The two Plasmodium actin isoforms were expressed, purified and crystallized in complex with the gelsolin G1 domain. Crystals diffracting to high resolution were obtained and data sets for actin I and II were collected to 1.19 and 2.2 Å resolution, respectively.
Keywords: actin, malaria, Plasmodium, protein–protein complex, circular-dichroism spectroscopy
Abstract
Malaria is a devastating disease caused by apicomplexan parasites of the genus Plasmodium that use a divergent actin-powered molecular motor for motility and invasion. Plasmodium actin differs from canonical actins in sequence, structure and function. Here, the purification, crystallization and secondary-structure analysis of the two Plasmodium actin isoforms are presented. The recombinant parasite actins were folded and could be purified to homogeneity. Plasmodium actins I and II were crystallized in complex with the gelsolin G1 domain; the crystals diffracted to resolutions of 1.19 and 2.2 Å and belonged to space groups P212121 and P21, respectively, each with one complex in the asymmetric unit.
1. Introduction
Actin is expressed at high levels in eukaryotic cells and is involved in crucial cellular functions such as cell motility (Mitchison & Cramer, 1996 ▶), intracellular transport (Goode et al., 2000 ▶) and cell division (Matsudaira, 1994 ▶). A number of human pathogens, including bacteria and parasites, are known to remodel intracellular actin networks in host cells (Cyrklaff et al., 2012 ▶; Gouin et al., 2005 ▶). Actin is one of the most conserved proteins at the amino-acid sequence level and its sequence is nearly identical between species as distant as yeast and man. Prototypical actins exist in two defined states: as monomeric actin (G-actin) and filamentous actin (F-actin). In higher eukaryotes, the transitions between and stability of these two states are regulated by approximately 150 actin-binding proteins (Lee & Dominguez, 2010 ▶).
Despite the overall conservation of actins across species, some evolutionarily distinct and clinically relevant parasites, such as Plasmodium (Wesseling et al., 1988 ▶), Toxoplasma (Dobrowolski et al., 1997 ▶), Trypanosoma (García-Salcedo et al., 2004 ▶) and Giardia (Paredez et al., 2011 ▶), have highly diverged actins. Among these, P. falciparum, the causative agent of malaria, is the most devastating human pathogen. In contrast to yeast or human actins, P. falciparum actin is divergent in function, regulation and polymerization properties, and presumably also in the structure of the filamentous actin (Schmitz et al., 2005 ▶; Schüler & Matuschewski, 2006 ▶; Sahoo et al., 2006 ▶). In addition, Plasmodium actin dynamics are controlled by a remarkably small set of regulatory proteins, most likely only ∼10–15 different G-actin- and F-actin-binding proteins (Sattler et al., 2011 ▶). Plasmodium species express two actin isoforms: actin I (ActI) and actin II (ActII). ActI is expressed ubiquitously, while ActII is present only during the sexual stages of the life cycle (Wesseling et al., 1988 ▶; Deligianni et al., 2011 ▶). The two parasite actins share only approximately 80% sequence identity with each other and with actins from higher eukaryotes (Wesseling et al., 1989 ▶; Gardner et al., 2002 ▶). ActI forms shorter and less stable filaments than muscle actin (Sahoo et al., 2006 ▶; Sibley, 2011 ▶) and the structure of these differs from canonical actin filaments (Schmitz et al., 2010 ▶). In Plasmodium, actin is involved in host-cell invasion by powering its motility, and jasplakinolide, which binds to actin monomers and affects filament dynamics in a complicated manner (Bubb et al., 2000 ▶), impairs the ability of the parasite to invade (Mizuno et al., 2002 ▶). Other components of the Plasmodium invasion machinery have been investigated as targets for structure-based drug design (Bosch et al., 2006 ▶, 2007 ▶, 2012 ▶; Kortagere et al., 2010 ▶). Detailed structural information on the central component, actin, will be valuable for further such studies.
In order to gain insight into the structural and biochemical properties of the two divergent parasite actins, we expressed and purified P. falciparum ActI (PfActI; PlasmoDB PF3D7_1246200) and P. berghei ActII (PbActII; PlasmoDB PBANKA_103010) from baculovirus-infected insect cells. Both recombinant actins were folded, as analyzed by circular-dichroism (CD) spectroscopy, and were successfully crystallized in the presence of the mouse gelsolin G1 domain.
