Abstract
Cohesion establishment and maintenance are carried out by proteins that modify the activity of Cohesin, an essential complex that holds sister chromatids together. Constituents of the replication fork, such as the DNA polymerase α-binding protein Ctf4, contribute to cohesion in ways that are poorly understood. To identify additional cohesion components, we analyzed a ctf4Δ synthetic lethal screen performed on microarrays. We focused on a subset of ctf4Δ-interacting genes with genetic instability of their own. Our analyses revealed that 17 previously studied genes are also necessary for the maintenance of robust association of sisters in metaphase. Among these were subunits of the MRX complex, which forms a molecular structure similar to Cohesin. Further investigation indicated that the MRX complex did not contribute to metaphase cohesion independent of Cohesin, although an additional role may be contributed by XRS2. In general, results from the screen indicated a sister chromatid cohesion role for a specific subset of genes that function in DNA replication and repair. This subset is particularly enriched for genes that support the S-phase checkpoint. We suggest that these genes promote and protect a chromatin environment conducive to robust cohesion.
INTRODUCTION
In budding yeast, Cohesin is a four subunit protein complex (Mcd1/Scc1, Scc3, Smc1, and Smc3) that depends on the activity of regulatory proteins for its chromosome association, activation, and destruction in each cell cycle (reviewed in Nasmyth, 2001). Sister chromatid association by Cohesin must be established during S phase (Skibbens et al., 1999; Toth et al., 1999) and is maintained until separation of sister chromatids at anaphase. Many studies indicate a role for the replication fork in establishment of robust sister chromatid cohesion, in addition to its traditional role of semiconservative DNA duplication. Several replication-associated proteins from budding yeast are known to support robust cohesion, including the products of nonessential genes CTF4, CTF8, CTF18, DCC1, TRF4, and TRF5 (Wang et al., 2000b; Hanna et al., 2001; Mayer et al., 2001). Null mutants for these genes lead to metaphase cohesion failure at frequencies of ∼25–35% in cells that are held at metaphase in the absence of microtubules.
These nonessential replication fork constituents represent several subcomplexes at the replication fork and contribute independent primary molecular functions that are not well understood. Ctf4 protein forms an association with DNA polymerase α (Formosa and Nittis, 1999) that may compete with binding of a chromatin remodeling subunit Cdc68/Pob3 (Wittmeyer and Formosa, 1997). Ctf18 protein is a component of an alternative RF-C complex in which it replaces Rfc1, and is joined by Ctf8 and Dcc1 subunits (Hanna et al., 2001; Mayer et al., 2001). The orthologous human RF-CCTF18 complex can load PCNA onto DNA and promote Polδ activity in vitro (Bermudez et al., 2003; Kanellis et al., 2003; Merkle et al., 2003). The nonessential proteins Trf4 and Trf5 together comprise an essential activity originally referred to as DNA polymerase σ (Wang et al., 2000b) (see also Read et al., 2002; Saitoh et al., 2002) and cooperate in a step required for S-phase progression as well as proper sister chromatid cohesion. In addition to these nonessential genes, partial loss of function for the essential budding yeast polymerase ε(POL2) causes a cohesion defect (Edwards et al., 2003). Furthermore, PCNA overexpression suppresses a conditional allele of the essential cohesion establishment factor Ctf7p, which encodes a histone acetyltransferase activity (Skibbens et al., 1999; Toth et al., 1999; Ivanov et al., 2002). Together, these studies provide strong support for the conclusion that the replication fork itself includes constituents important for conditions supporting robust sister chromatid cohesion.
High-fidelity DNA replication is ensured by the S-phase checkpoint, which slows the progress of DNA replication forks in the presence of limiting nucleotide substrate, prevents collapse of stalled replication forks, and coordinates the activity of DNA repair pathways within S phase to limit the effect of DNA lesions (Donaldson and Blow, 2001; Myung et al., 2001; D'Amours and Jackson, 2002; Melo and Toczyski, 2002; Nyberg et al., 2002). Most studies have focused on the role of this checkpoint in ensuring the linear structural integrity and correct nucleotide sequence of chromosomal DNA. However, recent work has also provided views of chromatin alteration seen at damage sites during S-phase checkpoint activation by cytological or by biochemical criteria. For example, the phosphorylation of histone H2AX at sites of DNA damage recruits repair proteins to cytologically visible “foci” in vertebrate cells (Celeste et al., 2002; Kobayashi et al., 2002; Goldberg et al., 2003; Stewart et al., 2003). Damage-induced foci are also visible in budding yeast (Lisby et al., 2001; Melo and Toczyski, 2002). Interestingly, foci containing some of the same repair proteins are detectable during S phase in human cells that have not been subjected to DNA-damaging agents (Costanzo et al., 2001; Maser et al., 2001; Mirzoeva and Petrini, 2003), suggesting that these repair proteins are used during S phase in unchallenged cell cycles.
