The GH42 β-galactosidase GanB from G. stearothermophilus has been crystallized in the primitive orthorhombic space group P212121. Complete diffraction data sets have been measured for the wild-type enzyme (2.45 Å resolution) and its catalytic mutant (E323A; 2.50 Å resolution) for use in a full three-dimensional structural analysis of the GanB protein.
Keywords: Geobacillus stearothermophilus, GH42, GanB, β-galactosidase
Abstract
Geobacillus stearothermophilus T-6 is a Gram-positive thermophilic soil bacterium that contains a multi-enzyme system for the utilization of plant cell-wall polysaccharides, including xylan, arabinan and galactan. The bacterium uses a number of endo-acting extracellular enzymes that break down the high-molecular-weight polysaccharides into decorated oligosaccharides. These oligosaccharides enter the cell and are further hydrolyzed into sugar monomers by a set of intracellular glycoside hydrolases. One of these intracellular degrading enzymes is GanB, a glycoside hydrolase family 42 β-galactosidase capable of hydrolyzing short β-1,4-galactosaccharides to galactose. GanB and related enzymes therefore play an important part in the hemicellulolytic utilization system of many microorganisms which use plant biomass for growth. The interest in the biochemical characterization and structural analysis of these enzymes stems from their potential biotechnological applications. GanB from G. stearothermophilus T-6 has recently been cloned, overexpressed, purified, biochemically characterized and crystallized in our laboratory as part of its complete structure–function study. The best crystals obtained for this enzyme belong to the primitive orthorhombic space group P212121, with average crystallographic unit-cell parameters of a = 71.84, b = 181.35, c = 196.57 Å. Full diffraction data sets to 2.45 and 2.50 Å resolution have been collected for both the wild-type enzyme and its E323A nucleophile catalytic mutant, respectively, as measured from flash-cooled crystals at 100 K using synchrotron radiation. These data are currently being used for the full three-dimensional crystal structure determination of GanB.
1. Introduction
β-Galactosidases (EC 3.2.1.23) are a large group of enzymes that hydrolyze the β-glycosidic bond between a terminal nonreducing d-galactose residue and other organic molecules. Based on their sequence similarity, β-galactosidases are classified into four main glycoside hydrolase (GH) families, 1, 2, 35 and 42, according to the CAZy database (Henrissat & Davies, 1997 ▶). These families have distinctly different sequences and exhibit different substrate specificities. In general, GH1 and GH2 β-galactosidases are found in mesophiles and demonstrate lactase activity, while enzymes belonging to families GH35 and GH42 are usually found in thermophiles and preferentially degrade β-1,4-linkages between two galactose moieties with a very low or a complete absence of activity towards lactose (Hinz et al., 2004 ▶; Ohtsu et al., 1998 ▶; Shipkowski & Brenchley, 2006 ▶; Yang et al., 2003 ▶; Schwab et al., 2010 ▶). Since the natural habitats of thermophilic microorganisms are poor in lactose, the native substrates for GH42 enzymes are thought to be galacto-oligomers derived from high-molecular-weight polymers composing the plant cell wall. Indeed, in many genetic systems GH42 β-galactosidases are located in proximity to GH53 galactanases in operons dedicated for the utilization of pectic type I galactan and arabinogalactan (Shipkowski & Brenchley, 2006 ▶; Daniel et al., 1997 ▶; Delangle et al., 2007 ▶; O’Connell Motherway et al., 2011 ▶).
Geobacillus stearothermophilus T-6 is a Gram-positive thermophilic soil bacterium that harbours a well regulated system for the utilization of plant cell-wall polysaccharides, including xylan, arabinan and galactan (Shulami et al., 1999 ▶, 2011 ▶; Tabachnikov & Shoham, 2013 ▶). The bacterium utilizes a limited number of endo-acting extracellular enzymes that break down the high-molecular-weight polysaccharides into decorated oligosaccharides. These oligosaccharides enter the cell via specialized ABC transporters (Rees et al., 2009 ▶) and are further hydrolyzed into sugar monomers by a set of intracellular glycoside hydrolases. For example, for the complete consumption of xylan, the bacterium secretes an extracellular xylanase (XT6; Gat et al., 1994 ▶; Teplitsky et al., 1997 ▶, 2000 ▶; Bar et al., 2004 ▶), which partially degrades xylan to decorated xylo-oligomers that are transported into the cell via the ABC transport system (Shulami et al., 2007 ▶). Inside the cell, the decorated xylo-oligomers are hydrolyzed by side-chain-cleaving enzymes, including arabinofuranosidases (Shallom, Belakhov, Solomon, Gilead-Gropper et al., 2002 ▶; Shallom, Belakhov, Solomon, Shoham et al., 2002 ▶; Hövel et al., 2003 ▶), glucuronidases (Teplitsky et al., 1999 ▶; Zaide et al., 2001 ▶; Golan et al., 2004 ▶; Shallom et al., 2004 ▶) and acetyl-esterases (Alalouf et al., 2011 ▶; Lansky et al., 2013 ▶), and finally by an intracellular xylanase (IXT6; Teplitsky et al., 2000 ▶; Solomon et al., 2007 ▶) and xylosidases (Bravman et al., 2003 ▶; Shallom et al., 2005 ▶; Brüx et al., 2006 ▶; Ben-David et al., 2007 ▶).