2. Materials and methods
2.1. Recombinant protein production and characterization
The mouse gelsolin cDNA was obtained from ImaGenes. Residues 50–174 (corresponding to the G1 domain) were cloned into the pETM22 vector (EMBL) as a fusion protein with thioredoxin. The final G1 protein contained an additional Gly-Pro sequence at the N-terminus. Thioredoxin-G1 was expressed in Escherichia coli Rosetta (DE3) cells (Novagen) at 293 K in ZYM-5052 autoinduction medium (Studier, 2005 ▶) and the cells were harvested after 24–46 h of agitation. The cell pellet was resuspended in lysis buffer A [20 mM Tris pH 8.0, 300 mM NaCl, 2 mM β-mercaptoethanol (β-ME), 20 mM imidazole] and lysed by sonication. After centrifugation at 30 000g, the clarified cell lysate containing thioredoxin-G1 was applied onto a nickel–nitriloacetate (Ni–NTA; Qiagen) column. The Ni–NTA beads were washed thoroughly with lysis buffer A and thioredoxin-G1 was eluted with 300 mM imidazole in lysis buffer A. Imidazole was removed by dialysis to lysis buffer A without imidazole and with the 2 mM β-ME replaced by 1 mM dithiothreitol at 277 K for 4 h and the fusion protein was cleaved with Tobacco etch virus (TEV) protease (van den Berg et al., 2006 ▶); the solution was then passed through an Ni–NTA column to remove the protease, the tag and any uncleaved fusion protein. Size-exclusion chromatography using a Superdex 75 16/60 column (GE Healthcare) in 20 mM Tris pH 8.0, 300 mM NaCl, 1 mM DTT was used as the final step and G1 eluted as a single peak from the size-exclusion column. The peak fractions were pooled, concentrated to 11–20 mg ml−1, frozen in liquid nitrogen and stored at 203 K. The cloning and expression details are summarized in Table 1 ▶.
Table 1. Recombinant protein production information.
In the protein sequences, the thioredoxin sequence is indicated in italics and the 6×His tag, the 3C/TEV protease cleavage sites and additional residues resulting from the cloning strategy are indicated in bold.
| Protein | Gelsolin G1 | PfActI | PbActII |
|---|---|---|---|
| Source organism | Mus musculus | P. falciparum | P. berghei |
| DNA source | Synthetic gene | Synthetic gene | Synthetic gene |
| Expression vector | pETM22 | pFastBac HTA | pFastBac Dual |
| Expression host | E. coli Rosetta (DE3) | S. frugiperda (Sf21) | S. frugiperda (Sf21) |
| Complete amino-acid sequence of the construct produced | MSDKIIHLTDDSFDTDVLKADGAILVDFWAEWCGPCKMIAPILDEIADEYQGKLTVAKLNIDQNPGTAPKYGIRGIPTLLLFKNGEVAATKVGALSKGQLKEFLDANLAGSGSGHM HHHHHHSSGLEVLFQ/GPMVVEHPEFLKAGKEPGLQIWRVEKFDLVPVPPNLYGDFFTGDAYVILKTVQLRNGNLQYDLHYWLGNECSQDESGAAAIFTVQLDDYLNGRAVQHREVQGFESSTFSGYFKSGLKYKKGGVASGF | MSYYHHHHHHDYDIPTTENLYFQ/GAMGEEDVQALVVDNGSGNVKAGVAGDDAPRSVFPSIVGRPKNPGIMVGMEEKDAFVGDEAQTKRGILTLKYPIEHGIVTNWDDMEKIWHHTFYNELRAAPEEHPVLLTEAPLNPKGNRERMTQIMFESFNVPAMYVAIQAVLSLYSSGRTTGIVLDSGDGVSHTVPIYEGYALPHAIMRLDLAGRDLTEYLMKILHERGYGFSTSAEKEIVRDIKEKLCYIALNFDEEMKTSEQSSDIEKSYELPDGNIITVGNERFRCPEALFQPSFLGKEAAGIHTTTFNSIKKCDVDIRKDLYGNIVLSGGTTMYEGIGERLTRDITTLAPSTMKIKVVAPPERKYSVWIGGSILSSLSTFQQMWITKEEYDESGPSIVHRKCF | MKHHHHHHSAGLEVLFQ/GPMPEESIALVVDNGSGMVKSGLAGDDAPKCVFPSIIGIPKMPNIMVGMEQKECYVGDEAQNKRGILTLKYPIEHGIVTNWDDMEKIWRHTFFNELRVSPEEHPVLLTEAPLNPKTNREKMTQIMFESFDVPAMYVSIQAILSLYASGRTTGIVLDSGDGVTHTVPIYEGYVLPHAINRTDMAGRDLTYYMMKLFTERGYTFTTTAEREIVRDIKEKLCYIALDYDEELKKSEERTEEVEEMYELPDGNLITVGSERFRCPEALFNPSLIGRECPGLHITAYQSIMKCDIDIRKELYNNIVLSGGTTMYNYIGERLTNEMTSLAPPSMKIKVIAPPERKYSVWIGGSILSSLSTFQKMWITKEEYDESGPSIVHRKCF |
