A recently published crystallographic model of GspB in complex with a disaccharide was proposed to provide insight into the interaction of the S. gordonii adhesin with the cell sialyl-T antigen from host cells. However, a close inspection of this model revealed that in this complex a buffer molecule was mistaken for the disaccharide. As a consequence, this complex provides no information regarding the interaction of GspB with host cell receptors.
Keywords: pathogen–host cell recognition, adhesion, misleading ligand identification, model error
Abstract
The structure of a complex between a fragment of the adhesin GspB from Streptococcus gordonii and a disaccharide (PDB entries 3qd1 and 4i8e) has recently been proposed to identify the binding site for the sialyl-T antigen recognized by GspB. This structure exhibits numerous unrealistic and unusual features such as an excessive number of van der Waals clashes and a lack of correlation between atomic structure and experimental electron density. Here, it is shown that the crystallographic data can be fully explained by an alternative model, namely replacing the disaccharide with a buffer molecule. The conclusion is that the experimental data are likely to contain no information regarding the carbohydrate receptor binding site in GspB or the interaction of GspB with host cell receptors.
1. Introduction
The serine-rich adhesin GspB from Streptococcus gordonii promotes the attachment of streptococci to host cells. Recently, the crystal structure of a GspB fragment that encompasses the so-called ligand-binding region of the cell-wall-anchored GspB protein has been published (GspB-BR, residues 233–617; Pyburn et al., 2011 ▶). Concomitantly with the publication in the journal PLoS Pathogens, atomic models of GspB have been deposited in the Protein Data Bank (PDB; Berman et al., 2007 ▶). Whereas two of these models describe the crystal structure of the GspB fragment by itself at resolutions of 1.4 and 1.9 Å (PDB entries 3qc5 and 3qc6), a third model reported at 1.9 Å resolution describes GspB in complex with a disaccharide that mimics the sialyl-T antigen recognized by GspB on host cells (PDB entry 3qd1, superseded by entry 4i8e). This latter complex is of key importance to the publication in PLoS Pathogens since additional mutational analyses focus on those residues that are close to the position of the disaccharide in the atomic model and do not investigate the entire surface of the GspB fragment (Pyburn et al., 2011 ▶).
The atomic coordinates of GspB in complex with a disaccharide (PDB entries 3qd1 and 4i8e) display a number of unusual features that raise questions concerning the correctness of the model. A first indication for this can be obtained from panel B of Fig. 4 in the PLoS Pathogens publication (Pyburn et al., 2011 ▶). This panel depicts a simulated-annealing OMIT map, but the electron density bears no resemblance to the suggested bound disaccharide. This is mirrored by a very low electron-density real-space correlation coefficient, as recently noticed with the program TWILIGHT (Pozharski et al., 2013 ▶; Weichenberger et al., 2013 ▶). Upon contacting the Editors of PLoS Pathogens, a formal correction has been issued on the PLoS Pathogens website. It is stated that the coordinates of the ligand only approximately indicate the position of a disaccharide bound on the surface of GspB. To account for this, the authors submitted a corrected coordinate file, PDB entry 4i8e, which supersedes the initial entry 3qd1. In this entry all occupancies of the atoms of the disaccharide are set to 0.0 without any further notable changes applied to the deposited coordinates.
Here, it is shown that the experimental crystallographic data associated with PDB entry 3qd1/4i8e can be comprehensively explained by an alternative atomic model. This improved model is obtained following the lead of standard crystallographic and physico-chemical indicators, namely the correlation between observed electron density and structure of the ligand, the geometry of non-bonded interactions and a scrutiny of the chemical composition of the crystallization solution. At the same time, however, this improved model indicates that the crystallographic data associated with PDB entry 3qd1/4i8e do not give any information on the binding of the serine-rich repeat adhesin GspB to carbohydrate-displaying host cell receptors.
2. Materials and methods
2.1. Model inspection and validation
Coordinates and structure-factor amplitudes were retrieved from the PDB server and visualized in the program COOT (Emsley & Cowtan, 2004 ▶, Berman et al., 2007 ▶). Inter-residue distances were calculated with the program CONTACT from the CCP4 program suite (Winn et al., 2011 ▶). Electron-density maps were either retrieved directly from the Electron Density Server from Uppsala (EDS; Kleywegt et al., 2004 ▶) or calculated with the program PHENIX (Adams et al., 2010 ▶).