We have recently characterized a 9.4 kb gene cluster, ganREFGBA, in G. stearothermophilus which encodes galactan-utilization elements (Tabachnikov & Shoham, 2013 ▶). In this cluster, the ganEFG genes encode an ATP-binding cassette sugar-transport system, the sugar-binding lipoprotein of which, GanE, has been shown to bind galacto-oligosaccharides. GanA is an extracellular GH53 β-1,4-galactanase which is active towards high-molecular-weight galactan and produces galactotetraose as the main product. GanB is a GH42 β-galactosidase capable of hydrolyzing short β-1,4-galactosaccharides to galactose. Detailed biochemical characterization of recombinant purified GanB has supported this role, demonstrating significant hydrolytic activity towards galactobiose and larger galacto-oligomers, with no detectable activity towards lactose. Applying both GanA and GanB to galactan resulted in the full degradation of the polymer into galactose, which is then metabolized into UDP-glucose via the Leloir pathway by the galKET gene products (Tabachnikov & Shoham, 2013 ▶).
As a key player in the biochemical hydrolysis of the polymeric β-1,4-galactosaccharides into galactose monomers, the GanB β-galactosidase and related enzymes play an important part in the hemicellulolytic utilization system of many microorganisms, which use plant biomass for growth. The interest in the biochemical characterization and structural analysis of these enzymes stems therefore not only from basic scientific interest but also from their numerous potential biotechnological applications. Two crystal structures of GH42 enzymes have been solved and reported to date: A4-β-gal from Thermus thermophilus A4 (Hidaka et al., 2002 ▶) and Bca-β-gal from Bacillus circulans sp. alkalophilus (Maksimainen et al., 2012 ▶). These enzymes were shown to have a homotrimeric biologically active unit, stabilized by multiple hydrogen bonds and salt bridges. The active site of each subunit contains a single Trp residue that belongs to the next subunit, which can form a hydrogen bond with a bound galactose via a bridged water molecule (Hidaka et al., 2002 ▶). Based on these data and the complementary biochemical experiments conducted in our laboratory, we rationally designed and produced a number of mutants of GanB, some of which already show promising biotechnological properties.
In the present report, we describe the crystallization and preliminary crystallographic characterization of the wild-type GanB enzyme (GanB-WT) and its nucleophile mutant, GanB-E323A, which was originally produced as a subject for glycosynthesis studies (Ben-David et al., 2007 ▶). Complete diffraction data sets to 2.45 and 2.50 Å resolution have been collected on both the wild-type (WT) enzyme and its E323A mutant, respectively. These data sets will be used for full three-dimensional structure determination of the GanB protein using molecular-replacement techniques for phasing and the available GH42 structures as search models.
2. Experimental
2.1. Production and purification of WT GanB and its E323A nucleophile mutant
The ganB gene was fused to a tag consisting of six histidine residues at its N-terminus and was cloned into the expression vector pET9d as described previously (Tabachnikov & Shoham, 2013 ▶). Site-directed mutagenesis of ganB was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, California, USA). The mutagenic primers for the mutation E323A were 5′-GCAGCCGTTTTTATTGATGGCCTGCACGCCGAGTTTAGTG-3′ and 5′-CACTAAACTCGGCGTGCAGGCCATCAATAAAAACGGCTGC-3′ (mutated nucleotides are shown in bold). The cloned His-tagged proteins were overproduced in Escherichia coli BL21(λDE3) (Stratagene, La Jolla, California, USA), using the T7 polymerase expression system and purified in one step using nickel-affinity chromatography as previously described for other His-tagged proteins from G. stearothermophilus (Shulami et al., 2011 ▶). The final protein sample was more than 95% pure, based on SDS–PAGE with a total yield of approximately 500 mg protein per litre of overnight culture.