PfActI and PbActII were expressed in Spodoptera frugiperda Sf21 cells (Invitrogen) at 300 K as described by Ignatev et al. (2012 ▶). A frozen cell pellet stored at 253 K was resuspended in lysis buffer B [20 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) pH 9.5, 250 mM NaCl, 5 mM CaCl2, 1 mM adenosine triphosphate (ATP), 2 mM β-ME, 10 mM imidazole] with 1× cOmplete EDTA-free protease inhibitor (Roche). 7–15 mg G1 was added to the cell suspension and the cells were lysed by sonication for 15 s. The cell lysate was centrifuged for 45 min at 43 000g and the supernatant was applied onto Ni–NTA beads and incubated for 30 min at 277 K. The beads were washed extensively with lysis buffer B, lysis buffer B with 20 mM imidazole, lysis buffer B with 25 mM imidazole and 300 mM NaCl, and finally with modified G-buffer (15 mM Tris pH 7.5, 0.2 mM CaCl2, 3 mM β-ME, 1 mM ATP, 50 mM NaCl, 20 mM imidazole). The protein complex was eluted with 300 mM imidazole in the modified G-buffer. For PfActI-G1, size-exclusion chromatography was carried out prior to TEV cleavage using a Superose 12 10/300 GL column (GE Healthcare) in the modified G-buffer without imidazole. The peak fractions from gel filtration for PfActI-G1 and the Ni–NTA-purified PbActII-G1 were incubated with TEV protease and 3C protease (Leong, 1999 ▶) on ice for 1 h, respectively. After cleavage, both proteins were passed through an Ni–NTA column before a final size-exclusion chromatography step with a Superose 12 10/300 GL column in the modified G-buffer without imidazole. Peak fractions from size-exclusion chromatography were pooled and concentrated to 5.1 and 5.7 mg ml−1 for PfActI–G1 and PbActII–G1, respectively. The expression-construct details for PfActI and PbActII are summarized in Table 1 ▶.
For secondary-structure and thermal stability analysis, uncomplexed PfActI and PbActII were expressed and purified as described previously (Ignatev et al., 2012 ▶). CD spectra from 260 to 195 nm were measured for both actins using an Applied Photophysics Chirascan-plus spectropolarimeter equipped with a thermal control unit (Quantum Northwest, TC125), a direct temperature probe and a 1 mm path-length quartz cuvette (Hellma). Spectra were recorded at a concentration of 0.4 mg ml−1 over the temperature range 287–355 K using a ramp of 1 K min−1 in the following buffer: 7 mM HEPES pH 7.5, 0.1 mM CaCl2, 0.4 mM ATP, 0.4 mM tris(2-carboxyethyl)phosphine. The DichroWeb server (Lobley et al., 2002 ▶) was used for secondary-structure determination using the CDSSTR algorithm and set4 optimized for 190–240 nm as a reference data set (Compton & Johnson, 1986 ▶).
2.2. Crystallization
PfActI and PbActII were crystallized in complex with G1 using the sitting-drop vapour-diffusion method in Corning 3556 96-well plates (Table 2 ▶). Initial screening was performed using a Mosquito nanodispenser at the Biocenter Finland crystallization core facilities at the University of Oulu and the Institute of Biotechnology, University of Helsinki by mixing equal volumes (0.1 µl) of protein and reservoir solutions and equilibrating against a reservoir volume of 50 µl at 293 K. Approximately 500–600 conditions were screened in the initial screens for both actin complexes and the following screens were used: PACT (Qiagen; PfActI–G1 only), JCSG (Qiagen), Helsinki Random I and II, Helsinki Complex and Helsinki Synergy. The initial hit for PfActI–G1 was obtained from the PACT screen (condition F4) and that for PbActII–G1 from Helsinki Complex (condition C6; Radaev et al., 2006 ▶). Optimization of the crystallization conditions was performed manually using a drop size of 0.5 + 0.5 µl (protein + reservoir) and varying the protein and precipitant as well as the salt/additive concentrations. PfActI crystals were obtained in 22%(w/v) polyethylene glycol (PEG) 3350, 0.2 M potassium thiocyanate, 0.1 M bis-tris propane pH 6.5. PbActII crystals were obtained in 18%(w/v) PEG 8000, 0.2 M ammonium sulfate, 0.1 M bis-tris propane pH 6.5. In both cases a single crystal was used for data collection.
