SpaA pilin, which forms the pilus shaft of SpaCBA pili in L. rhamnosus GG, was crystallized and X-ray diffraction data were collected to a resolution of 2.0 Å.
Keywords: Lactobacillus rhamnosus GG, SpaCBA pili, fimbria, adhesion, sortase, probiotics, bacterial surface proteins
Abstract
Lactobacillus rhamnosus GG, a widely used Gram-positive probiotic strain, is clinically well known for its perceived health-promoting effects. It has recently been shown to display proteinaceous pilus fibres (called SpaCBA) on its cell surface. Structurally, SpaCBA pili possess a characteristic three-pilin polymerized architecture, with repeating SpaA major pilins that form the backbone and two types of minor subunits (SpaB and SpaC). In this study, recombinant SpaA protein was purified, characterized and crystallized. The crystals diffracted to a resolution of 2.0 Å and belonged to space group C2, with unit-cell parameters a = 227.9, b = 63.2, c = 104.3 Å, β = 95.1°.
1. Introduction
Many bacteria assemble pili (or fimbriae) on their cell surface for facilitating host-cell adherence and promoting pathogenesis. While substantial structural information already exists for pili from Gram-negative bacteria, only recently has research effort been directed into pili from Gram-positive bacteria (for recent reviews, see Kang & Baker, 2012 ▶; Vengadesan & Narayana, 2011 ▶; Hendrickx et al., 2011 ▶; Kline et al., 2010 ▶; Waksman & Hultgren, 2009 ▶). The pili from Gram-positive bacteria are typified by a covalently bonded polymeric structure that consists of a major (or backbone) pilin subunit forming the shaft and two different minor (or ancillary) pilins, one situated at the pilus tip for adhesion and another at the pilus base for cell-wall anchoring (Ton-That et al., 2004 ▶). Most structural investigations conducted on Gram-positive pilins to date have focused primarily on various pathogenic species. Recently, it has emerged that pilus-like structures are also associated with a beneficial commensal Gram-positive bacterium, these being the so-called SpaCBA pili in the probiotic Lactobacillus rhamnosus GG strain (Kankainen et al., 2009 ▶).
Within the L. rhamnosus GG genome, the SpaCBA pilus genes are organized into an operon that encodes a major pilin (SpaA), two minor pilins (SpaB and SpaC) and a pilin-specific sortase for catalyzing pilus assembly (Kankainen et al., 2009 ▶). SpaCBA pili have a typical three-pilin architecture, with SpaA comprising the pilus backbone and SpaB and SpaC at the pilus base and tip, respectively (Kankainen et al., 2009 ▶). Interestingly, in contrast to the pili from Gram-positive pathogens, SpaC and, to a lesser extent, SpaB are found sporadically throughout the SpaCBA pilus backbone (Reunanen et al., 2012 ▶; Tripathi et al., 2013 ▶), which might enhance adherence to the intestinal mucosa and epithelial layer and thereby then extend the relative longevity and transient colonization of L. rhamnosus GG cells in the gut.
In order to better understand the structural features of the pili from beneficial commensal bacteria, we have initiated a structural study of the pilin constituents of the SpaCBA pilus. Here, we describe the purification, crystallization and preliminary crystallographic analysis of the L. rhamnosus GG SpaA backbone-pilin subunit.
2. Materials and methods
2.1. Cloning, expression and purification
The construction of the recombinant clone for L. rhamnosus GG SpaA pilin protein has been described previously (von Ossowski et al., 2010 ▶). Briefly, an expression plasmid construct (called pKTH5319) with the L. rhamnosus GG (ATCC 53103) spaA gene encoding the SpaA backbone pilin (residues 31–302) but lacking the N-terminal signal peptide and C-terminal LPXTG sorting motif regions was generated for intracellular protein production in the Escherichia coli BL21 (DE3) pLysS strain. Recombinant SpaA pilin was hexahistidine-tagged at the C-terminus but also had an extra seven N-terminal residues (MGRDPNS) and two additional residues before the C-terminal histidine tag (LEHHHHHH). All of these correspond to amino acids encoded by the pET28+ expression vector.