2.2. Crystallization experiments
Crystallization experiments were set up immediately after the last purification step of the recombinant GanB protein. The purified protein was concentrated to approximately 15–25 mg ml−1 using Centricon centrifugal concentrators (Millipore, Massachusetts, USA) and this protein solution was directly used for the crystallization experiments. All initial crystallization experiments were performed by the hanging-drop vapour-diffusion method using an extensive series of different factorial screens (Jancarik & Kim, 1991 ▶). In general, these initial conditions were based on commercially available sets of pre-set crystallization screens. Once positive results had been obtained (i.e. crystals or microcrystals), further refinement of these crystallization conditions was performed with specially prepared solutions, optimizing parameters such as pH, ionic strength, protein concentration, temperature and precipitating additives (Almog et al., 1993 ▶, 1994 ▶; Teplitsky et al., 1997 ▶, 1999 ▶, 2000 ▶; Bar et al., 2004 ▶; Lansky, Alalouf et al., 2013 ▶; Lansky, Salama et al., 2013 ▶).
The final GanB protein crystallization drops were prepared by mixing the concentrated protein solution (15–25 mg ml−1) with an equal amount of each of the specific screen conditions to give a final drop volume of 4.0 µl. Each of these protein drops was suspended over a 1.0 ml reservoir solution in 4 × 6 VDX crystallization plates (Hampton Research, California, USA), for a period of about 3–10 d at a constant temperature of 293 K. These initial screening experiments resulted practically in only one useful crystal form (TRP) which was found to be suitable for further crystallographic analysis (see below). Another crystal form, which appeared usually as a connected group of 10–15 very thin needles (TN), did not give sufficient X-ray diffraction for a reliable analysis and will not be discussed here.
The diffraction data measurements were performed at the European Synchrotron Research Facility (ESRF, Grenoble, France) and the Diamond Synchrotron Facility (England). Processing, reduction, indexing, integration and scaling of the diffraction data were conducted using the DENZO and SCALEPACK crystallographic programs (Otwinowski, 1993 ▶; Otwinowski & Minor, 1997 ▶) and the iMOSFLM and SCALA programs within the CCP4 crystallographic package (Winn et al., 2011 ▶).
3. Results and discussion
3.1. X-ray diffraction data for the GanB-WT crystals