2.2. Structure refinement
For validation purposes two separate refinement runs were performed in parallel with the program PHENIX using identical protocols (Adams et al., 2010 ▶). While in one refinement run the coordinates of PDB entry 4i8e were used as deposited with the Protein Data Bank (Berman et al., 2007 ▶), in a second parallel refinement the disaccharide molecule RMY that mimics the sialyl-T antigen was replaced by 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (commonly referred to as HEPES). All atom occupancies of the ligand HEPES were set to 1.0. The automated refinement included refinement of atom coordinates, real-space refinement and refinement of individual atom B factors. All atom B factors were set to a constant value of 30.0 Å2 prior to any refinement. TLS parameters were not refined. The refinement of the coordinates against the structure-factor amplitudes deposited with entry 4i8e converged after ten cycles.
3. Results
3.1. Quality of the deposited protein models
According to the published data, the deposited crystal structures were solved at resolutions of 1.4, 1.9 and 1.9 Å for PDB entries 3qc5, 3qc6 and 3qd1/4i8e, respectively (Pyburn et al., 2011 ▶). The crystal structure corresponding to PDB entry 3qc5 was solved first using a single Dy3+ derivative. PDB entry 3qc5 then served as a search model for solving entries 3qc6 and 3qd1/4i8e with molecular replacement (Pyburn et al., 2011 ▶). All models were refined to convergence and yielded R factors around 20% or lower (Pyburn et al., 2011 ▶). In each structure the protein chain appears well defined by its electron density. This is supported by the display of an OMIT map showing the positive difference electron density of a short protein chain segment (residues 515 to 520) from PDB entry 3qc5 in panel C of Fig. 2 of Pyburn et al. (2011 ▶). All data and model statistics reported in Table 1 of the PLoS Pathogens publication mirror general expectations for properly determined crystal structures (Pyburn et al., 2011 ▶). Unfortunately, this does not extend to the bound disaccharide in PDB entry 3qd1/4i8e.
3.2. Unusual ligand features in PDB entry 3qd1/4i8e
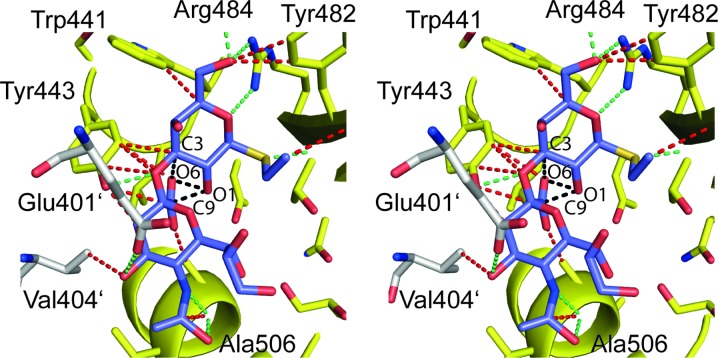
In the structure represented by PDB entry 3qd1/4i8e numerous unusually close contacts are present between the bound RMY disaccharide and the surrounding protein atoms (Fig. 1 ▶, Table 1 ▶). Close contacts such as those between atom C17 of the disaccharide and GspB atom Tyr443 OH (interatomic distance = 2.34 Å) and many others (Fig. 1 ▶, Table 1 ▶) physically translate into excessively high van der Waals repulsion energies, and outweigh by far any favorable protein ligand interaction energies. The same holds true for intramolecular van der Waals clashes. Close contacts such as those between atoms RMY O1 and RMY C9 (distance = 1.89 Å) or atoms RMY O1 and RMY O6 (2.02 Å) remove any inherent propensity of the ligand to adopt such a conformation in solution (Fig. 1 ▶).
Figure 1.
Stereo representation displaying all contacts smaller than 3.2 Å (green and red dashed lines, see also Table 1 ▶) between the bound disaccharide (residue RMY, in purple) and any protein atoms in PDB entry 3qd1/4i8e. Whereas contacts as those indicated in green between polar atoms could possibly represent hydrogen-bond interactions and were listed in Table 3 of the PLoS Pathogens publication, the close contacts drawn in red indicate van der Waals clashes and were omitted in the PLoS Pathogens publication (Pyburn et al., 2011 ▶). The contacts indicated with black dashed lines exemplify severe intramolecular clashes (for example distance RMY O1 to RMY C9 = 1.89 Å). Residues Glu401′ and Val404′ in white are from a neighboring molecule. All Figures were prepared with the program PyMOL (DeLano, 2003 ▶).
