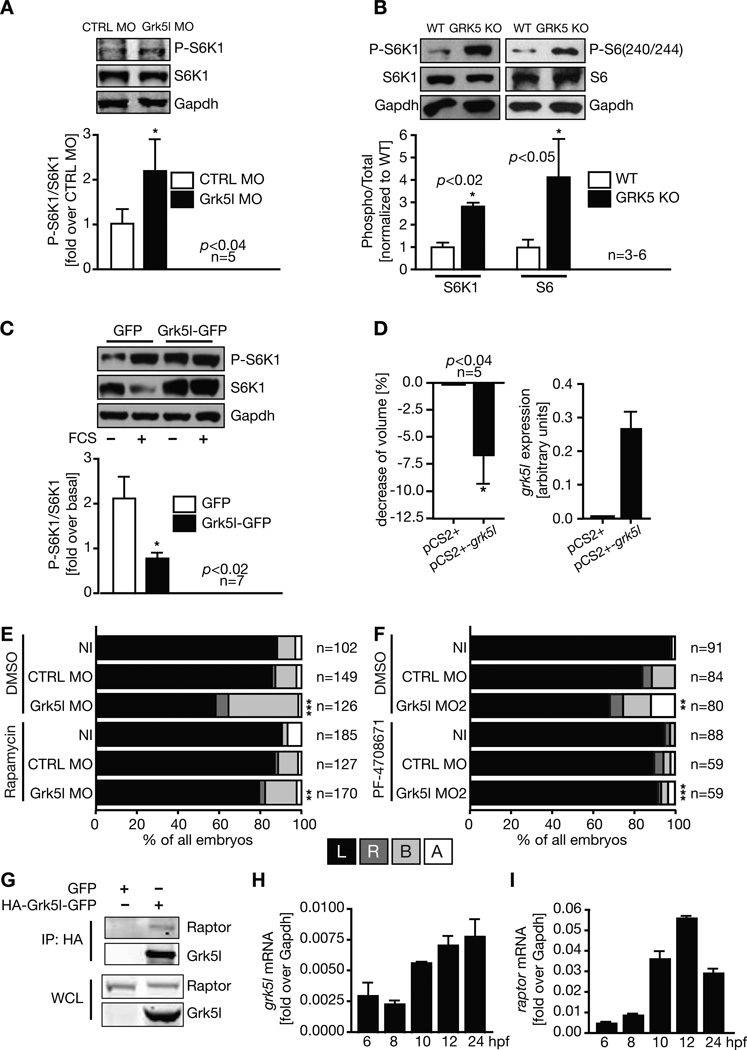
Recent studies on ciliary diseases including PKD and heterotaxy have highlighted the importance of the mammalian target of rapamycin complex 1 (mTORC1), which regulates cell growth (Zoncu et al., 2011). mTORC1 signaling has been connected to ciliary architecture and function and thus conditions which rely on cilia (Boehlke et al., 2010; DiBella et al., 2009; Yuan et al., 2012). We therefore investigated whether Grk5l influences mTORC1 activity by measuring phosphorylation of its target S6K1 in zebrafish embryos. We detected an increase in mTORC1 activity implicating a negative impact of Grk5l on mTORC1 activity (Figure 4A). This finding could be reproduced in hearts of young GRK5 knockout (KO) mice (Figure 4B). In addition, we found that transient overexpression of Grk5l in HEK293T cells caused a reduction of mTORC1 activity (Figure 4C) and a decrease in cell volume (Figure 4D). These results suggest that depletion of Grk5l might remove a dampening effect on mTORC1 activity and that an apparent mTORC1 overstimulation would be causal for the lateralization defect. To test this, we treated Grk5l KD embryos with the mTORC1 inhibitor rapamycin (rapa). Rapa rescued the lateralization phenotype of Grk5l KD embryos (Figure 4E and S4A), while control embryos were unaffected. Furthermore, a specific S6K1 inhibitor (Pearce et al., 2010; Rosner et al., 2012) could also rescue the phenotype (Figure 4F). We conclude from this that increased mTORC1 activity is causal for the asymmetry defects in the Grk5l KD embryos. To determine whether there may be a direct physical interaction, we performed co-immunoprecipitation assays in transfected as well as native cells. Raptor co-precipitated with overexpressed Grk5l (Figure 4G) as well as endogenous GRK5 (Figure S4B). Grk5l also co-localizes with the mTORC1 components mTOR, Raptor, and mLST8 in HEK293T cells (data not shown). These data are in line with our in vivo results and corroborate that Grk5l negatively regulates mTORC1 activity, probably through direct interaction with Raptor.