The most promising GanB microcrystals initially appeared from condition I28 of the commercially available Wizard Classic 1 crystal screen [Emerald BioSystems, Bedford, Massachusetts, USA; 20%(w/v) PEG 3000, 100 mM HEPES–NaOH pH 7.5, 200 mM NaCl]. This initial crystallization condition was then refined in order to obtain larger and better diffracting crystals. The best diffracting GanB crystals were obtained from a 4 µl drop produced by mixing 2 µl of the protein solution (18–22 mg ml−1) containing 50 mM Tris buffer pH 7.0, 100 mM NaCl, 0.02% sodium azide with 2 µl of the crystallization reservoir solution consisting of 16–18% PEG 3350, 250 mM NaCl, 100 mM HEPES buffer pH 7.0. These crystals grew to their full size after about 5–15 d. Under these conditions the GanB crystals usually appeared as a cluster of five to ten thin plates growing in different directions from a common origin (Fig. 1 ▶). Attempts were made to grow separated and thicker plates of the same form, but these efforts have so far been unsuccessful. The final shape of each of the crystals in the cluster looked usually like a thin rectangular plate (TRP), with typical dimensions of about 0.3 × 0.2 × 0.02 mm (Fig. 2 ▶).
Figure 1.
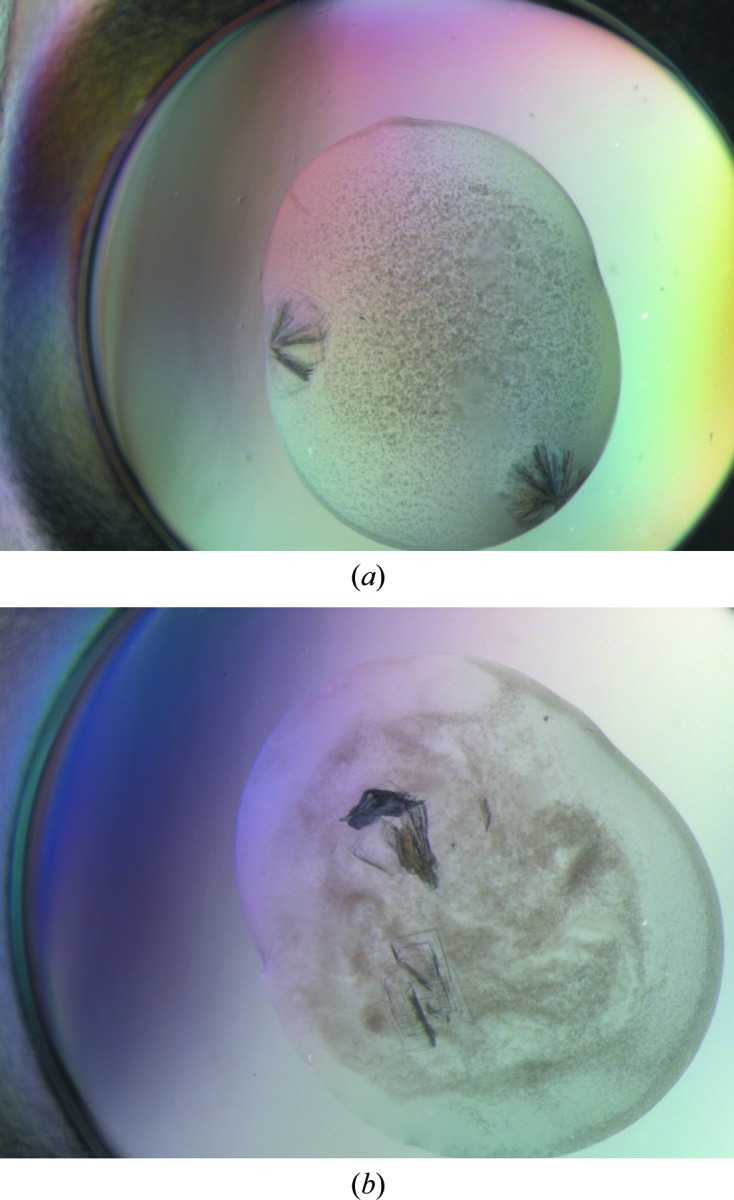
Typical crystals of GanB-WT of the TRP crystal habit. These crystals usually appear as a cluster of five to ten thin plates, growing in different directions from a common origin (a), or as layers of several thin plates mounted on each other via their wide faces (b).
Figure 2.
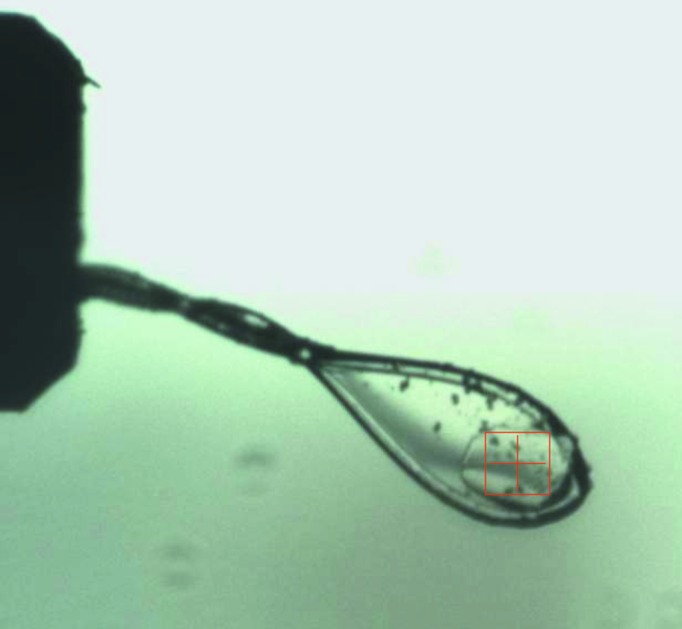
A single crystal of GanB-E323A (TRP habit) after separation from the originally grown cluster and mounting on a cryoloop. The loop is about 0.3 mm and the crystal dimensions are about 0.25 × 0.2 × 0.02 mm. The binocular square scale is 0.2 × 0.2 mm. Such a crystal was used for the full diffraction data collection at 2.5 Å resolution described here.
In order to characterize these crystals and use them to collect diffraction data, it was necessary to separate each of the single crystals from the multi-crystal cluster. This was performed by mechanical cutting of the cluster (using crystal-handling microtools) into separate plates, as close as possible to the cluster-connecting region, with special efforts to apply a minimal deformation force to the cluster and the crystals. Such separation of the multi-plate cluster into individual plates was followed by the isolation of each of the single plates into a different drop of the same solution content, from which the final crystal mounting was then performed.
