The crystal structure of a new P21 crystal form of the catalytic domain of mammalian casein kinase 1 δ is described and compared with previously deposited structures.
Keywords: casein kinase 1 δ, polymorphism
Abstract
Casein kinase 1 δ (CK1δ) is a regulatory enzyme in the mammalian circadian oscillator and represents a potential pharmacological target for modulating circadian rhythms. Crystal structures of four different polymorphs of CK1δ have previously been determined and this article reports the crystallization and structure determination of a new crystal form belonging to space group P21. Comparison of CK1δ crystal structures reveals few conformational differences within the C-terminal lobe, but more significant movements of the β-sheet region of the N-terminal lobe were observed.
1. Introduction
The seven mammalian isoforms of casein kinase 1 (α, β, γ1, γ2, γ3, δ and ∊) are Ser/Thr protein kinases that regulate diverse cellular processes including circadian oscillations of gene expression, Wnt and Hhg signaling and membrane trafficking (Knippschild et al., 2005 ▶; Vielhaber & Virshup, 2001 ▶; Price, 2006 ▶; Virshup et al., 2007 ▶). The casein kinase 1 δ and ∊ isoforms (CK1δ and CK1∊) share a nearly identical catalytic domain followed by a divergent C-terminal tail that undergoes autophosphorylation and serves as an autoinhibitory domain (Graves & Roach, 1995 ▶; Cegielska et al., 1998 ▶). The relationship between phosphoryl-group transfer by CK1δ/∊ and circadian rhythms was established by the identification of mutations in the catalytic domains of CK1δ and CK1∊ that lead to altered circadian-rhythm period length (Lowrey et al., 2000 ▶; Xu et al., 2005 ▶). CK1δ and CK1∊ phosphorylate key components of the circadian oscillator to control the nuclear accumulation of feedback proteins (Lee et al., 2009 ▶), making these enzymes potential targets for pharmacological modulation of circadian rhythms. Recent data indicate that CK1δ is the primary regulator of circadian-rhythm period length (Etchegaray et al., 2009 ▶, 2010 ▶).
The crystal structures of the catalytic domains of the fission yeast CK1δ homolog Cki (Xu et al., 1995 ▶) and rat CK1δ (Longenecker et al., 1996 ▶) were determined over 15 years ago, before the role of CK1δ in circadian rhythms was discovered. More recently, crystal structures of human CK1δ complexed with several inhibitors (Long et al., 2012a ▶,b ▶; Huang et al., 2012 ▶), as well as the first structures of CK1∊ (Long et al., 2012b ▶), have been reported. Here, we describe a P21 crystal form of the catalytic domain of mouse CK1δ (residues 1–299; identical in amino-acid sequence to rat and human CK1δ) and compare it with previously solved CK1δ structures.
2. Materials and methods
2.1. Protein expression, purification and crystallization
Amino acids 1–299 (which comprise the catalytic domain) are identical in human, mouse and rat CK1δ, so for simplicity all structures discussed will subsequently be referred to as CK1δ regardless of genetic source. The coding sequence for CK1δ (1–299) was amplified from the murine gene and cloned into the bacterial expression vector pLIC-HTK, which encodes an N-terminal 6×His tag and a tobacco etch virus (TEV) protease cleavage site, using ligation-independent cloning (Doyle, 2005 ▶). After observing inefficient tag cleavage owing to autophosphorylation of the serine in the TEV recognition site (ENLYFQ/S), site-directed mutagenesis (Liu & Naismith, 2008 ▶) was used to change the serine to glycine, resulting in a construct that underwent complete TEV protease digestion.
Rosetta2 (DE3) pLysS competent Escherichia coli cells (Novagen) were transformed with the CK1δ (1–299) expression plasmid and grown in Terrific broth at 310 K to an OD600 of 0.8, at which point the temperature was lowered to 289 K and expression was induced by addition of IPTG to a final concentration of 1 mM. Cells were grown for 18 h post-induction, harvested by centrifugation and frozen at 193 K. Pellets were resuspended in lysis buffer consisting of 20 mM Tris pH 8.0, 500 mM NaCl, 1 mM tris-(2-carboxyethyl)phosphine, 1 mM phenylmethanesulfonyl fluoride, 1 µg ml−1 pepstatin A, 1 µg ml−1 leupeptin, 50 U ml−1 Benzonase nuclease (Novagen), lysed by sonication and clarified by centrifugation at 20 000g for 20 min. The clarified extract was applied onto a Co2+-charged 5 ml HiTrap IMAC FF column (GE Biosciences) and 6×His-CK1δ (1–299) was eluted with buffer consisting of 20 mM Tris pH 8.0, 500 mM NaCl, 500 mM imidazole. After exchanging the buffer into 50 mM Tris pH 7.5, 200 mM NaCl, 5 mM octyl-β-d-glucoside, 1 mM EDTA, 1 mM DTT using a HiTrap Desalting column (GE Biosciences), 6×His-CK1δ (1–299) was treated with ProTEV Plus protease (Promega) to remove the N-terminal 6×His tag. Finally, CK1δ (1–299) was purified using a HiLoad Superdex 200 16/60 size-exclusion column (GE Biosciences) equilibrated with 50 mM Tris pH 7.5, 200 mM NaCl, 5 mM octyl-β-d-glucoside, 1 mM EDTA, 1 mM DTT.