For recombinant SpaA protein production, each litre of Luria–Bertani (LB) growth medium supplemented with 50 µg ml−1 kanamycin was inoculated with a 10 ml aliquot of overnight-grown culture and incubated at 310 K until it reached an OD600 nm of ∼0.6. Recombinant protein expression in E. coli cells was then induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, after which the culture was grown overnight at 303 K. Cells were harvested by centrifugation, resuspended in a lysis buffer consisting of 40 mM NaH2PO4 pH 7.4, 150 mM NaCl and EDTA-free protease-inhibitor cocktail tablets (Roche) and then disrupted by sonication on ice. To separate soluble proteins from cellular debris, the cell lysate was then centrifuged at 48 400g for 20 min at 277 K.
For SpaA purification, the cell-free lysate was loaded onto a 5 ml NiCl2-charged HiTrap chelating HP column (GE Healthcare) that had previously been equilibrated with buffer A (40 mM NaH2PO4 pH 7.4, 150 mM NaCl). Resin-bound SpaA protein was then eluted with buffer B (40 mM NaH2PO4 pH 7.4, 150 mM NaCl, 250 mM imidazole) using a linear gradient equivalent to ten column volumes. SpaA-containing elution fractions were determined by SDS–PAGE, pooled and dialyzed against 20 mM HEPES pH 7.0, 150 mM NaCl, 1 mM EDTA. The dialyzed protein solution was then concentrated using an Amicon ultrafiltration device fitted with a 10 kDa molecular-weight cutoff and subsequently purified further by a Sephacryl S-200 26/60 gel-filtration column (GE Healthcare) equilibrated with dialysis buffer (see above).
2.2. Crystallization
Purified recombinant SpaA protein in dialysis buffer (see above) was concentrated to ∼70 mg ml−1 as determined by UV absorbance at 280 nm using a theoretical extinction coefficient of 39 880 M −1 cm−1 (ProtParam tool on the ExPASy Proteomics Server; http://www.expasy.org) and used for crystallization trials. At first, 576 crystallization screening conditions were tested using the vapour-diffusion method at 295 K with a Mosquito Crystal (TTP LabTech Ltd) automated liquid microdispensing system. Microcrystals appeared under several screening conditions. Flower-like crystals of a size suitable for diffraction were observed after 3 weeks in drops containing 0.1 M sodium acetate and 25% PEG 2000 as well as those containing 0.2 M trisodium citrate and 20% PEG 3350. Further optimization of the latter condition produced the best crystals after equilibration for 4 d at 295 K. The drops consisted of 1 µl protein solution mixed with an equal volume of reservoir solution. The optimized reservoir solutions consisted of 0.35 M trisodium citrate and 14% PEG 3350.
2.3. Data collection
Initial X-ray diffraction data from SpaA crystals were collected to a resolution of 2.6 Å on laboratory X-ray equipment comprising a Xenocs GeniX3D Cu HF (High Flux) microbeam X-ray generator operating at 50 kV and 0.6 mA equipped with a MAR 345 image-plate detector (MAR Research). Higher resolution data were collected to 2.0 Å on the BM14 beamline at the ESRF, Grenoble, France. The synchrotron data were collected from a single crystal soaked in 20%(v/v) PEG 400 cryoprotectant solution using a MAR 225 CCD area detector (MAR Research) over a range of 360° with 0.25° oscillation steps, 182 mm crystal-to-detector distance and 10 s exposure. X-ray diffraction data were processed using the HKL-2000 suite of programs (Otwinowski & Minor, 1997 ▶).