Table 1. Fifteen close protein–ligand distances in PDB entries 3qd1/4i8e and in the corrected HEPES-bound model.
| Ligand RMY† | Ligand HEPES | ||||
|---|---|---|---|---|---|
| Ligand atom | Protein atom | Distance (Å)‡ | Ligand atom | Protein atom§ | Distance (Å) |
| C17 | Tyr443 OH | 2.34¶ | N4 | Glu401′ OE2 | 2.98 |
| O2 | Tyr443 OH | 2.46 | O2S | Arg484 NH1 | 3.05 |
| N1 | Ala506 O | 2.48 | C8 | Glu401′ OE2 | 3.19†† |
| O13 | Ala506 O | 2.58 | C2 | Glu401′ OE2 | 3.40 |
| S1 | Thr483 N | 2.58 | C9 | Tyr443 OH | 3.43 |
| C18 | Ala506 O | 2.71¶ | C2 | Tyr443 OH | 3.48 |
| O5 | Arg484 NH2 | 2.72 | C5 | Glu401′ OE2 | 3.48 |
| C13 | Tyr443 OH | 2.76¶ | C3 | Glu401′ OE2 | 3.52 |
| O5 | Tyr482 CD2 | 2.80¶ | C6 | Glu401′ OE2 | 3.53 |
| O7 | Ala506 CB | 2.85¶ | C3 | Ala506 O | 3.56 |
| O5 | Arg449 O | 2.91 | O3S | Trp441 CE3 | 3.60 |
| O8 | Val404′ CG2 | 2.99¶ | C7 | Glu401′ OE2 | 3.62 |
| O2 | Tyr443 CE2 | 3.00¶ | O3S | Arg484 NH1 | 3.63 |
| C17 | Glu401′ OE2 | 3.03¶ | O1S | Tyr443 CE2 | 3.70 |
| C3 | Tyr443 CE2 | 3.05¶ | O8 | Asn428′ N | 3.72 |
Distances are listed in ascending order. Only the 15 closest contacts between any ligand and protein atom are listed. Distances between ligand and solvent molecules are not shown as well as unusually close intraresidue distances in the case of the RMY ligand.
Atoms from symmetry related molecules are marked with primes.
Distances not listed in Table 3 of the PLoS Pathogens publication (Pyburn et al., 2011 ▶).
The only distance qualifying as a close contact in the HEPES complex.
In support of their structural model, the authors report in the PLoS Pathogens publication a table with potential hydrogen-bonding distances (Table 3 in Pyburn et al., 2011 ▶). The reported distances match those in the models deposited with the Protein Data Bank (Table 1 ▶). However, these distances do not provide a comprehensive picture of the intermolecular interactions, since the authors do not list the numerous van der Waals clashes between atoms that are even closer than those putatively involved in hydrogen-bond formation (Table 1 ▶).
In PDB entry 3qd1 all ligand occupancies are set to 0.15. This appears unusual for a 1.9 Å resolution crystal structure that visualizes the specific interaction between a protein and its biological ligand. This could hint that the ligand only binds with low affinity as would be expected, for example, for the non-specific binding of a fortuitously bound molecule such as a buffer molecule. It is also possible that during the handling of the crystals, such as the soaking with cryoprotectants, the ligand partially dissociated from the ligand-binding site. As a consequence of the fact that the deposited structure is presented as a crystal-wide averaged structure derived from 15% of molecules with a bound disaccharide and 85% of molecules with no ligand bound, it could be expected that residues that line the binding site display some conformational heterogeneity. This could possibly also explain the occurrence of unrealistic close atomic contacts in PDB entry 3qd1. However, in contrast to the bound disaccharide, all surrounding residues are well defined in the electron-density maps and do not appear to display alternative side-chain orientations. Therefore, the close contacts in PDB entry 3qdi do not appear to originate from a combined description of occupied and unoccupied binding sites in a single atomic model.
In PDB entry 4i8e, which supersedes entry 3qd1, all ligand occupancies are set to 0.0. Although the title of entry 4i8e reads: GspB plus α-2,3-sialyl (1-thioethyl) galactose, the fact that all ligand atom occupancies are set to 0.0 means that the ligand does not contribute to the crystal scattering. Hence, the experimental structure-factor amplitudes recorded from these crystals and deposited with this entry don’t contain any information regarding the location of the ligand. Entry 4i8e must therefore be considered a purely theoretical ligand-protein model not based on any crystallographic evidence. The presence of entries such as 4i8e presents a challenge to the database to alert its users to the questionable nature of such entries.