Table 2. Crystallization details of the PfActI–G1 and PbActII–G1 complexes.
| PfActI–G1 | PbActII–G1 | |
|---|---|---|
| Method | Sitting-drop vapour diffusion | Sitting-drop vapour diffusion |
| Plate type | Corning 3556 | Corning 3556 |
| Temperature (K) | 293 | 293 |
| Protein concentration (mg ml−1) | 5.1 | 5.7 |
| Buffer composition of protein solution | 15 mM Tris pH 7.5, 0.2 mM CaCl2, 3 mM β-ME, 1 mM ATP, 50 mM NaCl | 15 mM Tris pH 7.5, 0.2 mM CaCl2, 3 mM β-ME, 1 mM ATP, 50 mM NaCl |
| Composition of reservoir solution | 22%(w/v) PEG 3350, 0.2 M potassium thiocyanate, 0.1 M bis-tris propane pH 6.5 | 18%(w/v) PEG 8000, 0.2 M ammonium sulfate, 0.1 M bis-tris propane pH 6.5 |
| Volume and ratio of drop | 0.5 µl protein + 0.5 µl reservoir | 0.5 µl protein + 0.5 µl reservoir |
| Volume of reservoir (µl) | 50 | 50 |
2.3. Diffraction data collection and analysis
Prior to flash-cooling in liquid nitrogen, the PfActI crystal was soaked in a cryosolution consisting of 22%(w/v) PEG 3350, 10%(w/v) PEG 400, 5 mM Tris pH 7.5, 50 mM NaCl, 0.2 mM CaCl2, 0.5 mM ATP and the PbActII crystal in 23%(w/v) PEG 8000, 10%(w/v) PEG 400, 5 mM Tris pH 7.5, 0.5 mM ATP, 100 mM HEPES pH 7.5, 50 mM NaCl, 200 mM ammonium sulfate. X-ray diffraction data for PfActI were collected with a MAR Mosaic 225 detector on beamline BL14.1 at BESSY (Berlin, Germany; Mueller et al., 2012 ▶). The diffraction data were processed with XDS and scaled with XSCALE (Kabsch, 2010 ▶) using XDSi (Kursula, 2004 ▶). The PbActII data were collected on a Bruker MICROSTAR copper-anode X-ray generator equipped with a PLATINUM135 CCD detector. The images were integrated and scaled in PROTEUM 2.0 using SAINT and SADABS with the narrow-frame algorithm. Details of the data-collection and processing statistics are given in Table 3 ▶.
Table 3. Data-collection and processing information.
Values in parentheses are for the outer shell.
| Crystal | PfActI–G1 | PbActII–G1 |
|---|---|---|
| Diffraction source | BL14.1, BESSY | Bruker MICROSTAR rotating anode |
| Wavelength (Å) | 0.91841 | 1.54 |
| Temperature (K) | 100 | 100 |
| Detector | MAR Mosaic 225 | PLATINUM135 CCD |
| Rotation range per image (°) | 0.5 | 0.5 |
| No. of frames | 360 | 944 |
| Space group | P212121 | P21 |
| Unit-cell parameters (Å, °) | a = 40.34, b = 57.90, c = 111.59, α = β = γ = 90 | a = 64.25, b = 60.91, c = 75.52, α = γ = 90, β = 97.24 |
| Resolution range (Å) | 45–1.19 (1.25–1.19) | 31.9–2.20 (2.25–2.20) |
| Total No. of reflections | 1269750 (162217) | 105649 (3271) |
| No. of unique reflections | 174587 (23705) | 29458 (1410) |
| Completeness (%) | 100 (100) | 99.4 (95.2) |
| Multiplicity | 7.2 (6.8) | 3.6 (2.3) |
| 〈I/σ(I)〉† | 10.9 (1.07) | 5.6 (0.9) |
| R meas ‡ | 0.126 (1.194) | 0.156 (0.786) |
| CC1/2 § (%) | 99.9 (38.3) | 98.6 (56.8) |
| Overall B factor from Wilson plot (Å2) | 15.9 | 27.6 |
The mean I/σ(I) in the outer shell falls below 2.0 at approximately 1.32 Å for PfActI–G1 and 2.5 Å for PbActII–G1.