Biochemical connection between the Cohesin subunit SMC1, S-phase checkpoint activity, and DNA repair has been observed. In human cells, after DNA damage the Cohesin subunit Smc1 is recruited to repair foci (Kim et al., 2002a). Smc1 is also phosphorylated by the ATM checkpoint kinase (Kim et al., 2002b; Yazdi et al., 2002). Optimal Smc1 phosphorylation requires Nbs1 (Kim et al., 2002b), a constituent of MRN (Mre11, Rad50, and Nbs1). MRN is a well-studied protein complex required for double-strand break repair and is conserved across Eukaryotae (reviewed in D'Amours and Jackson, 2002; Wyman and Kanaar, 2002; Bradbury and Jackson, 2003). In human cells, subunits of MRN are found in S phase and damage-induced foci (Wang et al., 2000a; Kim et al., 2002b; Nakanishi et al., 2002; Tauchi et al., 2002; Yazdi et al., 2002; Zhao et al., 2002; Mirzoeva and Petrini, 2003). A highly conserved complex found in budding yeast is referred to as MRX (XRS2 is the Nbs1 homologue; reviewed in D'Amours and Jackson, 2002). The Mre11–Rad50 complex exhibits structural similarity to Cohesin (Jessberger, 2002; Wyman and Kanaar, 2002; Milutinovich and Koshland, 2003). These observations have led recently to a speculative model in which Mre11–Rad50 complex might directly support double-strand break repair by promoting sister chromatid association (Wyman and Kanaar, 2002). In this model, the double-strand break ends would be held in proximity to one another, as well as to a sister chromatid, promoting proximity to a homologous recombination target (Wyman and Kanaar, 2002). However, to date there has been no direct in vivo evidence for sister chromatid association provided by the MRX complex.
To identify additional genes with roles in sister chromatid cohesion, we performed a synthetic lethal screen using ctf4Δ. We took advantage of a recently developed approach (Ooi et al., 2003; Pan, Yuan, Xiang, Cheng, Wang, Sookhai-Mahadeo, Boone, Hieter, Spencer, and Boeke, unpublished data) to identify double mutant combinations with reduced viability by using a microarray hybridization assay. This approach is made possible by the collection of knockout strains containing oligonucleotide tags unique to each deletion (Giaever et al., 2002). Confirmed ctf4Δ interactions from the microarray data increased the number of synthetic lethal combinations known from four to 26 nonessential pairs. In our analysis of this list of ctf4Δ-interacting mutations, we focused on mutants that exhibited genetic instability themselves. In total, we identified 17 new genes contributing to cohesion, all of which had been previously studied for other phenotypes. Fifteen of the 17 genes function in DNA replication and repair. One of these was XRS2, a constituent of MRX. Specific analysis of the role of MRX confirmed a contribution to metaphase sister chromatid cohesion, which seems to modulate Cohesin function, rather than act through an independent additive pathway. The list of 17 newly identified cohesion genes exhibits enrichment for roles in the S-phase checkpoint. We propose that the S-phase checkpoint contributes to robust sister chromatid cohesion, as well as ensuring the linear integrity of chromosomal DNA and high-fidelity replication of nucleotide sequence.