3.3. Missing correlation between the ligand-binding model and the experimental crystallographic data
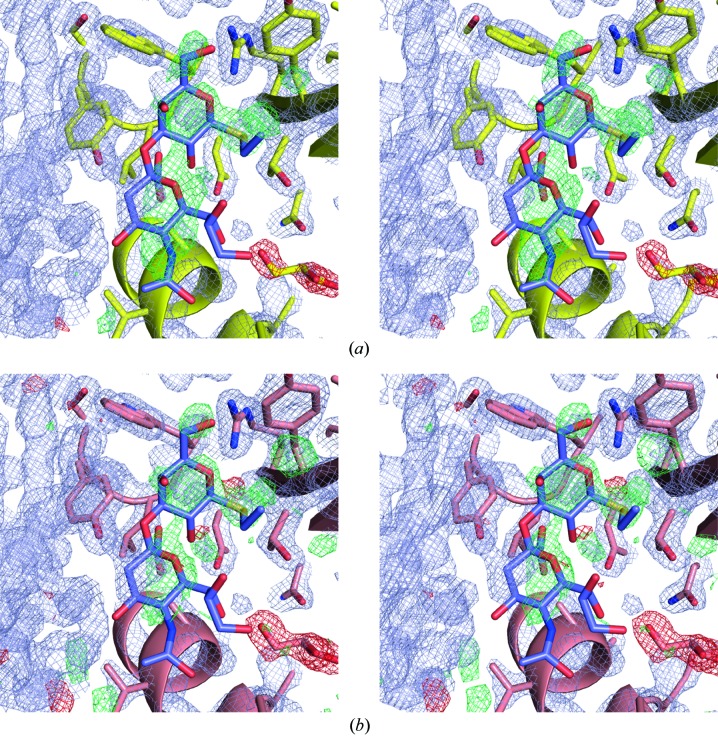
Inspection of electron-density maps from the EDS for either entry 3qd1 or 4i8e reveals strong residual positive density at the position of the bound ligand in both entries (Fig. 2 ▶). However, the shape of the positive difference electron density does not resemble the structure of the bound disaccharide and the highest difference peaks are located in between the positions of the ligand atoms. This makes it unlikely that the residual positive electron density is caused by a systematic underestimation of the ligand atom occupancies. Moreover, the residual positive difference density is even more pronounced in the case of PDB entry 3qd1 where the ligand atom occupancies were set to 0.15 than in PDB entry 4i8e where all ligand atom occupancies were set to 0.0 (Fig. 2 ▶).
Figure 2.
(a) Stereoview of residual positive difference electron density at the site of the bound disaccharide in PDB entry 3qd1. The 2m F o − D F c electron density (in bluish gray) is contoured at 1.8σ. The difference electron density m F o − D F c is shown in green and red and contoured at 4.0 and −4.0σ, respectively. The bound disaccharide is depicted in mauve. (b) Residual positive difference electron density at the site of the bound disaccharide in PDB entry 4i8e. The electron-density maps are color-coded as in panel (a). Please note that in panel (b) the positive and negative difference electron density is contoured at 3.0σ and −3.0σ, respectively. All electron-density maps were downloaded from the EDS (Kleywegt et al., 2004 ▶).
The misfit between electron density and refined ligand structure in entry 3qd1 is also apparent from the real-space correlation coefficient (RSCC) reported by the EDS (Kleywegt et al., 2004 ▶). The RSCC value for residue RMY is as low as 0.34 in entry 3qd1 and is lower than the RSCC value of the poorest defined protein residue, namely Gly324 (RSCC = 0.57). Unfortunately, the fact that all atom occupancies of the ligand RMY were set to 0.0 in entry 4i8e leads to an erroneous report of an RSCC value of 1.03 by the EDS for this entry (Kleywegt et al., 2004 ▶). It has been emphasized before that atom occupancies of 0.0 hamper the validation of protein and protein–ligand structures since they can cause unpredictable results in many widely used validation and visualization programs (Rupp, 2012 ▶).
The combined observations above suggest that the disaccharide modeled into the electron density of PDB entry 3qd1/4i8e might not represent the ligand bound to this site in the crystals. As mentioned in the introduction, a first indication for this was directly obtained from the simulated-annealing OMIT map shown in panel B of Fig. 4 in the PLoS Pathogens publication (Pyburn et al., 2011 ▶).