Several such GanB-WT crystals of the TRP habit were used for a detailed crystallographic characterization under cryogenic conditions (90–100 K). Each of these different crystals produced a full data set, which was processed and examined in order to determine the optimal strategy for X-ray diffraction data measurement. These experiments were performed using X-ray synchrotron radiation (λ = 0.9334 Å) and a CCD area detector (ADSC Q210) on the ID14-1 beamline of the ESRF synchrotron facility. The crystal-cooling procedure used for these experiments included a short soaking of the selected crystal (for about 20–30 s) in a cryoprotecting solution containing 87% of the original crystallization reservoir solution (18% PEG 3350, 250 mM NaCl, 100 mM HEPES pH 7.0) and 13%(v/v) glycerol. The pre-soaked crystals were then submitted to immediate flash-cooling, directly within a cold nitrogen-gas stream (100 K, Oxford Cryosystems). The observed diffraction pattern of these crystals indicated that the crystals belong to a primitive orthorhombic crystal system (space group P212121), with average crystallographic unit-cell dimensions of a = 71.84, b = 181.35, c = 196.57 Å. Different crystals of this crystallization batch gave similar unit-cell dimensions with an overall deviation from the average values of less than 0.4%. Although the observed diffraction data of these crystals exceeded usually the 2.9 Å resolution limit (Figs. 3 ▶ and 4 ▶), a significant variation has been observed in the quality of these data. The variations were especially noticeable in the overall mosaicity of the data and the differences between the diffraction resolution limits along different axes and in different orientations of the same crystal. These crystal-dependent differences could probably be accounted for, at least partially, by the different deformation forces that each of the crystals experienced during the mechanical crystal separation process.
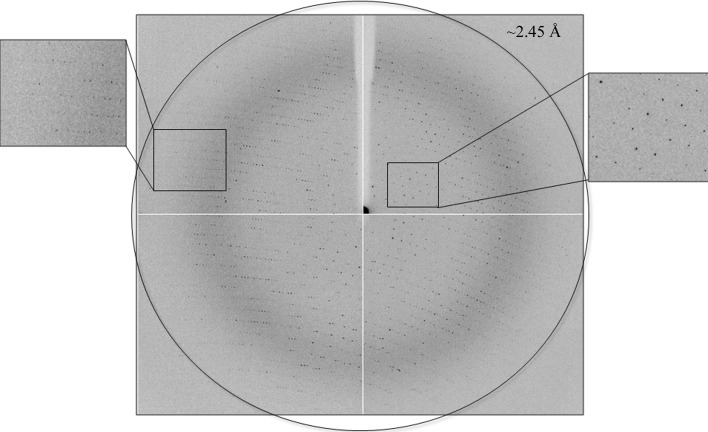
Figure 3.
X-ray diffraction pattern of GanB-WT obtained using a synchrotron-radiation source (ID14-1, ESRF). The outer circle corresponds to a 2.45 Å resolution limit. The insets represent magnified views of the sections indicated by the corresponding squares (right, low resolution; left, high resolution).
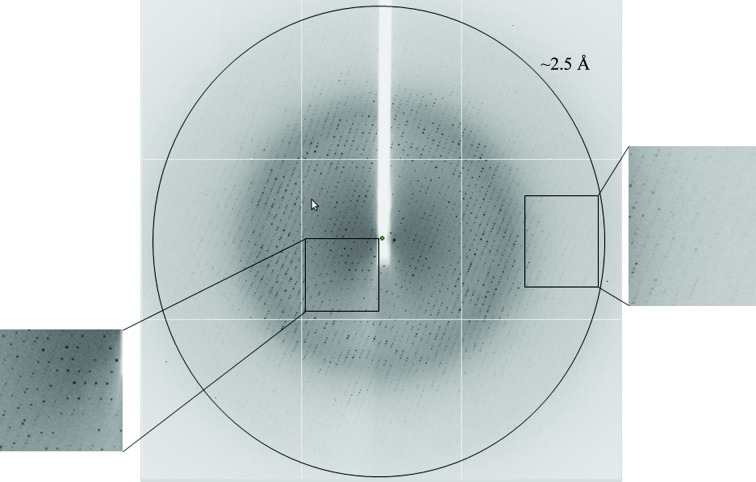
Figure 4.
X-ray diffraction pattern of GanB-E323A obtained using a synchrotron-radiation source (I04, Diamond). The outer circle corresponds to a 2.50 Å resolution limit. The insets represent magnified views of the sections indicated by the corresponding squares (left, low resolution; right, high resolution).
One of the GanB-WT crystals (of the TRP crystal habit) was used for a full high-resolution X-ray diffraction data measurement at 2.45 Å resolution (Fig. 3 ▶) using the same separation and cooling procedures as described above. An oscillation data set (Δϕ = 0.5°, 10 s exposure, 360 frames, 180°) was measured on the ID14-1 beamline at the ESRF (λ = 0.9334 Å, 100 K). The raw CCD diffraction images were processed with the DENZO and SCALEPACK software packages (Otwinowski, 1993 ▶). A total of 531 503 accepted reflections [F > 0σ(F)] were measured in the 49.0–2.45 Å resolution range and resulted in 91 957 independent reflections with 96.5% completeness to 2.45 Å resolution and 88.6% completeness for the highest resolution shell (2.58–2.45 Å). The overall multiplicity in the data set was 5.8, the overall mosaicity was 0.71°, the average 〈I/σ(I)〉 was 11.7 and the final R merge for the whole data set was 11.3% (34.8% for the highest resolution shell). These parameters confirm that the final data set used represents a full diffraction data set of reasonable crystallographic quality (Table 1 ▶), especially considering that these crystals originated from a cluster and a significant mechanical force had been applied to them during the separation process.