MATERIALS AND METHODS
Synthetic Lethality Screen
Sample Generation. A pool of 5916 heterozygous diploid knockout strains obtained in arrayed form from Research Genetics was modified by introduction of a yeast haploid selection marker can1Δ::MFA1pr-HIS3 (similar to Tong et al., 2001). This modified pool (gift of X. Pan, Baltimore, MD) was transformed with a query polymerase chain reaction (PCR) construct containing a ctf4Δ::natMX drug resistance cassette with 2-kb flank on either side. Then, 5 × 105 ctf4Δ::natMX ykoΔ::kanMX double mutants were selected on YPD + 200 μg/ml G418 (Cellgro, Herndon, VA) and 100 μg/ml clonNAT (Hans-Knöll Institute für Naturstoff-Forschung, Jena, Germany), scraped together, and a 2.5 × 108 cell aliquot was grown in liquid sporulation medium for 7 d. The sporulated culture was plated on haploid selection media (SC-HIS–ARG + 50 μg/ml canavanine [Sigma-Aldrich, St. Louis, MO]) + G418 + NAT) to select for ∼106 MATa ctf4Δ::natMX ykoΔ::kanMX double mutants. In all defined (SC) media where G418 or clonNAT selections were applied, ammonium sulfate was replaced by monosodium glutamic acid (1 g/l) as in Tong et al., 2001). Transformation of a query construct containing ura3Δ::natMX was performed in parallel to serve as a control. Control and experimental transformations were performed in duplicate. These experiments were performed early during optimization of this method. Its optimization and evaluation are described in depth elsewhere (Ooi et al., 2003; Pan, Yuan, Xiang, Cheng, Wang, Sookhai-Mahdeo, Boone, Hieter, Spencer, and Boeke, unpublished data).
Tag Microarray Hybridization. Genomic DNA was prepared from 2 × 108 haploid double mutants and 0.2 μg was used as template for PCR amplification of the UPTAGs or DOWNTAGs in separate reactions. PCR was performed using biotinylated primers as described previously (Giaever et al., 2002). The resulting labeled UPTAGs or DOWNTAGs were separated from unincorporated primers on Microcon YM-10 columns, combined, and hybridized to Affymetrix Tag3 arrays as described previously (Giaever et al., 2002), except that four blocking primers were used. Washing, staining, and scanning were performed as described previously (Winzeler et al., 1999).
Signal Ratio Determination. Signal intensity values from the Affymetrix.cel files were imported into R (www.r-project.org). By using the BioConductor (www.BioConductor.org) Affy package, data from the two experimental and two control chips were transformed (log2) and quantile normalized. Hybridization signals of probes from complementary strands were correlated, but more weakly than expected (0.80 < r < 0.85). Strand asymmetry has also been noted by Cutler et al. (2001). Perfect match (PM) or complementary perfect match (CPM) data were chosen for use based on population signal intensity histograms exhibiting strongest separation between nonessential (absent) and essential (present) tag distributions: the CPM value for UPTAGs and PM value for DOWNTAGs were used. Four pairwise ratios were generated (URA3A:CTF4A, URA3A:CTF4B, URA3B:CTF4A, and URA3B:CTF4B). These were averaged to provide a single UPTAG and single DOWNTAG ratio for each knockout. Oligonucleotide tag sequences present in yeast sometimes differ from their design sequences, presumably due to synthesis errors. Some tags will therefore exhibit poor hybridization, but most often only the UPTAG or DOWTAG of any given construct is affected. To filter out the effect of these tag mutations, the larger experiment:control tag ratio was chosen to represent a single ratio for each knockout. Then, 124 tags (2% of 6129 total) exhibiting high variation on the URA3 chips were filtered out. The remaining data were sorted by ratio, and a gene list containing 45 tag-marked deletion strains with ratios ≥21.1 was identified (see Supplemental Table 1). In replicates of the control experiment, ratios above 21.1 were rarely observed (27 genes, or 0.5% of the data), and this cutoff was chosen to limit false positive observations for this study.
Gene List Adjustment. Eight open reading frame deletions from the list of 45 were disqualified from further analysis for reasons listed below. Thus, the total number of genes identified by the array data is stated as 37. YPR133W-a and YPR134W are immediately adjacent to CTF4 (YPR135W), within the extent of the left homology flank present on the transforming DNA fragment. Their tag underrepresentation in the experimental sample is due to tag loss during transformation with the ctf4Δ query construct. The adjacent gene to the right (RRP9) is essential. YDR410C (STE14) was not present in the MATa collection (Research Genetics) from which haploids were picked for further confirmation. YER014C-A (BUD25) was present but did not grow on attempted recovery. YBR140C (IRA1) and YDR232W (HEM1) were false positive essential genes and were dropped. Two examples of deletion mutations affecting a single gene product were encountered: ypl017cΔ overlaps ctf19Δ, and ycl060cΔ plus ycl061cΔ together comprise the MRC1 gene deletion (Saccharomyces Genome Database at www.yeastgenome.org). In each case, the related pairs exhibited indistinguishable interaction, a-like faker, and cohesion phenotypes, and were therefore assumed to be identical for the purposes of this study.