3.4. An alternative model readily explains the crystallographic data
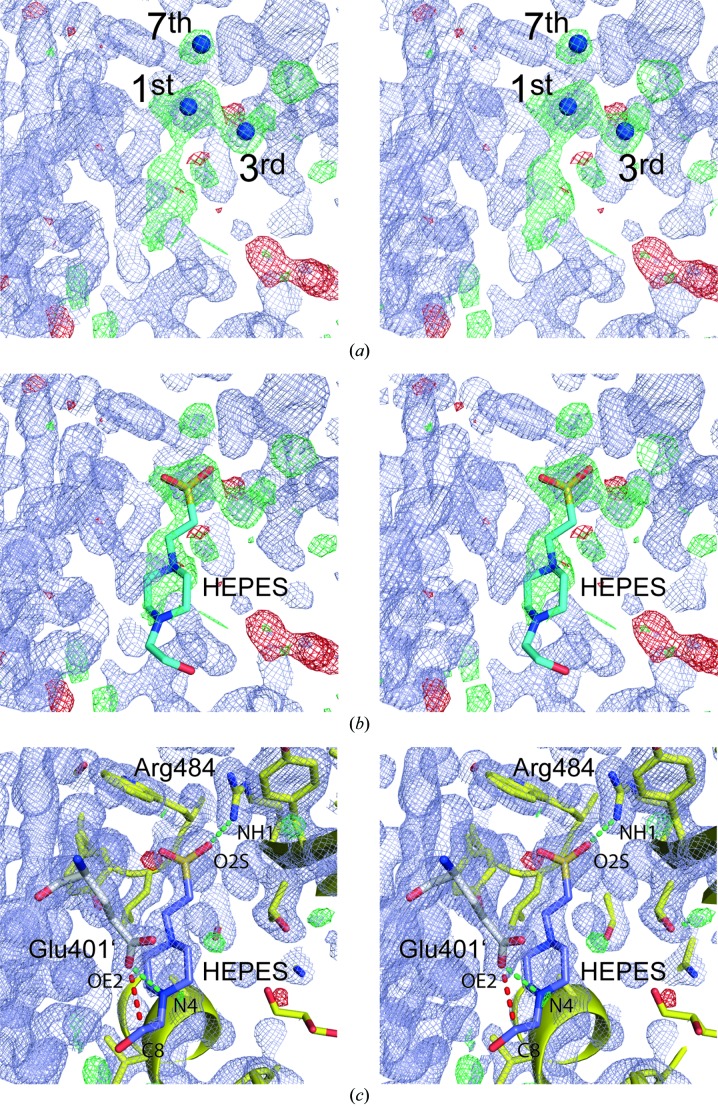
Inspection of the electron-density maps from the EDS for PDB entries 3qd1 and 4i8e provide important clues regarding the identity of the bound ligand (Kleywegt et al., 2004 ▶). Thus, the highest positive density peaks in the difference-density maps of entries 3qd1 (peak height 9.4σ) and 4i8e (8.4σ) coincide and are located within the outline of the modeled disaccharide, however, at the same time, do not overlap with the position of any of the disaccharide atoms. In case of entry 4i8e, the third and seventh highest positive difference densities are also located close by (Fig. 3a ▶). Furthermore, a broader shaped positive density feature higher than 3σ extends from the highest positive difference density peak. All these features can be fully explained by placing a HEPES molecule into the difference electron density (Fig. 3b ▶). HEPES was present at concentrations of 100 mM in the crystallization setup (Pyburn et al., 2011 ▶). In the modeled HEPES molecule, the position of the sulfur atom coincides with the highest positive density peak. The additionally annotated third and seventh highest peaks in Fig. 3(a) ▶ can easily be explained by protein-bound water molecules, which bridge between the sulfonic acid group of HEPES and protein atoms. The crystallographic refinement of the HEPES complex strongly supports the correct identification of the ligand in the crystals associated with PDB entry 3qd1/4i8e (Table 2 ▶). Although refinement only results in very moderate improvements in the global indicators R factor and R free, the visual inspections of electron-density maps show that the structure of the bound HEPES molecule correlates extremely well with the 2m F o − D F c density (Fig. 3 ▶ c). This is also in line with an RSCC value of 0.928 reported for the ligand HEPES in the program PHENIX in comparison to an RSCC value of 0.658 for the worst protein residue (Gly324).