Table 1. Representative parameters from the crystallographic data measurement of GanB-WT and its catalytic mutant GanB-E323A.
Values in parentheses are for the last resolution shell.
| Protein | GanB-E323A | GanB-WT |
|---|---|---|
| Beamline | I04, Diamond | ID14-1, ESRF |
| Wavelength (Å) | 0.9763 | 0.9334 |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å, °) | a = 71.73, b = 181.31, c = 197.66, α = β = γ = 90.00 | a = 71.84, b = 181.35, c = 196.57, α = β = γ = 90.00 |
| No. of reflections (total/unique) | 285572/87146 | 531503/91957 |
| Multiplicity | 3.3 | 5.8 |
| 〈I/σ(I)〉 | 13.9 | 11.7 |
| Mosaicity (°) | 0.58 | 0.71 |
| Resolution (Å) | 71.7–2.50 (2.56–2.50) | 48.9–2.45 (2.58–2.45) |
| Completeness (%) | 97.4 (96.6) | 96.5 (88.6) |
| R merge † (%) | 6.1 (19.8) | 11.3 (34.8) |
R
merge = 
 , where I
i(hkl) is an individual intensity measurement and 〈I(hkl)〉 is the average intensity for this reflection, with summation over all data.
, where I
i(hkl) is an individual intensity measurement and 〈I(hkl)〉 is the average intensity for this reflection, with summation over all data.
A rough calculation of the specific ratio of volume/protein (V M) was performed in order to estimate the number of protein monomers per crystallographic asymmetric unit. The volume of the GanB-WT (TRP form) crystallographic unit cell, as determined from the mean value of the unit-cell parameters at 100 K, is 2.57 × 106 Å3. Assuming that the VM and the overall solvent content values here are within the normal range observed for soluble proteins (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶), there should be two to four GanB monomers (each monomer of 686 amino-acid residues; molecular weight 81 056 Da; Tabachnikov & Shoham, 2013 ▶) in the crystallographic asymmetric unit. With two molecules in the P212121 asymmetric unit (eight in the unit cell), the calculated V M is 3.96 Å3 Da−1 and the corresponding solvent content in the crystals is 69.0%. With three molecules in the asymmetric unit (12 in the unit cell), the calculated V M is 2.64 Å3 Da−1 and the corresponding solvent content in the crystals is 53.5%. With four molecules in the asymmetric unit (16 in the unit cell), the calculated V M is 1.98 Å3 Da−1 and the corresponding solvent content in the crystals is 38.0%. Although the medium value of V M seems to be statistically more probable (i.e. three monomers per asymmetric unit), all three V M values have been observed for soluble proteins. Self-rotation calculations confirmed that there is no additional (non-crystallographic) symmetry within the crystallographic asymmetric unit and indicated that the number of monomers per asymmetric unit is closer to three. The actual content of the GanB TRP asymmetric unit (and the unit cell) will only be unequivocally resolved when the crystallographic protein structure is fully determined.
3.2. X-ray diffraction data for the GanB-E323A crystals
The nucleophile catalytic mutant of GanB presented above, GanB-E323A, was crystallized in the same conditions as described for the WT enzyme and the resulting TRP crystals were confirmed to be highly isomorphous to the corresponding GanB-WT crystals (space group P212121; average unit-cell parameters a = 71.73, b = 181.31, c = 197.66 Å). One of these GanB-E323A crystals was used for the measurement of a complete oscillation data set to 2.50 Å resolution (Fig. 4 ▶) at 100 K using X-ray synchrotron radiation (λ = 0.9763 Å) and an ADSC Q315r area detector on the I04 beamline of the Diamond Light Source facility (Oxford, England). This data set was collected over an oscillation range of 90° (ϕ start 167°, ϕ end 257°) with a Δϕ of 0.5°, an exposure time of 0.5 s and a crystal-to-detector distance of 378.51 mm. The raw CCD diffraction images were processed with the iMOSFLM and SCALA programs, as available within the CCP4 crystallographic package (Winn et al., 2011 ▶). A total of 285 572 accepted reflections [F > 0σ(F)] were measured in the 71.7–2.50 Å resolution range, resulting in 87 146 independent reflections with 97.4% completeness to 2.50 Å resolution. The overall multiplicity in the data set was 3.3, the overall mosaicity was 0.58°, the average 〈I/σ(I)〉 was 13.9 and the final R merge was 6.1%. These parameters confirm that this data set is complete and sufficient for structural analysis, and seems to be even better and more reliable than the WT crystallographic data presented above (Table 1 ▶).