R meas is the redundancy-independent merging R factor, as defined by Diederichs & Karplus (1997 ▶) and Weiss & Hilgenfeld (1997 ▶). For PfActII-G1, R meas has been estimated by multiplying the R int values from SAINT (0.133 and 0.591 for the whole data set and the outer shell, respectively) by the factor [N/(N − 1)]1/2, where N is the data multiplicity.
CC1/2 is defined as the correlation coefficient between two random half data sets, as described by Karplus & Diederichs (2012 ▶).
3. Results and discussion
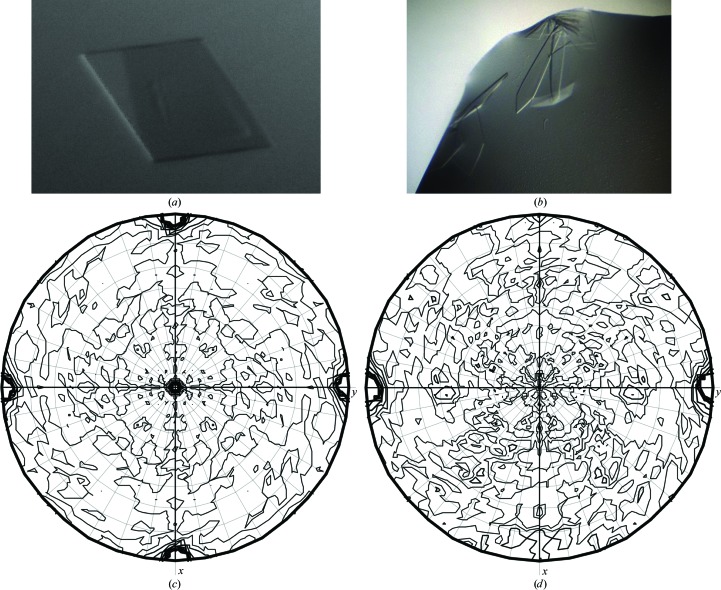
Recombinant PfActI and PbActII were successfully expressed using a baculovirus expression system and were purified to homogeneity as a complex with G1 in two steps by affinity and size-exclusion chromatography. Crystallization trials were performed using different protein concentrations and commercially available and homemade crystallization screens. For both actin–G1 complexes several initial hits were observed in different conditions containing PEGs and various salts and/or additives. The most promising-looking conditions were used for further optimization to achieve data-collection quality crystals. For PfActI and PbActII, crystals with maximum dimensions of 200 and 300 µm, respectively, appeared in 2–3 d. Both displayed plate-like morphology, although the PfActI crystals were slightly thicker (Figs. 1 ▶ a and 1 ▶ b).
Figure 1.
Diffraction analysis of the Plasmodium actin–G1 crystals. (a) A single crystal of the PfActI–G1 complex grown in 22%(w/v) PEG 3350, 0.2 M potassium thiocyanate, 0.1 M bis-tris propane pH 6.5. The longest edge of the crystal is approximately 200 µm. (b) Plate-like crystals of the PbActII–G1 complex grown in 18%(w/v) PEG 8000, 0.2 M ammonium sulfate, 0.1 M bis-tris propane pH 6.5. The longest edge of the largest crystal is approximately 300 µm. (c) Self-rotation function calculated from the PfActI–G1 data (P222) using MOLREP at χ = 180°. (d) Self-rotation function calculated from the PbActII–G1 data (P2) at χ = 180°. There were no additional peaks indicating noncrystallographic symmetry in either crystal form in addition to the crystallographic peaks.