Figure 4. Grk5l limits mTOR signaling.
A, Western blot analysis of zebrafish embryos (6–8 ss) for S6K1 phosphorylation. Representative images of a single blot shown.
B, Western blot of mouse heart lysates of wild-type (WT) and GRK5 KO mice for phosphorylation of S6K1 and its target ribosomal protein S6.
C, Overexpression of Grk5l-GFP in cells attenuates TORC1 activity towards S6K1.
D, Cell volume of Grk5l transfected cells (left, mean ± SEM). Cells were controlled for grk5l expression (right graph, means ± SD, n=2–3).
E, Reversal of the Grk5l lateralization phenotype by rapa. Analysis of spaw in the presence of DMSO or rapa from 6 to 10 hpf. CTRL MODMSO vs Grk5l MODMSO: p<0.0001; Grk5l MODMSO vs Grk5l MORapa: p=0.0033.
F, Inhibition of S6K1 activity by PF-470871 reverses the spaw misexpression upon Grk5l KD. CTRL MODMSO vs Grk5l MO2DMSO: p=0.0063; Grk5l MO2DMSO vs Grk5l MO2PF: p<0.0001.
G, Co-immunoprecipitation of transfected GFP or HA- Grk5l-GFP and endogenous Raptor using a HA-antibody. IP, immunoprecipitation; WCL, whole cell lysate.
H, Relative expression levels of grk5l obtained by qPCR (mean ± SD, n=2).
I, raptor mRNA levels (mean ± SD, n=2).
See also Figures S2, S3, and S4.
We also analyzed the spatial expression patterns of grk5l and raptor and found them to be very similar (Figure S2). Moreover, mRNA levels of both grk5l as well as raptor substantially increase at 10 hpf (Figure 4H, I), when the KV starts to form, the essential structure for symmetry breaking. We thus reasoned that a rescue of the Grk5l KD phenotype might also be achieved by inhibition of mTORC1 from 10 hpf on, which is what we observed (Figure S4A). Moreover, randomization of laterality could also be reversed by rapa, when Grk5l was knocked down specifically in the KV (Figure S4C). We conclude that Grk5l is needed to fine-tune mTORC1 activity at the level of the temporal organ of laterality in order to accomplish successful symmetry breaking.
Previously, it has always been assumed that GRK5 would be dispensable for embryonic development, primarily because GRK5 KO mice are viable (Gainetdinov et al., 1999). Yet, combined loss of GRK5 and GRK6 results in embryonic lethality (Table S1). A detailed characterization of this lethality remains to be undertaken and is subject of future studies in our lab.
In this study, we have used zebrafish to delineate the importance of vertebrate GRK5 during development. Similar to GRK5 KO mice, depletion of Grk5l in zebrafish does not necessarily result in embryonic lethality, but in a complex, often overlooked embryonic phenotype. Similarly, heterotaxy can have a plethora of mild phenotypes that are easily missed. Future studies will thus be required to illuminate if and how GRK5 would influence symmetry breaking in mammals.
In summary, we here provide evidence that vertebrate GRK5 possesses physiological relevance during development of the heart. We found that the close homolog in zebrafish, Grk5l, is required to steer lateralization in zebrafish. By that it ensures proper cardiogenesis and may potentially prevent congenital heart defects, which are common complications in situs anomalies. Mechanistically, Grk5l confines mTORC1 signaling at the level of the temporal organ of asymmetry and controls ciliary morphology. Moreover, Grk5l potentially influences heart morphology by affecting expression of cardiac genes. The well-characterized role vertebrate GRK5 plays in the adult heart thus needs to be expanded to very early steps during development, when the body plan and thus a properly functioning heart is defined.