The two complete data sets described above will now be used for full three-dimensional structural analysis of both GanB-WT and GanB-E323A. A search of the Protein Data Bank (PDB) showed that a full three-dimensional structure is available for a reasonably homologous protein (33% amino-acid sequence identity), a native β-galactosidase from the extreme thermophile T. thermophilus A4 (A4-β-Gal; 1.6 Å resolution; PDB entry 1kwg; Hidaka et al., 2002 ▶). This structure should provide a good reference model for a molecular-replacement crystallographic calculation, expected to solve the phase problem and provide a good starting model for the structural analysis of the GanB proteins. Such analyses are currently in progress in our laboratory.
Acknowledgments
This work was supported by the Israel Science Foundation Grants 500/10 and 152/11, the I-CORE Program of the Planning and Budgeting Committee, the Ministry of Environmental Protection and the Grand Technion Energy Program (GTEP) and comprises part of The Leona M. and Harry B. Helmsley Charitable Trust reports on Alternative Energy series of the Technion, Israel Institute of Technology and the Weizmann Institute of Science. NEC thanks the UK Engineering and Physical Sciences Research Council (EPSRC) Grants EP/G027005 for financial support. We thank the staff at the European Synchrotron Research Facility (ESRF, ID14-1 beamline) for their helpful support in the X-ray synchrotron data measurement and analysis. YS holds the Erwin and Rosl Pollak Chair in Biotechnology at the Technion.
References
- Alalouf, O., Balazs, Y., Volkinshtein, M., Grimple, Y., Shoham, G. & Shoham, Y. (2011). J. Biol. Chem. 286, 41993–42001. [DOI] [PMC free article] [PubMed]
- Almog, O., Greenblatt, H. M., Spungin, A., Ben-Meir, D., Blumberg, S. & Shoham, G. (1993). J. Mol. Biol. 230, 342–344. [DOI] [PubMed]
- Almog, O., Klein, D., Braun, S. & Shoham, G. (1994). J. Mol. Biol. 235, 760–762. [DOI] [PubMed]
- Bar, M., Golan, G., Nechama, M., Zolotnitsky, G., Shoham, Y. & Shoham, G. (2004). Acta Cryst. D60, 545–549. [DOI] [PubMed]
- Ben-David, A., Bravman, T., Balazs, Y. S., Czjzek, M., Schomburg, D., Shoham, G. & Shoham, Y. (2007). Chembiochem, 8, 2145–2151. [DOI] [PubMed]
- Bravman, T., Zolotnitsky, G., Belakhov, V., Shoham, G., Henrissat, B., Baasov, T. & Shoham, Y. (2003). Biochemistry, 42, 10528–10536. [DOI] [PubMed]
- Brüx, C., Ben-David, A., Shallom-Shezifi, D., Leon, M., Niefind, K., Shoham, G., Shoham, Y. & Schomburg, D. (2006). J. Mol. Biol. 359, 97–109. [DOI] [PubMed]
- Daniel, R. A., Haiech, J., Denizot, F. & Errington, J. (1997). J. Bacteriol. 179, 5636–5638. [DOI] [PMC free article] [PubMed]
- Delangle, A., Prouvost, A.-F., Cogez, V., Bohin, J.-P., Lacroix, J.-M. & Hugouvieux Cotte-Pattat, N. (2007). J. Bacteriol. 189, 7053–7061. [DOI] [PMC free article] [PubMed]
- Gat, O., Lapidot, A., Alchanati, I., Regueros, C. & Shoham, Y. (1994). Appl. Environ. Microbiol. 60, 1889–1896. [DOI] [PMC free article] [PubMed]
- Golan, G., Shallom, D., Teplitsky, A., Zaide, G., Shulami, S., Baasov, T., Stojanoff, V., Thompson, A., Shoham, Y. & Shoham, G. (2004). J. Biol. Chem. 279, 3014–3024. [DOI] [PubMed]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol. 7, 637–644. [DOI] [PubMed]
- Hidaka, M., Fushinobu, S., Ohtsu, N., Motoshima, H., Matsuzawa, H., Shoun, H. & Wakagi, T. (2002). J. Mol. Biol. 322, 79–91. [DOI] [PubMed]
- Hinz, S. W., van den Brock, L. A., Beldman, G., Vincken, J. P. & Voragen, A. G. (2004). Appl. Microbiol. Biotechnol. 66, 276–284. [DOI] [PubMed]
- Hövel, K., Shallom, D., Niefind, K., Belakhov, V., Shoham, G., Baasov, T., Shoham, Y. & Schomburg, D. (2003). EMBO J. 22, 4922–4932. [DOI] [PMC free article] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst. 24, 409–411.