Purified CK1δ 1–299 was concentrated to 5 mg ml−1 for crystallization using a 10 000 molecular-weight cutoff Vivaspin centrifugal filter unit (GE Healthcare). Crystals were grown using the hanging-drop vapor-diffusion method by equilibrating a drop consisting of 1 µl CK1δ (1–299) in 50 mM Tris pH 7.5, 200 mM NaCl, 5 mM octyl-β-d-glucoside, 1 mM EDTA, 1 mM DTT mixed with 1 µl reservoir solution over a 0.7 ml reservoir consisting of 100 mM succinic acid pH 7.0, 15%(w/v) PEG 3350. Crystals appeared after 4 d at room temperature and reached maximum dimensions of 25 × 25 µm.
2.2. Data collection, structure determination and refinement
Crystals were cryoprotected in mother liquor containing 25%(v/v) glycerol, mounted and flash-cooled in liquid N2. Diffraction data were collected at 100 K on Advanced Photon Source beamline 21-ID-G to a resolution of 2.41 Å. Data were indexed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure of CK1δ (1–299) was solved by molecular replacement using Phaser (McCoy et al., 2007 ▶) with chain A of PDB entry 3uzp (apo CK1δ 1–294; Long et al., 2012a ▶) as a search model. Iterative model building using Coot (Emsley et al., 2010 ▶) and refinement using PHENIX (Adams et al., 2010 ▶) led to the final model with an R work of 21.0% and an R free of 23.7%. Individual isotropic B factors were used throughout the refinement process. Noncrystallographic symmetry was not used in either the positional or the thermal refinement. Data-collection and refinement statistics are presented in Table 1 ▶. MolProbity (Chen et al., 2010 ▶) was used for model validation. Intermolecular interfaces were analyzed with the PISA server (Krissinel & Henrick, 2007 ▶). The coordinates and structure factors have been deposited in the RCSB Protein Data Bank with PDB code 4jjr.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P21 |
| Wavelength (Å) | 0.97857 |
| Unit-cell parameters (Å, °) | a = 52.15, b = 102.25, c = 64.59, β = 101.53 |
| Resolution (Å) | 40.66–2.41 (2.49–2.41) |
| Total reflections | 196881 (18518) |
| Unique reflections | 25572 (2405) |
| Multiplicity | 7.7 (7.7) |
| Completeness (%) | 99.2 (92.7) |
| R merge † | 0.094 (0.681) |
| 〈I/σ(I)〉 | 15.0 (2.8) |
| Optical resolution‡ (Å) | 1.84 |
| Wilson B factor (Å2) | 41.9 |
| Refinement statistics | |
| R work § | 0.210 (0.276) |
| R free | 0.237 (0.313) |
| No. of protein atoms | 4726 |
| No. of waters | 87 |
| R.m.s.d. bond lengths (Å) | 0.004 |
| R.m.s.d. bond angles (°) | 0.96 |
| Average B factors (Å2) | |
| Protein atoms | 32.2 |
| Waters | 35.4 |
| Ramachandran plot statistics, residues in¶ (%) | |
| Favored region | 96.93 |
| Allowed region | 3.07 |
| Outlier region | 0.0 |
R
merge = 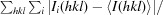
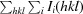 , where 〈I(hkl)〉 is the mean intensity of symmetry-related reflections Ii(hkl).
, where 〈I(hkl)〉 is the mean intensity of symmetry-related reflections Ii(hkl).
Calculated using SFCHECK (Vaguine et al., 1999 ▶).
R
work = 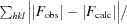
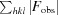 , where F
obs and F
calc are the experimental and calculated structure-factor amplitudes, respectively. R
free was calculated as for R
work but using a random 5% of the data that were excluded from refinement.