Figure 3.
(a) Stereoview of the electron-density map of Fig. 2(b) ▶ with the atomic model removed and using identical contour levels. Since this map was calculated from PDB entry 4i8e in which all disaccharide atom occupancies are set to 0.0, the map technically qualifies as an OMIT electron-density map. The first, third and seventh highest positive difference density peaks identified with the program COOT are marked. Their peak heights correspond to 8.4, 6.6 and 5.6σ, respectively. (b) Placing a buffer molecule into the difference density map nicely explains the features in this map. In particular the position of the sulfur atom present in HEPES coincides with the highest difference density peak in panel (a). (c) Following a single round of refinement, the HEPES molecule becomes well defined by its electron density and contacts protein residues via two salt bridges via Arg484 and Glu401′ from a symmetry-related molecule (green dashed lines). The red dashed line indicates a close contact (distance = 3.19 Å, Table 1 ▶) Electron-density maps are displayed at 1.6σ (in bluish gray, 2m F o − D F c map), 3.8σ (green, m F o − D F c map) and −3.8σ (red, m F o − D F c map).
Table 2. Juxtaposition of the crystallographic refinement statistics for PDB entry 4ie8 and for the corrected HEPES-bound model.
| Model | Entry 4i8e | Corrected model |
|---|---|---|
| Key feature | Contains a protein-bound disaccharide molecule named RMY with atom occupancies set to 0.0 | Includes a HEPES molecule at the position of the disaccharide in entry 4i8e with atom occupancies of 1.0 |
| Structure factors | 4ie8-sf.cif | 4ie8-sf.cif |
| Starting atom coordinates | 4ie8.pdb | 4ie8.pdb† |
| Starting B factors | All atom B factors were set 30.0 Å2 at the start of the refinement | All atom B factors were set 30.0 Å2 at the start of the refinement |
| R free flags | As present in 4ie8-sf.cif | As present in 4ie8-sf.cif |
| Ligand | RMY | HEPES |
| R work (%) | 16.29 | 16.21 |
| R free (%) | 20.86 | 20.73 |
| R.m.s.d. bonds (Å) | 0.022 | 0.022 |
| R.m.s.d. angles (°) | 1.505 | 1.396 |
| Ligand RSCC value | n.a.‡ | 0.928 for HEPES |
| Lowest protein residue RSCC value | 0.623 for Gly324 | 0.658 for Gly324 |
| Average isotropic B factor (Å2) | 33.9 | 33.7 |
| Average isotropic B factor of ligand atoms (Å2) | n.a.‡ | 54.3 |
In the case of the HEPES complex, the molecule RMY was replaced by HEPES, and all ligand atom occupancies were set to 1.0. In addition, one solvent molecule was deleted and two new solvent molecules were introduced.
All interactions observed between the HEPES molecule and GspB agree with physicochemical expectations. When crystallized by itself, HEPES forms a zwitterion (Sledz et al., 2009 ▶; Wouters et al., 1996 ▶). It carries a negative charge at the sulfonate group, and it is presently under debate whether the positive countercharge is located at either atom N1 or N4 (Sledz et al., 2009 ▶; Wouters et al., 1996 ▶). The buffering qualities of HEPES in the range pH 6.8 to 8.2 (pK a = 7.5) originate from the protonation/deprotonation of either atom N1 or N4, whereas the sulfonate group always carries a negative charge in this pH range (Wouters et al.; 1996 ▶, Sledz et al., 2009 ▶). In the HEPES GspB complex, HEPES participates in direct protein contacts via two different sites (Table 1 ▶, Fig. 3c ▶). These interactions can easily be interpreted as salt bridges, namely one salt bridge being formed between the sulfonate group of HEPES and Arg484 (distance HEPES O2S to Arg484 NH1 = 3.05 Å, Table 1 ▶ and Fig. 3c ▶) and one between atom N4 of HEPES and Glu401′ (distance HEPES N4 to Glu401′ OE2 = 2.98 Å) from a symmetry related GspB molecule. HEPES also participates in numerous additional polar interactions, namely with solvent molecules that bridge between HEPES and protein atoms (data not shown). With the exception of a single 3.19 Å distance involving poorly defined atom HEPES C8 (Table 1 ▶, Fig. 3c ▶), no unusual close contacts are observed between HEPES and GspB.