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Lansky, S., Alalouf, O., Solomon, V., Alhassid, A., Govada, L., Chayan, N. E., Belrhali, H., Shoham, Y. & Shoham, G. (2013). Acta Cryst. F69, 430–434. [DOI] [PMC free article] [PubMed]
- Lansky, S., Salama, R., Solomon, V. H., Belrhali, H., Shoham, Y. & Shoham, G. (2013). Acta Cryst. F69, 695–699. [DOI] [PMC free article] [PubMed]
- Maksimainen, M., Paavilainen, S., Hakulinen, N. & Rouvinen, J. (2012). FEBS J. 279, 1788–1798. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- O’Connell Motherway, M., Fitzgerald, G. F. & van Sinderen, D. (2011). Microb. Biotechnol. 4, 403–416. [DOI] [PMC free article] [PubMed]
- Ohtsu, N., Motoshima, H., Goto, K., Tsukasaki, F. & Matsuzawa, H. (1998). Biosci. Biotechnol. Biochem. 62, 1539–1545. [DOI] [PubMed]
- Otwinowski, Z. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 56–62. Warrington: Daresbury Laboratory.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Rees, D. C., Johnson, E. & Lewinson, O. (2009). Nature Rev. Mol. Cell Biol. 10, 218–227. [DOI] [PMC free article] [PubMed]
- Schwab, C., Sørensen, K. I. & Gänzle, M. G. (2010). Syst. Appl. Microbiol. 33, 300–307. [DOI] [PubMed]
- Shallom, D., Belakhov, V., Solomon, D., Gilead-Gropper, S., Baasov, T., Shoham, G. & Shoham, Y. (2002). FEBS Lett. 514, 163–167. [DOI] [PubMed]
- Shallom, D., Belakhov, V., Solomon, D., Shoham, G., Baasov, T. & Shoham, Y. (2002). J. Biol. Chem. 277, 43667–43673. [DOI] [PubMed]
- Shallom, D., Golan, G., Shoham, G. & Shoham, Y. (2004). J. Bacteriol. 186, 6928–6937. [DOI] [PMC free article] [PubMed]
- Shallom, D., Leon, M., Bravman, T., Ben-David, A., Zaide, G., Belakhov, V., Shoham, G., Schomburg, D., Baasov, T. & Shoham, Y. (2005). Biochemistry, 44, 387–397. [DOI] [PubMed]
- Shipkowski, S. & Brenchley, J. E. (2006). Appl. Environ. Microbiol. 72, 7730–7738. [DOI] [PMC free article] [PubMed]
- Shulami, S., Gat, O., Sonenshein, A. L. & Shoham, Y. (1999). J. Bacteriol. 181, 3695–3704. [DOI] [PMC free article] [PubMed]
- Shulami, S., Raz-Pasteur, A., Tabachnikov, O., Gilead-Gropper, S., Shner, I. & Shoham, Y. (2011). J. Bacteriol. 193, 2838–2850. [DOI] [PMC free article] [PubMed]
- Shulami, S., Zaide, G., Zolotnitsky, G., Langut, Y., Feld, G., Sonenshein, A. L. & Shoham, Y. (2007). Appl. Environ. Microbiol. 73, 874–884. [DOI] [PMC free article] [PubMed]
- Solomon, V., Teplitsky, A., Shulami, S., Zolotnitsky, G., Shoham, Y. & Shoham, G. (2007). Acta Cryst. D63, 845–859. [DOI] [PubMed]
- Tabachnikov, O. & Shoham, Y. (2013). FEBS J. 280, 950–964. [DOI] [PubMed]
- Teplitsky, A., Feinberg, H., Gilboa, R., Lapidot, A., Mechaly, A., Stojanoff, V., Capel, M., Shoham, Y. & Shoham, G. (1997). Acta Cryst. D53, 608–611. [DOI] [PubMed]
- Teplitsky, A., Shulami, S., Moryles, S., Shoham, Y. & Shoham, G. (2000). Acta Cryst. D56, 181–184. [DOI] [PubMed]
- Teplitsky, A., Shulami, S., Moryles, S., Zaide, G., Shoham, Y. & Shoham, G. (1999). Acta Cryst. D55, 869–872. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yang, T.-C., Hu, R.-M., Hsiao, Y.-M., Weng, S.-F. & Tseng, Y.-H. (2003). J. Mol. Microbiol. Biotechnol. 6, 145–154. [DOI] [PubMed]
- Zaide, G., Shallom, D., Shulami, S., Zolotnitsky, G., Golan, G., Baasov, T., Shoham, G. & Shoham, Y. (2001). Eur. J. Biochem. 268, 3006–3016. [DOI] [PubMed]