, where F
obs and F
calc are the experimental and calculated structure-factor amplitudes, respectively. R
free was calculated as for R
work but using a random 5% of the data that were excluded from refinement.
Calculated using the MolProbity web server (Chen et al., 2010 ▶).
3. Results and discussion
3.1. Structure of CK1δ (1–299)
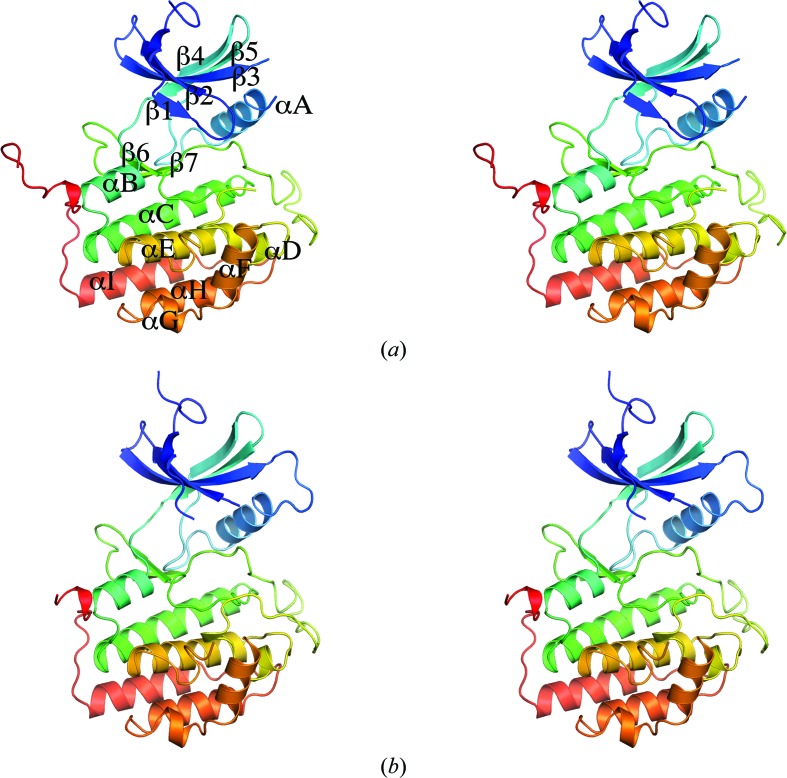
CK1δ (1–299) crystallized in space group P21 with two molecules in the asymmetric unit. The N-terminal lobe of the canonical protein kinase fold comprises a five-stranded antiparallel β-sheet (β1–β5) and a single α-helix (αA), while the larger C-terminal lobe is predominantly helical (Fig. 1 ▶). Residues not modelled owing to poor or absent electron density were residues 1–2, 43–46, 171–173 and 217–222 in chain A and residues 17–20, 218–223 and 294–299 in chain B. Several residues in the middle of loop L-EF (in the C-terminal lobe) are missing from the model in both chains; it has previously been suggested that this loop is involved in the recognition and binding of substrate and it has been found to be partially disordered in most previously determined CK1δ structures (Longenecker et al., 1996 ▶). The missing residues 17–20 in chain B form part of the P-loop (loop L-1,2), which is presumably more flexible in the absence of ligand in the ATP-binding cleft. The backbones of chains A and B have a root-mean-square displacement (r.m.s.d.) of 0.82 Å (calculated over 273 Cα atoms).
Figure 1.
Crystal structure of P21 CK1δ (1–299). (a) Stereoview of chain A of P21 CK1δ (1–299) colored from blue at the N-terminus to red at the C-terminus, with β-strands and α-helices labeled as in Longenecker et al. (1996 ▶). (b) Stereoview of chain B of P21 CK1δ (1–299) colored from blue at the N-terminus to red at the C-terminus. The figures were prepared using MacPyMOL (Schrödinger LLC).
3.2. Comparisons to previously determined CK1δ structures
Apo CK1δ has previously been crystallized in two different space groups: P212121 (CK1δ 1–317; PDB entries 1cki and 1ckj; Longenecker et al., 1996 ▶) and P1 (CK1δ 1–294; PDB entry 3uys; Long et al., 2012a ▶). There are four deposited structures of CK1δ (1–294) with inhibitors bound in the ATP-binding site: two different crystal forms (P21 and P1) of a complex with the inhibitor PF670462 (PDB entries 3uzp and 3uyt; Long et al., 2012a ▶), a C2 complex with the inhibitor PF4800567 (PDB entry 4hgt; Long et al., 2012b ▶) and a P21 complex with a pyrimidinyl pyrrolopyridinone inhibitor (PDB entry 4hnf; Huang et al., 2012 ▶). The conformation adopted by the two lobes of the catalytic domain in P21 CK1δ (1–299) is essentially the same as that seen in the previously deposited structures, with overall Cα r.m.s.d. values for both chains of P21 CK1δ (1–299) with all chains of previous CK1δ structures ranging from 0.6 to 1.3 Å. All of the loops that are not modeled in our structure are also missing in at least some of the previously determined structures, suggesting that they are intrinsically flexible and disordered in the absence of bound substrate.