Data sets were collected for the PfActI and PbActII crystals and were processed to resolutions of 1.19 and 2.2 Å, respectively (Table 3 ▶). The PfActI crystal had the P222 lattice, and the most likely space group based on the systematic absences is P212121. The unit-cell parameters of a = 40.34, b = 57.90, c = 111.59 Å and a Matthews coefficient (Matthews, 1968 ▶) of 2.4 Å3 Da−1 suggest the presence of one PfActI–G1 complex in the asymmetric unit, with 48% solvent content. The PbActII crystal belonged to space group P21, with unit-cell parameters a = 64.25, b = 60.91, c = 75.52 Å, β = 97.24°. A Matthews coefficient of 2.6 Å3 Da−1 indicates the presence of one actin–gelsolin complex in the asymmetric unit, with a solvent content of 52%. The data showed no signs of twinning or pseudotranslational symmetry, and a self-rotation function calculated using MOLREP (Vagin & Teplyakov, 2010 ▶) was consistent with the presence of one complex in the asymmetric unit in both cases (Figs. 1 ▶ c and 1 ▶ d). Subsequently, molecular-replacement solutions have been found for both actin–G1 data sets and the complex structures are currently under refinement. The PbActII–G1 structure was solved using the α-actin–G1 complex (PDB entry 1p8z; Irobi et al., 2003 ▶) as a search model. For PfActI–G1, the PbActII–G1 structure was used as the search model. The log-likelihood gain values from Phaser (McCoy et al., 2007 ▶) were 4800 for PbActII–G1 and 1100 for PfActI–G1. The rotation-function and translation-function Z scores were 45 and 22, respectively, for PbActII–G1 and 19 and 28, respectively, for PfActI–G1.
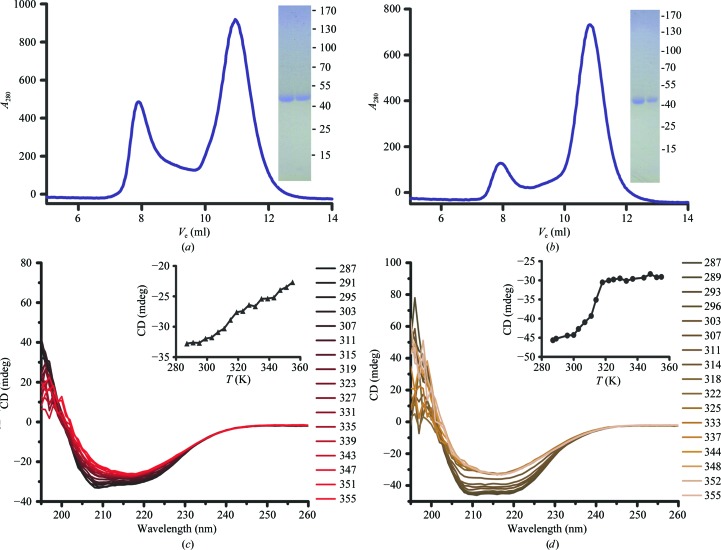
Uncomplexed PfActI and PbActII were also purified to homogeneity in soluble monomeric form and used for CD spectroscopy (Figs. 2 ▶ a and 2 ▶ b). The CD spectra of both PfActI (Fig. 2 ▶ c) and PbActII (Fig. 2 ▶ d) indicated that they were folded. PfActI contains 57% α-helix and 22% β-sheet and PbActII consists of 47% α-helix and 27% β-sheet. The secondary-structure contents are similar to those of previously determined actin crystal structures, which generally consist of 41–46% α-helix and 18–19% β-strand [PDB entries 3el2 (Nair et al., 2008 ▶), 1j6z (Otterbein et al., 2001 ▶), 3cip (Baek et al., 2008 ▶), 1nwk (Graceffa & Dominguez, 2003 ▶) and 2hf3 (Rould et al., 2006 ▶)]. Both parasite actins start to denature at 298–303 K (Figs. 2 ▶ c and 2 ▶ d). Instead of simply losing their secondary structure, both actins form some kind of β-aggregate upon heating and denaturation. For PbActII, a T m of 308 K could be determined, while for PfActI it was not possible to reliably determine a single T m. Both parasite actins are significantly less stable than muscle actin, for which melting temperatures of ∼333 K have been determined (Perieteanu et al., 2008 ▶; Levitsky et al., 2008 ▶).
Figure 2.
Purification and CD analysis of the Plasmodium actins. (a, b) Size-exclusion chromatograms and Coomassie-stained denaturing gels of the peak fractions eluting at 10–12 ml of (a) PfActI and (b) PbActII. The positions of molecular-weight markers are indicated in kDa. (c, d) Thermal denaturation CD curves of PfActI (c) and PbActII (d). Spectra from 260 to 195 nm were measured at 4 K intervals from 287 to 355 K. In the insets at the upper right of (c) and (d), the change in CD signal at 208 nm has been plotted.