4. Discussion
Following these observations, the Editors of PLoS Pathogens were contacted, and the ensuing correspondence presumably led to the replacement of PDB entry 3qd1 with 4i8e. In 4i8e the authors changed the occupancies of the disaccharide from 0.15 to 0.0 and also removed two atoms from the original entry (one solvent molecule and a C-terminal OXT atom of Gly603). No crystallographic refinement was performed and all statistical values and structure description parameters are identical in PDB entries 3qd1 and 4i8e. In a correction statement issued on the website of PLoS Pathogens, the authors state that ‘while additional electron density only appeared in this location when GspB was crystallized in the presence of this glycan, the authors would like to clarify that these crystallographic data only suggest a possible gross location of the glycan-binding site and do not explicitly reveal the precise position of all atoms of the glycan or the atomic details of the receptor–GspB interaction’ (http://www.plospathogens.org/article/info:doi/10.1371/journal.ppat.1002112).
It is true that no density for a HEPES molecule is apparent at this position in PDB entries 3qc5 and 3qc6 although the data sets were obtained from crystals also grown in HEPES buffer conditions (0.1 M) with similar reported pHs (7.5) albeit in different space groups (orthorhombic in the case of 3qc5 and 3qc6 and monoclinic in the case of 3qd1/4i8e). HEPES is bound within a crystal packing contact and therefore space-group differences can easily abolish ligand binding. Even though, a superficial inspection of the molecular packing of the molecules in PDB entries 3qc5, 3qc6 and 3qd1/4i8e shows that a similar packing contact is formed in all three crystals, it is quite likely that small shifts in the positions of the protein molecules preclude binding of HEPES to one site and enables it to bind to another. Most likely however, the differential behavior of HEPES is caused by small pH shifts. The pH values commonly reported for a crystallization condition unfortunately often correspond to the pH values of the buffer stock solutions used for preparing the crystallization solution without taking into account pH shifts that are caused by the addition of precipitants and additives (Jancarik & Kim, 1991 ▶).
One strength of crystallography is that at a resolution of 1.9 Å, electron-density maps provide very detailed and unbiased clues regarding the content of crystals. In fact, the entire GspB structure was built based on this principle, namely model building by alternating between rounds of refinement and manual correction following the guidance from electron-density maps. Here, it is shown that very similar crystallographic considerations readily lead to an improved ligand–protein model that better explains the experimental crystallographic data associated with PDB entries 3qd1/4i8e. Whereas an originally modeled disaccharide showed multiple unlikely features resulting in severe van der Waals clashes and in a failure to explain the electron density present at this site in the crystal, replacing the disaccharide by a HEPES molecule completely resolved all these issues. The interactions in which HEPES participates, fully agree with the physicochemical properties of HEPES, and following a single round of crystallographic refinement, HEPES becomes well defined by its electron density. The RSCC value increases from 0.34 for the disaccharide to 0.928 for the modeled HEPES molecule (Table 2 ▶). Since both HEPES (100 mM) and the disaccharide (1 mM) were present in the crystallization buffer, good crystallographic practice makes it imperative to place a HEPES molecule at this position rather than a disaccharide.
It is not possible to define an absolute RSCC threshold value that either validates or dismisses the placement of a ligand. However, clues regarding appropriate values can be derived from regions with poorly defined protein residues. In crystal structures, one or two surface loops are, on average, always poorly defined because of high inherent flexibility. Therefore, these loops cannot be built with confidence and in the case of entry 3qd1 this appears to be so for the loop region around Gly324 in GspB (data not shown). Gly324 displays the lowest RSCC value of the entire chain, namely 0.57. However, the RSCC value of the ligand RMY is even considerably lower, namely 0.34. For any protein–ligand complex, it must be considered as extremely unlikely that a ligand that can only be modeled with an RSCC value of almost half the value of the poorest protein residue will be able to provide reliable insight into atomic details of a biologically relevant recognition process.
Although the presence of erroneous protein–ligand complexes in the PDB is recognized as a significant problem (Pozharski et al., 2013 ▶), these errors are very difficult to identify during the peer review process since, in general, coordinates and structure-factor amplitudes are not made available to reviewers. A step in the right direction is the emerging requirement of some journals to provide a PDB summary validation report upon submission of a manuscript (Baker et al., 2010 ▶; Read et al., 2011 ▶). In case of protein–ligand complexes, the inclusion of an OMIT map either in the main text or in the supplementary material of a publication should also be required. As recently proposed, the reviewing process would certainly also benefit from the provision of electronic excerpts of electron-density maps that describe the binding site (Pozharski et al., 2013 ▶; Weichenberger et al., 2013 ▶). A simple list of all ligand–protein interactions with distances below an arbitrary threshold of 3.5 Å also greatly helps to assess the plausibility of a ligand-binding model. Such a list should be free of all biological expectations and should also include contacts with neighboring molecules, if applicable.