Alignment of P21 CK1δ (1–299) with other CK1δ structures by superposition of the C-terminal lobe (residues 88–281) indicates that the conformation of the C-terminal lobe is well conserved in all structures but that there is movement of the N-terminal domain relative to the C-terminal domain and conformational changes within the N-terminal domain are observed in different structures. Differences observed in the C-terminal lobe are mainly confined to the extreme C-terminus and a region of the activation loop (Figs. 2 ▶ and 3 ▶). In some structures, smaller displacements of part of loop L-EF and at the N-terminus of αF (residues 210–225) can be seen (Fig. 3 ▶).
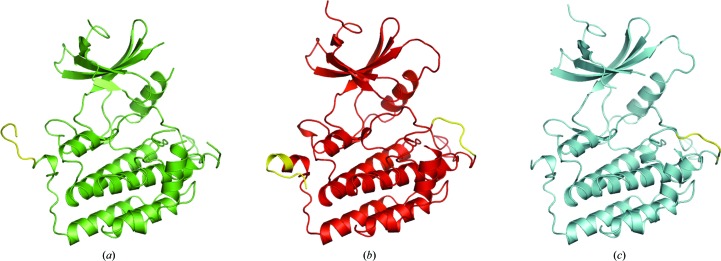
Figure 2.
Conformational differences in the C-terminal lobe. (a) P21 CK1δ (1–299) chain A (green), with residues 294–299 at the C-terminus colored yellow. (b) PDB entry 1cki chain A (red), with the C-terminal residues and the variable-conformation region of the activation loop highlighted in yellow. (c) The activation loop of P21 CK1δ (1–299) chain B (cyan) adopts the more common conformation seen in other CK1δ structures, rather than the alternative conformation seen in PDB entry 1cki.
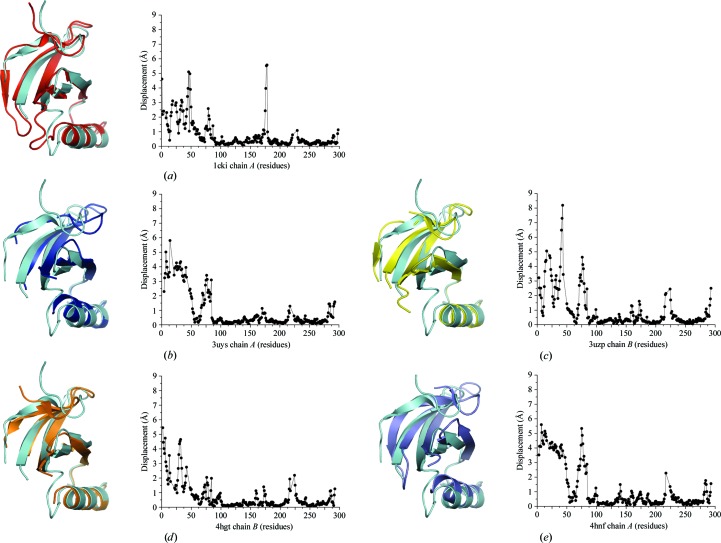
Figure 3.
Conformational differences in the N-terminal lobe. Chains were aligned by the residues of the C-terminal lobe (amino acids 88–281). The C-terminus of helix αA aligns well in all structures, but rotation of the β-sheet, and in some cases pivoting of αA, are observed between different structures. R.m.s.d. values were calculated using Chimera (Pettersen et al., 2004 ▶). (a) Superposition of CK1δ (1–299) chain B (cyan) with 1cki chain A (red). (b) Superposition of CK1δ (1–299) chain B (cyan) with 3uys chain A (blue). (c) Superposition of CK1δ (1–299) chain B (cyan) with 3uzp chain B (yellow). (d) Superposition of CK1δ (1–299) chain B (cyan) with 4hgt chain B (orange). (e) Superposition of CK1δ (1–299) chain B (cyan) with 4hnf chain A (purple).