To conclude, we have collected high-resolution crystallographic data for the two actin isoforms of the malaria parasite. Structure determination and refinement are ongoing and we expect the crystal structures to elucidate the molecular basis for the enigmatic functional properties of these parasite actins. The results will serve as a basis for understanding the structural differences between evolutionarily distant actins. In addition, we hope that the structures can be useful for the design of specific inhibitors for use as lead compounds for drug design against one of the most devastating global health threats.
Acknowledgments
We thank Dr Juha Kallio for helping with the synchrotron data collection. We are grateful to the support staff at BESSY beamline 14.1. This work was financially supported by the Academy of Finland, the Sigrid Jusélius Foundation, the Finnish Cultural Foundation and the German Ministry for Education and Research (BMBF).
References
- Baek, K., Liu, X., Ferron, F., Shu, S., Korn, E. D. & Dominguez, R. (2008). Proc. Natl Acad. Sci. USA, 105, 11748–11753. [DOI] [PMC free article] [PubMed]
- Berg, S. van den, Löfdahl, P. A., Härd, T. & Berglund, H. (2006). J. Biotechnol. 121, 291–298. [DOI] [PubMed]
- Bosch, J., Buscaglia, C. A., Krumm, B., Ingason, B. P., Lucas, R., Roach, C., Cardozo, T., Nussenzweig, V. & Hol, W. G. J. (2007). Proc. Natl Acad. Sci. USA, 104, 7015–7020. [DOI] [PMC free article] [PubMed]
- Bosch, J., Paige, M. H., Vaidya, A. B., Bergman, L. W. & Hol, W. G. J. (2012). J. Struct. Biol. 178, 61–73. [DOI] [PMC free article] [PubMed]
- Bosch, J., Turley, S., Daly, T. M., Bogh, S. M., Villasmil, M. L., Roach, C., Zhou, N., Morrisey, J. M., Vaidya, A. B., Bergman, L. W. & Hol, W. G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 4852–4857. [DOI] [PMC free article] [PubMed]
- Bubb, M. R., Spector, I., Beyer, B. B. & Fosen, K. M. (2000). J. Biol. Chem. 275, 5163–5170. [DOI] [PubMed]
- Compton, L. A. & Johnson, W. C. (1986). Anal. Biochem. 155, 155–167. [DOI] [PubMed]
- Cyrklaff, M., Sanchez, C. P., Frischknecht, F. & Lanzer, M. (2012). Trends Parasitol. 28, 479–485. [DOI] [PubMed]
- Deligianni, E., Morgan, R. N., Bertuccini, L., Kooij, T. W., Laforge, A., Nahar, C., Poulakakis, N., Schüler, H., Louis, C., Matuschewski, K. & Siden-Kiamos, I. (2011). Cell. Microbiol. 13, 1714–1730. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Dobrowolski, J. M., Niesman, I. R. & Sibley, L. D. (1997). Cell Motil. Cytoskeleton, 37, 253–262. [DOI] [PubMed]
- García-Salcedo, J. A., Pérez-Morga, D., Gijón, P., Dilbeck, V., Pays, E. & Nolan, D. P. (2004). EMBO J. 23, 780–789. [DOI] [PMC free article] [PubMed]
- Gardner, M. J. et al. (2002). Nature (London), 419, 498–511.
- Goode, B. L., Drubin, D. G. & Barnes, G. (2000). Curr. Opin. Cell Biol. 12, 63–71. [DOI] [PubMed]
- Gouin, E., Welch, M. D. & Cossart, P. (2005). Curr. Opin. Microbiol. 8, 35–45. [DOI] [PubMed]
- Graceffa, P. & Dominguez, R. (2003). J. Biol. Chem. 278, 34172–34180. [DOI] [PubMed]
- Ignatev, A., Bhargav, S. P., Vahokoski, J., Kursula, P. & Kursula, I. (2012). PLoS One, 7, e33586. [DOI] [PMC free article] [PubMed]
- Irobi, E., Burtnick, L. D., Urosev, D., Narayan, K. & Robinson, R. C. (2003). FEBS Lett. 552, 86–90. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Kortagere, S., Welsh, W. J., Morrisey, J. M., Daly, T., Ejigiri, I., Sinnis, P., Vaidya, A. B. & Bergman, L. W. (2010). J. Chem. Inf. Model. 50, 840–849. [DOI] [PMC free article] [PubMed]
- Kursula, P. (2004). J. Appl. Cryst. 37, 347–348.