Experimental Procedures
Zebrafish and mouse experiments
Zebrafish were maintained under standard conditions at a 14 hours light and 10 hours dark cycle. Fertilized eggs were injected and allowed to develop at 28.5 °C. Mice lacking GRK5 and/or GRK6 were generated as described previously (Gainetdinov et al., 2003; Gainetdinov et al., 1999) and bred from heterozygous breeding pairs. Tissue for Western blotting was harvested from 3 to 5 months old animals. Housing of mice was under a 12 h light/dark cycle in a SPF facility. All husbandry and experiments described here were approved by German authorities or by the IACUC at Duke University. For further details regarding material and methods please refer to extended experimental procedures.
Supplementary Material
Highlights.
GRK5 has a role during early embryonic development by guiding symmetry breaking.
GRK5 modulates cilia length in the organ of laterality by negatively regulating mTOR.
GRK5 regulates heart development.
ACKNOWLEDGEMENTS
We thank the Zebrafish International Resource Center and many colleagues for providing reagents, Sabrina Matysik for technical assistance, Cornelia Donow for help with the MO binding assay and Jim Burris and Julia Schäfer for fish maintenance. This work was supported in part by: FP7 of the European Commission (MP), Deutsche Stiftung für Herzforschung (MP), NIH grants NS-019576 and MH-073853 (MGC). During the initial stages of this work MP was the recipient of a Marie Curie Outgoing International Fellowship of the European Commission.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, Beyer T, Janusch H, Hamann C, Godel M, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron A, Xu X, Lin X. Wnt/beta-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer's vesicle. Development. 2012;139:514–524. doi: 10.1242/dev.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EP, Bittner HB, Akhter SA, Koch WJ, Davis RD. Myocardial function in hearts with transgenic overexpression of the G protein-coupled receptor kinase 5. The Annals of thoracic surgery. 2001;71:1320–1324. doi: 10.1016/s0003-4975(00)01754-9. [DOI] [PubMed] [Google Scholar]
- Chen M, Philipp M, Wang J, Premont RT, Garrison TR, Caron MG, Lefkowitz RJ, Chen W. G Protein-coupled receptor kinases phosphorylate LRP6 in the Wnt pathway. J Biol Chem. 2009;284:35040–35048. doi: 10.1074/jbc.M109.047456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Park A, Sun Z. Zebrafish Tsc1 reveals functional interactions between the cilium and the TOR pathway. Hum Mol Genet. 2009;18:595–606. doi: 10.1093/hmg/ddn384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart AD, Duncan SJ, Penn RB, Benovic JL, Lefkowitz RJ, Koch WJ. Hybrid transgenic mice reveal in vivo specificity of G protein-coupled receptor kinases in the heart. Circ Res. 2000;86:43–50. doi: 10.1161/01.res.86.1.43. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- Fujinaga M, Hoffman BB, Baden JM. Receptor subtype and intracellular signal transduction pathway associated with situs inversus induced by alpha 1 adrenergic stimulation in rat embryos. Dev Biol. 1994;162:558–567. doi: 10.1006/dbio.1994.1109. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Bohn LM, Sotnikova TD, Cyr M, Laakso A, Macrae AD, Torres GE, Kim KM, Lefkowitz RJ, Caron MG, et al. Dopaminergic supersensitivity in G protein-coupled receptor kinase 6-deficient mice. Neuron. 2003;38:291–303. doi: 10.1016/s0896-6273(03)00192-2. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Bohn LM, Walker JK, Laporte SA, Macrae AD, Caron MG, Lefkowitz RJ, Premont RT. Muscarinic supersensitivity and impaired receptor desensitization in G protein-coupled receptor kinase 5-deficient mice. Neuron. 1999;24:1029–1036. doi: 10.1016/s0896-6273(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Gold JI, Gao E, Shang X, Premont RT, Koch WJ. Determining the absolute requirement of g protein-coupled receptor kinase 5 for pathological cardiac hypertrophy: short communication. Circ Res. 2012;111:1048–1053. doi: 10.1161/CIRCRESAHA.112.273367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ma J, Liu X, Zhang Y, Luo L. Retinoic acid signaling sequentially controls visceral and heart laterality in zebrafish. J Biol Chem. 2011a;286:28533–28543. doi: 10.1074/jbc.M111.244327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZM, Gold JI, Koch WJ. G protein-coupled receptor kinases in normal and failing myocardium. Frontiers in bioscience : a journal and virtual library. 2011b;17:3047–3060. doi: 10.2741/3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe KM, Thiberge SY, Bisher ME, Burdine RD. Imaging cilia in zebrafish. Methods Cell Biol. 2010;97:415–435. doi: 10.1016/S0091-679X(10)97022-2. [DOI] [PubMed] [Google Scholar]
- Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini JS, Raake P, Vinge LE, DeGeorge BR, Jr, Chuprun JK, Harris DM, Gao E, Eckhart AD, Pitcher JA, Koch WJ. Uncovering G protein-coupled receptor kinase-5 as a histone deacetylase kinase in the nucleus of cardiomyocytes. Proc Natl Acad Sci U S A. 2008;105:12457–12462. doi: 10.1073/pnas.0803153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michal AM, So CH, Beeharry N, Shankar H, Mashayekhi R, Yen TJ, Benovic JL. G Protein-coupled receptor kinase 5 is localized to centrosomes and regulates cell cycle progression. J Biol Chem. 2012;287:6928–6940. doi: 10.1074/jbc.M111.298034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Takeda S, Tanaka Y, Belmonte JC, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Oteiza P, Koppen M, Concha ML, Heisenberg CP. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–2813. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Alton GR, Richter DT, Kath JC, Lingardo L, Chapman J, Hwang C, Alessi DR. Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1) Biochem J. 2010;431:245–255. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- Philipp M, Fralish GB, Meloni AR, Chen W, MacInnes AW, Barak LS, Caron MG. Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol Biol Cell. 2008;19:5478–5489. doi: 10.1091/mbc.E08-05-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont ME, Basson CT, Benson DW, Jr, Gelb BD, Giglia TM, Goldmuntz E, McGee G, Sable CA, Srivastava D, Webb CL. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:3015–3038. doi: 10.1161/CIRCULATIONAHA.106.183056. [DOI] [PubMed] [Google Scholar]
- Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annual review of physiology. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- Premont RT, Macrae AD, Aparicio SA, Kendall HE, Welch JE, Lefkowitz RJ. The GRK4 subfamily of G protein-coupled receptor kinases. Alternative splicing, gene organization, and sequence conservation. J Biol Chem. 1999;274:29381–29389. doi: 10.1074/jbc.274.41.29381. [DOI] [PubMed] [Google Scholar]
- Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288:1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Choi DJ, Rahman NU, Akhter SA, Lefkowitz RJ, Koch WJ. Receptor-specific in vivo desensitization by the G protein-coupled receptor kinase-5 in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:9954–9959. doi: 10.1073/pnas.93.18.9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M, Schipany K, Hengstschlager M. p70 S6K1 nuclear localization depends on its mTOR-mediated phosphorylation at T389, but not on its kinase activity towards S6. Amino Acids. 2012;42:2251–2256. doi: 10.1007/s00726-011-0965-4. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Concordet JP, Ingham PW. Regulation of left-right asymmetries in the zebrafish by Shh and BMP4. Dev Biol. 1999;210:277–287. doi: 10.1006/dbio.1999.9214. [DOI] [PubMed] [Google Scholar]
- Sorriento D, Santulli G, Fusco A, Anastasio A, Trimarco B, Iaccarino G. Intracardiac injection of AdGRK5-NT reduces left ventricular hypertrophy by inhibiting NF-kappaB-dependent hypertrophic gene expression. Hypertension. 2010;56:696–704. doi: 10.1161/HYPERTENSIONAHA.110.155960. [DOI] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. American journal of medical genetics Part C, Seminars in medical genetics. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- Yuan S, Li J, Diener DR, Choma MA, Rosenbaum JL, Sun Z. Target-of-rapamycin complex 1 (Torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc Natl Acad Sci U S A. 2012;109:2021–2026. doi: 10.1073/pnas.1112834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo A, Iaconis D, Barra A, Cantone A, Messaddeq N, Capasso G, Dolle P, Igarashi P, Franco B. Kidney-specific inactivation of Ofd1 leads to renal cystic disease associated with upregulation of the mTOR pathway. Hum Mol Genet. 2010;19:2792–2803. doi: 10.1093/hmg/ddq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.