Residues 294–299 at the C-terminus of chain A in P21 CK1δ (1–299) adopt an extended conformation that interfaces with the N-terminal lobe of chain B in the asymmetric unit (Figs. 2 ▶ a and 5a). This is markedly different from the structure adopted by the corresponding residues in PDB entry 1cki chain A (Longenecker et al., 1996 ▶), in which these residues form an extra turn of helix and then point in a different direction that is not involved in intermolecular contacts in the crystal (Figs. 2 ▶ b and Fig. 3 ▶ a). In all other deposited CK1δ chains these residues are not present in the model, either because electron density was not visible or because they were not present in the crystallized construct.
Two conformations have previously been observed for residues 171–173 in the activation loop (Figs. 2 ▶ b, 2 ▶ c and 3 ▶ a). One conformation has been observed in chain A of PDB entries 1cki and 1ckj (Longenecker et al., 1996 ▶), in which these residues bulge out and away from helix αD (Fig. 2 ▶ b) and are stabilized by an extensive hydrogen-bonding network involving hydrogen bonds from the backbone amide of Asn170 to the carbonyl of Gly187, from the backbone amide of Asn172 to the Oδ atom of Asn170, from the side chain of Arg172 to the carbonyl of Leu173 and from the side chain of Lys154 to the Oδ atom of Asn172. In most CK1δ structures, including all chains of PDB entries 3uzp, 3uyt, 4hgt and 4hnf, as well as chain B of PDB entries 1cki and 1ckj, this region of the activation loop is found in a different conformation in which the backbone amide of Lys171 forms a hydrogen bond to the carbonyl of Gly187. This conformation permits divalent anions to bind via electrostatic interactions with Lys154, Arg127 and Lys171. This is one of four observed anion-binding sites that have potential roles as phosphate-recognition sites for substrate binding and autoinhibition (Longenecker et al., 1996 ▶). In PDB entry 1ckj chain B, which was solved from crystals soaked in 17 mM sodium tungstate, a WO4 2− ion is bound to this site with a refined occupancy of 0.1 (Longenecker et al., 1996 ▶); in PDB entries 3uys chains A and B and 3uyt chain A (which were crystallized from sodium sulfate), an SO4 2− ion with a refined occupancy of 1.0 occupies the same binding site (Long et al., 2012a ▶). Chain B of P21 CK1δ (1–299) adopts the more common conformation seen in the anion-bound structures (Fig. 2 ▶ c), and in P21 CK1δ (1–299) chain A residues 171–173 were not modeled owing to poor electron density.
Significant movement of the N-terminal domain β-sheet can be seen when aligning different CK1δ structures by the C-terminal domain (Fig. 3 ▶). As helix αA in the N-terminal lobe (residues 49–59 superimposes well in most structures (Figs. 3 ▶ a, 3 ▶ c and 3 ▶ d), these movements represent rotations of the β-sheet within the N-terminal lobe rather than movement of the entire N-terminal lobe relative to the C-terminal lobe. The greatest displacements are observed for β1–β3 (residues 1–50) and β4–β5 (residues 68–83); however, the extent of movement varies greatly among the different structures (Fig. 3 ▶).
In PDB entries 3uzp (Fig. 3 ▶ c) and 4hgt (Fig. 3 ▶ d), the β-sheet undergoes a clockwise rotation that brings β1 and β2 towards the C-terminal lobe to form a more closed conformation over the inhibitor bound in the ATP-binding cleft and results in a large displacement of the C-terminal end of β3. In PDB entries 3uys (Fig. 3 ▶ b) and 4hnf (Fig. 3 ▶ e) helix αA pivots on its C-terminal end, rotating along with the β-sheet to adopt a more open conformation in which β1 and β2 move farther away from the ATP-binding cleft. The largest movement of the β-sheet can be seen when comparing PDB entry 4hnf and P21 CK1δ (1–299) (Fig. 3 ▶ e). β1 is displaced by 4.3 Å and β2 by 3.5 Å. The C-terminus of β3 in PDB entry 4hnf has shifted to where the N-terminus of β4 in P21 CK1δ (1–299) is. Loop L-3,4 is shifted by 5.5 Å between the two structures (Gly75 Cα displacement). Despite these conformational changes, the hinge connecting the two lobes is virtually unmoved as Cα of Leu85 is displaced by 0.4 Å between the two structures. These movements of the N-terminal lobe lead to small displacements of the catalytically important ATP-binding residue Lys38 (Fig. 4 ▶), but the conformation of the putative catalytic base Glu128 is essentially unchanged.
Figure 4.