- Lee, S. H. & Dominguez, R. (2010). Mol. Cells, 29, 311–325. [DOI] [PMC free article] [PubMed]
- Leong, L. E. (1999). Mol. Biotechnol. 12, 269–274. [DOI] [PubMed]
- Levitsky, D. I., Pivovarova, A. V., Mikhailova, V. V. & Nikolaeva, O. P. (2008). FEBS J. 275, 4280–4295. [DOI] [PubMed]
- Lobley, A., Whitmore, L. & Wallace, B. A. (2002). Bioinformatics, 18, 211–212. [DOI] [PubMed]
- Matsudaira, P. (1994). Semin. Cell Biol. 5, 165–174. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mitchison, T. J. & Cramer, L. P. (1996). Cell, 84, 371–379. [DOI] [PubMed]
- Mizuno, Y., Makioka, A., Kawazu, S., Kano, S., Kawai, S., Akaki, M., Aikawa, M. & Ohtomo, H. (2002). Parasitol. Res. 88, 844–848. [DOI] [PubMed]
- Mueller, U., Darowski, N., Fuchs, M. R., Förster, R., Hellmig, M., Paithankar, K. S., Pühringer, S., Steffien, M., Zocher, G. & Weiss, M. S. (2012). J. Synchrotron Rad. 19, 442–449. [DOI] [PMC free article] [PubMed]
- Nair, U. B., Joel, P. B., Wan, Q., Lowey, S., Rould, M. A. & Trybus, K. M. (2008). J. Mol. Biol. 384, 848–864. [DOI] [PMC free article] [PubMed]
- Otterbein, L. R., Graceffa, P. & Dominguez, R. (2001). Science, 293, 708–711. [DOI] [PubMed]
- Paredez, A. R., Assaf, Z. J., Sept, D., Timofejeva, L., Dawson, S. C., Wang, C.-J. R. & Cande, W. Z. (2011). Proc. Natl Acad. Sci. USA, 108, 6151–6156. [DOI] [PMC free article] [PubMed]
- Perieteanu, A. A., Sweeting, B. & Dawson, J. F. (2008). Biochemistry, 47, 9688–9696. [DOI] [PubMed]
- Radaev, S., Li, S. & Sun, P. D. (2006). Acta Cryst. D62, 605–612. [DOI] [PubMed]
- Rould, M. A., Wan, Q., Joel, P. B., Lowey, S. & Trybus, K. M. (2006). J. Biol. Chem. 281, 31909–31919. [DOI] [PubMed]
- Sahoo, N., Beatty, W., Heuser, J., Sept, D. & Sibley, L. D. (2006). Mol. Biol. Cell, 17, 895–906. [DOI] [PMC free article] [PubMed]
- Sattler, J. M., Ganter, M., Hliscs, M., Matuschewski, K. & Schüler, H. (2011). Eur. J. Cell Biol. 90, 966–971. [DOI] [PubMed]
- Schmitz, S., Grainger, M., Howell, S., Calder, L. J., Gaeb, M., Pinder, J. C., Holder, A. A. & Veigel, C. (2005). J. Mol. Biol. 349, 113–125. [DOI] [PubMed]
- Schüler, H. & Matuschewski, K. (2006). Trends Parasitol. 22, 146–147. [DOI] [PubMed]
- Schmitz, S., Schaap, I. A., Kleinjung, J., Harder, S., Grainger, M., Calder, L., Rosenthal, P. B., Holder, A. A. & Veigel, C. (2010). J. Biol. Chem. 285, 36577–36585. [DOI] [PMC free article] [PubMed]
- Sibley, L. D. (2011). Immunol. Rev. 240, 72–91. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst. 30, 203–205.
- Wesseling, J. G., Smits, M. A. & Schoenmakers, J. G. (1988). Mol. Biochem. Parasitol. 30, 143–153. [DOI] [PubMed]
- Wesseling, J. G., Snijders, P. J., van Someren, P., Jansen, J., Smits, M. A. & Schoenmakers, J. G. (1989). Mol. Biochem. Parasitol. 35, 167–176. [DOI] [PubMed]