In the PLoS Pathogens publication, the placement of the disaccharide provided a starting point for additional functional experiments (Pyburn et al., 2011 ▶). However, in the absence of any crystallographic evidence regarding the identification of the carbohydrate-binding site, the entire body of functional data might require a reassessment. Such erroneous crystallographic binding-site models have the potential to harm an entire field (Petsko, 2007 ▶). Biochemists that have experimental data that do not agree with the receptor-binding model will have difficulty in publishing these, and no scientist will be able to secure research grants to describe structurally the correct binding site since the issue is considered somehow solved, even if it is clearly not.
It is not clear why this misinterpretation occurred since such an error seems easily avoidable. Whatever the reasons are, once such erroneous models are discovered they should be swiftly corrected. Such corrections are best done by the authors with some incentives from the journals that published these findings. This could for example now be swiftly achieved for a similar ligand-protein complex structure published by the same group with again unrealistic and highly implausible interatomic distances between 1.9 and 2.6 Å (data not shown, PDB entry 3a2s, Tanabe et al., 2010 ▶).
Up to March 2013, the PDF file for the article that describes the GspB structure (Pyburn et al., 2011 ▶) did not include the formal correction issued on the PLoS Pathogens website in December 2012. The present contribution emphasizes how important it is to discuss such controversial crystallographic analyses in a fully open and transparent manner.
Acknowledgments
I thank all members of my group as well as colleagues for enlightening discussions on PDB entries 3qd1 and 4i8e. YAM acknowledges support from the collaborative research project SFB796.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Baker, E. N., Dauter, Z., Einspahr, H. & Weiss, M. S. (2010). Acta Cryst. D66, 115. [DOI] [PubMed]
- Berman, H., Henrick, K., Nakamura, H. & Markley, J. L. (2007). Nucleic Acids Res. 35, D301–D303. [DOI] [PMC free article] [PubMed]
- DeLano, W. (2003). The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific LLC.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Kleywegt, G. J., Harris, M. R., Zou, J., Taylor, T. C., Wählby, A. & Jones, T. A. (2004). Acta Cryst. D60, 2240–2249. [DOI] [PubMed]
- Petsko, G. A. (2007). Genome Biol. 8, 103. [DOI] [PMC free article] [PubMed]
- Pozharski, E., Weichenberger, C. X. & Rupp, B. (2013). Acta Cryst. D69, 150–167. [DOI] [PubMed]
- Pyburn, T. M., Bensing, B. A., Xiong, Y. Q., Melancon, B. J., Tomasiak, T. M., Ward, N. J., Yankovskaya, V., Oliver, K. M., Cecchini, G., Sulikowski, G. A., Tyska, M. J., Sullam, P. M. & Iverson, T. M. (2011). PLoS Pathog. 7, e1002112. [DOI] [PMC free article] [PubMed]
- Read, R. J., Adams, P. D., Arendall, W. B., 3rd, Brunger, A. T., Emsley, P., Joosten, R. P., Kleywegt, G. J., Krissinel, E. B., Lutteke, T., Otwinowski, Z., Perrakis, A., Richardson, J. S., Sheffler, W. H., Smith, J. L., Tickle, I. J., Vriend, G. & Zwart, P. H. (2011). Structure, 19, 1395–1412. [DOI] [PMC free article] [PubMed]
- Rupp, B. (2012). Acta Cryst. F68, 366–376. [DOI] [PMC free article] [PubMed]
- Sledz, P., Minor, T. & Chruszcz, M. (2009). Acta Cryst. E65, o3027–o3028. [DOI] [PMC free article] [PubMed]
- Tanabe, M., Nimigean, C. M. & Iverson, T. M. (2010). Proc. Natl Acad. Sci. USA, 107, 6811–6816. [DOI] [PMC free article] [PubMed]
- Weichenberger, C. X., Pozharski, E. & Rupp, B. (2013). Acta Cryst. F69, 195–200. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wouters, J., Häming, L. & Sheldrick, G. (1996). Acta Cryst. C52, 1687–1688.