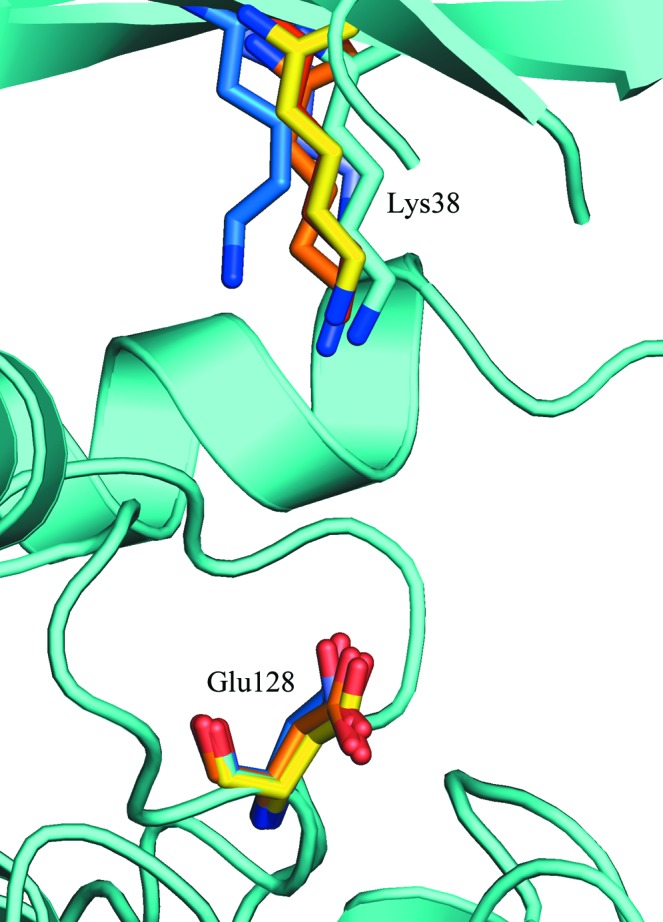
Catalytically important residues. Lys38 and Glu128 from 1cki chain A (red), 3uys chain A (blue), 3uzp chain B (yellow), 4hgt chain B (orange) and 4hnf chain A (purple) are superimposed on the structure of CK1δ (1–299) chain B (cyan) after alignment by the C-terminal lobes as in Fig. 3 ▶. The conformation of the putative general base Glu128 does not change much between structures, but the conformational changes in the β-sheet shown in Fig. 3 ▶ lead to small shifts in the position of the ATP-binding residue Lys38.
3.3. Dimer interface and crystal packing
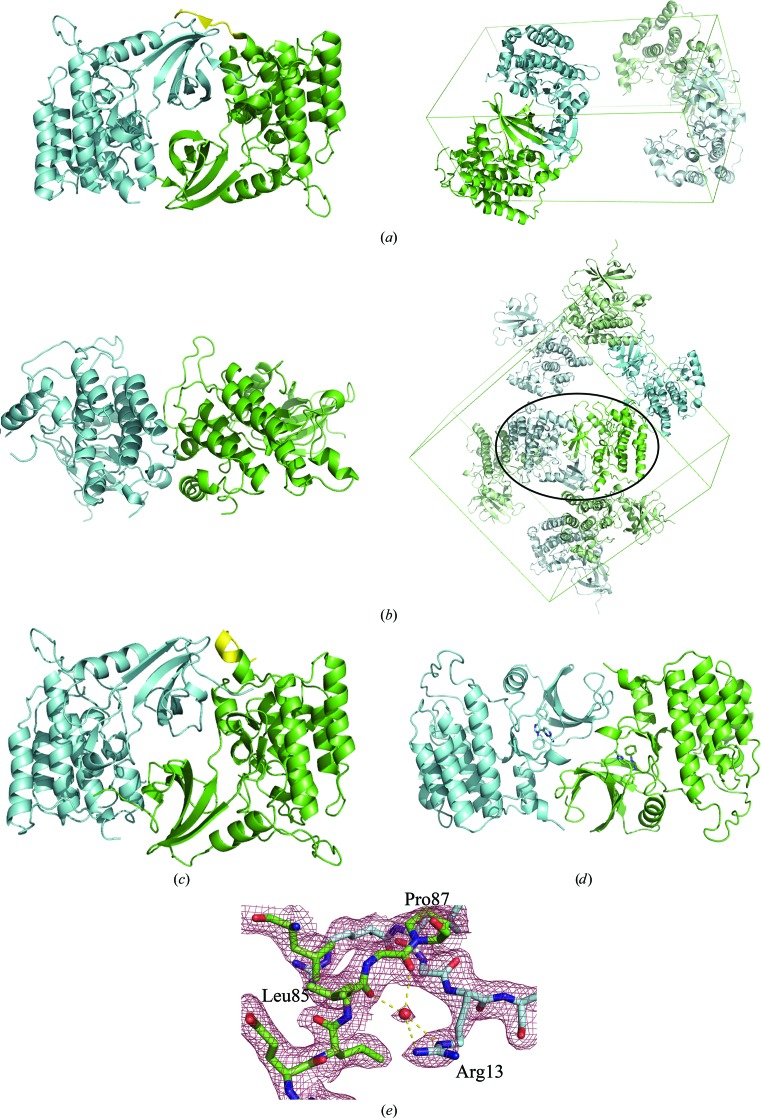
Even though CK1δ (1–299) is a monomer in solution, the two molecules in the asymmetric unit of the P21 CK1δ (1–299) structure share an extensive dimer interface that buries a total of 3530 Å2, involving 51 residues on each monomer and containing 15 hydrogen bonds. The N-terminal lobe of each molecule fits between the N-terminal lobe and C-terminal lobe of its partner (Fig. 5 ▶ a). An approximate noncrystallographic twofold axis is at the center of an interface involving the side chains of Arg13, Lys14, Ile15 (on L-1,2), Leu25 (on β2) and Glu34 (on β3), as well as the backbone carbonyls of Leu85 and Pro87 (on the hinge connecting the two lobes). The side chain of Arg13 extends partially into the ATP-binding groove between the two lobes in the partner molecule to interact through a bridging water molecule with the backbone carbonyls of Leu85 and Pro87 (Fig. 5 ▶ e). This penetration into the ATP-binding groove makes this crystal form unsuitable for soaking experiments for inhibitor screening. Side chains on helix αB in the C-terminal lobe interact with side chains on β4–β5 in the N-terminal lobe of the other molecule and the C-terminus of chain A interacts with strands β4 and β5 as well as the N-terminus of chain B. Although there are no intermolecular contacts between residues in the two C-terminal lobes, the N-terminal ends of helices αF in the C-terminal lobes are pointing towards each other, forming a semicircle.
Figure 5.