2.4. Mass spectrometry analysis
The bands were excised from an SDS–PAGE gel and exposed to repeated dehydration and rehydration cycles, with a 2:1 mixture of acetonitrile (ACN) and 50 mM ammonium bicarbonate (ABC) for 5 min and then with 25 mM ABC for 2 min. The gel pieces were then incubated overnight with trypsin (20 µg ml−1 in 25 mM ABC) at 310 K. The trypsinized sample was analyzed using an ABSCIEX Triple TOF (time-of-flight) 5600 mass spectrometer system equipped with an ESI (electrospray ionization) source. Protein identification was achieved by the Mascot search engine (Matrix Science) using the NCBI nonredundant protein databases.
2.5. Dynamic light scattering
Polydispersity and the oligomeric state of purified SpaA were analyzed by dynamic light scattering (DLS) using a Zetasizer Nano ZS90 (Malvern Instruments Ltd) equipped with a 633 nm laser and a temperature-controlled measuring chamber. SpaA protein (1 mg ml−1) in 20 mM HEPES pH 7.0, 150 mM NaCl, 1 mM EDTA was used in DLS measurements. Before measuring the scattering, the protein solution was passed through a 0.2 µm filter. DLS measurements were performed using polystyrene cuvettes at 298 K and the experimental data obtained were analyzed using the Zetasizer software (Malvern Instruments Ltd).
3. Results and discussion
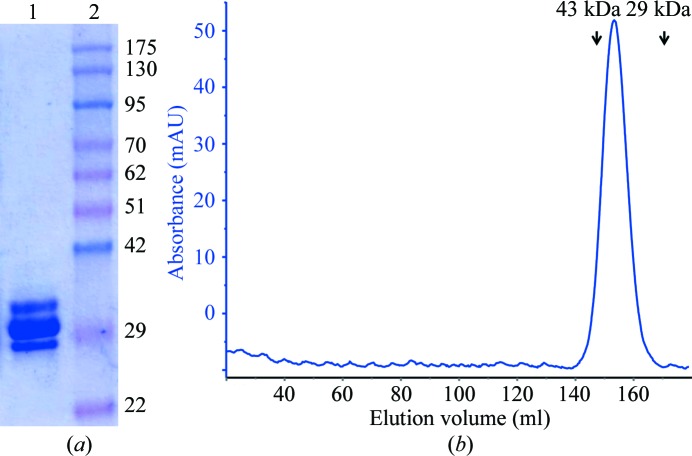
SpaA protein was purified in a highly soluble form by Ni2+-chelating affinity and gel-filtration chromatography. The recombinant SpaA appeared to migrate as three bands near a molecular weight of 30 kDa (Fig. 1 ▶ a). We could observe only a single protein peak in the gel-filtration profile, corresponding to the molecular weight of monomeric SpaA protein (∼32 kDa), even for a small-volume sample (100 µl; Fig. 1 ▶ b). This is also consistent with the results of DLS experiments, which showed that the purified SpaA is monodisperse with an average particle size equivalent to its monomeric molecular weight. Nonetheless, when trypsin-digested proteins extracted from the SDS–PAGE gel were analyzed by ESI Triple TOF MS, each of the three protein bands was identifiable as L. rhamnosus GG SpaA pilin (Supplementary Table S11). The reason for the aberrant electrophoretic migration pattern of purified SpaA is not clear. It could be related to intramolecular isopeptide bonds that stabilize the pilin structures (Kang et al., 2007 ▶). Such an interpretation was given for a similar feature in an earlier report (von Ossowski et al., 2010 ▶).
Figure 1.
Purification of recombinant L. rhamnosus GG SpaA. (a) Coomassie Blue-stained recombinant L. rhamnosus GG SpaA protein on a 12% SDS–PAGE gel. Lane 1, purified recombinant SpaA from gel-filtration chromatography; lane 2, molecular-weight markers (labelled in kDa). (b) Sephacryl S-200 (26/60) column (GE Healthcare) size-exclusion profile of SpaA. The protein peak elutes at a volume of 154 ml, which is close to the calculated weight of the monomeric protein. The positions of eluted peaks corresponding to the standards (ovalbumin, 43 kDa; carbonic anhydrase, 29 kDa) are marked with arrows.