Asymmetric units and crystal packing in CK1δ structures. (a) Left, the asymmetric unit of P21 CK1δ (1–299). Residues 293–299 of chain A are shown in yellow. Right, the unit cell of P21 CK1δ (1–299). The symmetry-related asymmetric unit is shown in pale green/cyan. (b) Left, the asymmetric unit of PDB entry 1cki. Right, the unit cell of PDB entry 1cki. Symmetry-related asymmetric units are shown in pale green/cyan. The circled crystal-packing interface corresponds to the dimer interface observed in P21 CK1δ (1–299). (c) Closer view of the two 1cki monomers related by the circled crystal contact. The C-terminus of chain A (yellow) adopts a different conformation than in the asymmetric unit of P21 CK1δ (1–299). (d) The asymmetric unit of PDB entry 3uzp, which despite crystallizing in the same space group has a completely different dimer interface to P21 CK1δ (1–299) owing to the presence of the inhibitor PF670462 in the ATP-binding cleft (shown as a stick model), which blocks some of the interactions between monomers observed in the P21 CK1δ (1–299) asymmetric unit. (e) Close-up of the dimer interface in P21 CK1δ (1–299), showing the interaction between the side chain of Arg13 and the hinge region of the other molecule. Unweighted 2F o − F c electron density is shown contoured at 1.0σ.
Although this is a new crystal form, closer inspection revealed that the interface between monomers in P21 CK1δ (1–299) was nearly the same as a crystal-packing interface observed in the P212121 crystal form of apo CK1δ (1–317) (Figs. 5 ▶ b and 5 ▶ c; PDB entry 1cki and 1ckj; Longenecker et al., 1996 ▶), with the exception of the interactions made by residues 294–299 at the C-terminus of chain A with the N-terminal lobe of chain B. In PDB entry 1cki this crystal contact involves 50 residues on chain A and 49 on chain B, buries a total of 3380 Å2 and involves 18 hydrogen bonds and one salt bridge, in contrast to the interface between molecules in the asymmetric unit that mainly involves the C-terminal lobes and spans only 920 Å2 (Fig. 5 ▶ b). A similar dimer interface to that observed in P21 CK1δ (1–299) was described for two molecules of CK1δ (1–342) related to one another by a crystallographic twofold axis in a C2221 crystal structure (coordinates not deposited; Longenecker et al., 1998 ▶). Although this is an extensive dimer interface that occurs in at least three different crystal forms, size-exclusion chromatography indicates that CK1δ is a monomer in solution. It is unlikely that this dimer interface is biologically relevant, as this interface blocks access of ATP and peptide substrate to the active site.