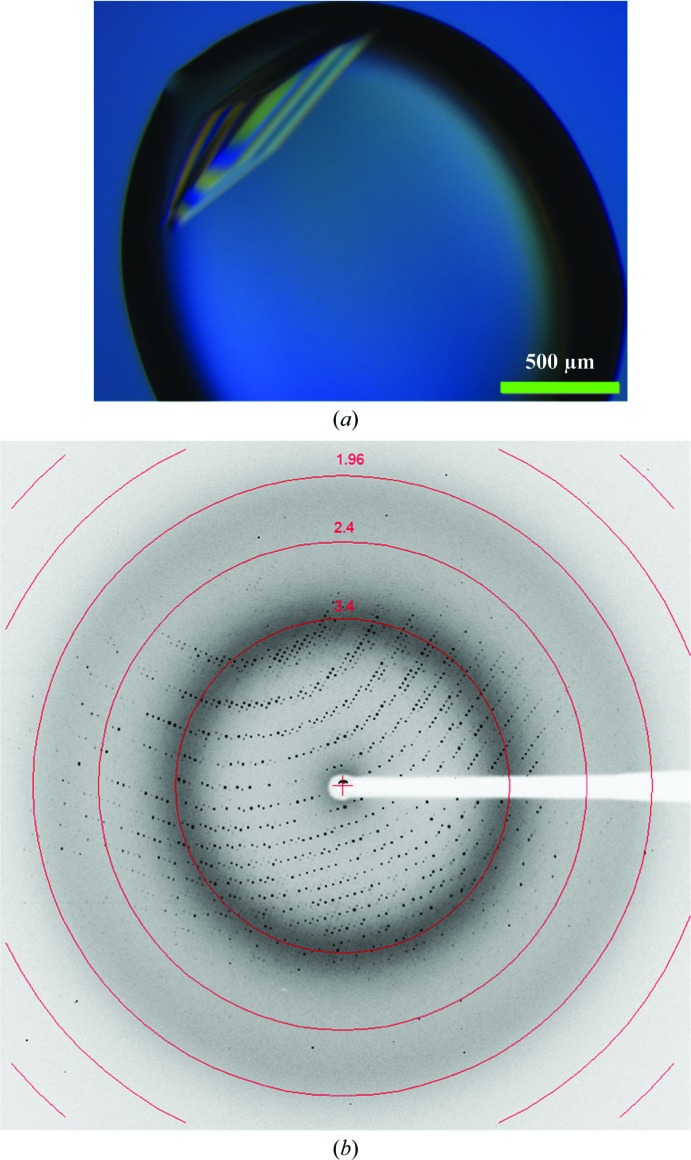
When screened for crystal growth, purified recombinant SpaA protein could be crystallized at room temperature by hanging-drop vapour diffusion under several conditions. Further optimization of one of these conditions to 0.35 M trisodium citrate, 14% PEG 3350 led to the best crystals (Fig. 2 ▶ a), which diffracted to 2.6 Å resolution on a home X-ray source and 2.0 Å resolution at the ESRF (Fig. 2 ▶ b).
Figure 2.
X-ray diffraction of an SpaA crystal. (a) Crystal of SpaA grown in 0.35 M trisodium citrate, 14% PEG 3350 using the hanging-drop vapour-diffusion method. (b) Diffraction image of an SpaA crystal collected on BM14 beamline at the ESRF.
SpaA crystals belonged to the monoclinic space group C2, with unit-cell parameters a = 227.9, b = 63.2, c = 104.3 Å, β = 95.1° (Table 1 ▶). Calculation of the Matthews coefficient (V M = 2.44 Å3 Da−1) (Matthews, 1968 ▶) suggests the presence of five molecules in the asymmetric unit, which corresponds to a solvent content of 50%. Since SpaA has a limited sequence identity of less than 25% to known pilin structures in the Protein Data Bank (PDB), our initial attempts to obtain phases by molecular replacement were not successful. Moreover, as methionine or cysteine residues are not present in SpaA, phase calculation by selenium or sulfur single-wavelength anomalous dispersion (SAD) was not feasible. We are instead currently attempting to solve the phasing problem through the use of heavy-atom derivatization.
Table 1. Data-collection and processing statistics for SpaA.
Values in parentheses are for the outermost resolution shell.
| No. of crystals | 1 |
| Beamline | BM14, ESRF |
| Wavelength (Å) | 0.97625 |
| Detector | MAR CCD 225 |
| Crystal-to-detector distance (mm) | 182 |
| Rotation range per image (°) | 0.25 |
| Total rotation range (°) | 360 |
| Exposure time per image (s) | 10 |
| Resolution range (Å) | 40–2.0 (2.03–2.00) |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 227.9, b = 63.2, c = 104.3, β = 95.1 |
| Total No. of measured intensities | 642704 |
| Unique reflections | 94497 |
| Multiplicity | 6.8 (4.5) |
| Mean I/σ(I) | 34.2 (3.5) |
| Completeness (%) | 95.2 (75.1) |
| R merge (%) | 8.7 (38.1) |
Supplementary Material
Supplementary material file. DOI: 10.1107/S1744309113024676/ft5038sup1.pdf
Acknowledgments
This investigation was supported by the Regional Centre for Biotechnology (RCB) and the Department of Biotechnology (DBT). VK and DS thank Professor Dinakar Salunke for reviews and support. VK and DS acknowledge Dr Hassan Belrhali and Dr Babu Manjashetty at the ESRF BM14 beamline, Madhava Rao at the in-house X-ray facility and Vishakha Choudhary at the Mass Spectrometry facility for their help during data collection. Recombinant cloning performed at the University of Helsinki was financed by an Academy of Finland general research grant (118165) and the Center of Excellence in Microbial Food Safety (CoE-MiFoSa) research program (141140), which was initiated and funded as part of the Research Program on Nutrition, Foods and Health (ELVIRA) by the Academy of Finland.
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: FT5038).
References
- Hendrickx, A. P., Budzik, J. M., Oh, S.-Y. & Schneewind, O. (2011). Nature Rev. Microbiol. 9, 166–176. [DOI] [PubMed]
- Kang, H. J. & Baker, E. N. (2012). Curr. Opin. Struct. Biol. 22, 200–207. [DOI] [PubMed]
- Kang, H. J., Coulibaly, F., Clow, F., Proft, T. & Baker, E. N. (2007). Science, 318, 1625–1628. [DOI] [PubMed]
- Kankainen, M. et al. (2009). Proc. Natl Acad. Sci. USA, 106, 17193–17198.
- Kline, K. A., Dodson, K. W., Caparon, M. G. & Hultgren, S. J. (2010). Trends Microbiol. 18, 224–232. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Ossowski, I. von, Reunanen, J., Satokari, R., Vesterlund, S., Kankainen, M., Huhtinen, H., Tynkkynen, S., Salminen, S., de Vos, W. M. & Palva, A. (2010). Appl. Environ. Microbiol. 76, 2049–2057. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Reunanen, J., von Ossowski, I., Hendrickx, A. P., Palva, A. & de Vos, W. M. (2012). Appl. Environ. Microbiol. 78, 2337–2344. [DOI] [PMC free article] [PubMed]
- Ton-That, H., Marraffini, L. A. & Schneewind, O. (2004). Mol. Microbiol. 53, 251–261. [DOI] [PubMed]
- Tripathi, P., Beaussart, A., Alsteens, D., Dupres, V., Claes, I., von Ossowski, I., de Vos, W. M., Palva, A., Lebeer, S., Vanderleyden, J. & Dufrêne, Y. F. (2013). ACS Nano, 7, 3685–3697. [DOI] [PubMed]
- Vengadesan, K. & Narayana, S. V. (2011). Protein Sci. 20, 759–772. [DOI] [PMC free article] [PubMed]
- Waksman, G. & Hultgren, S. J. (2009). Nature Rev. Microbiol. 7, 765–774. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material file. DOI: 10.1107/S1744309113024676/ft5038sup1.pdf