A different P21 crystal form has previously been reported for the CK1δ (1–294)–inhibitor structures 3uzp (Long et al., 2012a ▶) and 4hgt (Huang et al., 2012 ▶). As in the P21 CK1δ (1–299) structure, the N-terminal lobe of one monomer sits between the N-terminal and C-terminal lobes of its partner, but does not penetrate as deeply into the ATP-binding groove owing to the presence of the inhibitor molecule (Fig. 5 ▶ d). As a result, the interactions making up the dimer interface in these structures are completely different from those in P21 CK1δ (1–299), as are the relative orientations of the two molecules to one another.
Supplementary Material
PDB reference: casein kinase 1 δ, 4jjr
Acknowledgments
The authors wish to thank Donald Ronning and members of his research group for assistance with data collection and helpful discussions. This work was supported by start-up funds from the University of Toledo and a grant from the deArce Memorial Endowment Fund in Support of Biomedical Research (to JJB) and Undergraduate Summer Research Awards (to LM and EAM). Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Cegielska, A., Gietzen, K. F., Rivers, A. & Virshup, D. M. (1998). J. Biol. Chem. 273, 1357–1364. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Doyle, S. A. (2005). Methods Mol. Biol. 310, 107–113. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Etchegaray, J.-P., Machida, K. K., Noton, E., Constance, C. M., Dallmann, R., Di Napoli, M. N., DeBruyne, J. P., Lambert, C. M., Yu, E. A., Reppert, S. M. & Weaver, D. R. (2009). Mol. Cell. Biol. 29, 3853–3866. [DOI] [PMC free article] [PubMed]
- Etchegaray, J.-P., Yu, E. A., Indic, P., Dallmann, R. & Weaver, D. R. (2010). PLoS One, 5, e10303. [DOI] [PMC free article] [PubMed]
- Graves, P. R. & Roach, P. J. (1995). J. Biol. Chem. 270, 21689–21694. [DOI] [PubMed]
- Huang, H. et al. (2012). ACS Med. Chem. Lett. 3, 1059–1064. [DOI] [PMC free article] [PubMed]
- Knippschild, U., Gocht, A., Wolff, S., Huber, N., Löhler, J. & Stöter, M. (2005). Cell. Signal. 17, 675–689. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lee, H., Chen, R., Lee, Y., Yoo, S. & Lee, C. (2009). Proc. Natl Acad. Sci. USA, 69, 6–9.
- Liu, H. & Naismith, J. H. (2008). BMC Biotechnol. 8, 91. [DOI] [PMC free article] [PubMed]
- Long, A., Zhao, H. & Huang, X. (2012a). J. Med. Chem. 55, 956–960. [DOI] [PubMed]
- Long, A., Zhao, H. & Huang, X. (2012b). J. Med. Chem. 55, 10307–10311. [DOI] [PubMed]
- Longenecker, K. L., Roach, P. J. & Hurley, T. D. (1996). J. Mol. Biol. 257, 618–631. [DOI] [PubMed]
- Longenecker, K. L., Roach, P. J. & Hurley, T. D. (1998). Acta Cryst. D54, 473–475. [DOI] [PubMed]
- Lowrey, P. L., Shimomura, K., Antoch, M. P., Yamazaki, S., Zemenides, P. D., Ralph, M. R., Menaker, M. & Takahashi, J. S. (2000). Science, 288, 483–492. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed]
- Price, M. A. (2006). Genes Dev. 20, 399–410. [DOI] [PubMed]
- Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999). Acta Cryst. D55, 191–205. [DOI] [PubMed]
- Vielhaber, E. & Virshup, D. M. (2001). IUBMB Life, 51, 73–78. [DOI] [PubMed]
- Virshup, D. M., Eide, E. J., Forger, D. B., Gallego, M. & Harnish, E. V. (2007). Cold Spring Harb. Symp. Quant. Biol. 72, 413–420. [DOI] [PubMed]
- Xu, R., Carmel, G., Sweet, R. M., Kuret, J. & Cheng, X. (1995). EMBO J. 14, 1015–1023. [DOI] [PMC free article] [PubMed]
- Xu, Y., Padiath, Q. S., Shapiro, R. E., Jones, C. R., Wu, S. C., Saigoh, N., Saigoh, K., Ptáček, L. J. & Fu, Y.-H. (2005). Nature (London), 434, 640–644. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: casein kinase 1 δ, 4jjr