Random Spore Analysis
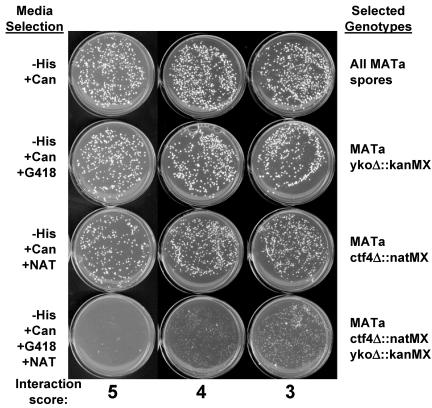
To individually assay the growth of double mutants, strains YJH96 (MATα ctf4::natMX can1 mfa1::MFA1pr-HIS3) and YJH97 (MATα ctf18::natMX can1 mfa1::MFA1pr-HIS3) were mated to candidate MATa deletion mutants, and diploids were selected on YPD+G418+clonNAT. The diploids were transferred to sporulation medium and incubated for 5 d at 25°C. A swatch of sporulated yeast was resuspended in 500 μl of sterile distilled H2O (dH2O), and 10, 20, 20, and 40 μl were plated on haploid selection media with drug additions to select the four different classes of haploids shown in Figure 1. Drugs added to haploid selection media (SC-HIS-ARG + 50 mg/ml Canavanine [selects all MATa haploids]) were 200 mg/ml G418 (selects MATa yko::kanMX single mutants), 100 mg/ml clonNAT (selects MATa ctf4 or ctf18 single mutants), or G418+clonNat (selects MATa double mutants). Synthetic interactions were scored by comparing colony growth of the double mutants to the single mutants.
Figure 1.
Confirmation of synthetic lethal or synthetic fitness interactions. Each genetic interaction predicted by the microarray experiment was independently tested using random spores derived from crosses containing the MATa-specific marker MFA1pr-HIS3 and can1R to select haploids as described previously (Tong et al., 2001), except that all manipulations were performed manually. Each mutant was given a synthetic interaction score based on the growth on the double mutant selection plate after 42 h at 30°C, taking into account a comparison with the single mutant control plates. 5, synthetic lethal interaction; 4, 3, decreasing synthetic fitness interactions; 2, 1, little or no interaction. In the examples shown, ctf4Δ::natMX is the query allele, and the interacting genes are mrc1Δ::kanMX (5), mcm21Δ::kanMX (4), and chl4Δ::kanMX (3).
Genetic Instability: the a-Like Faker Assay
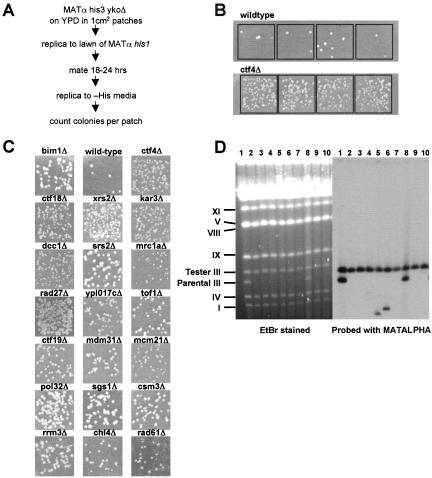
On each master plate, three controls [wild-type MATα (BY4742), wild-type MATa (BY4741), and MATα bim1::kanMX] and 12 independent MATα deletion colonies were patched in 1-cm2 squares on YPD and incubated overnight at 30°C. One milliliter containing 5 × 107 cells of mating tester YFS475 (MATα his1) was evenly spread and dried on fresh YPD plates. Patches were transferred onto this mating tester lawn by replica plating, followed by incubation at 30°C overnight (18–24 h). The mated lawn was then replica plated to SC-6 (synthetic complete medium lacking uracil, lysine, adenine, histidine, tryptophan, and leucine) and incubated for 2 d at 30°C to select His+ products. The results were scored by comparing the number of colonies per patch to the wild-type MATα control patch for that plate. Strains that showed an increase in the production of mated products were retested as described above for four independent single colonies of each mutant, except that strain YFS773 (MATα his5) was used as the mating tester. To quantitate the phenotype for a given mutant, a calculated median of the number of colonies in the four patches was divided by the median number of colonies for wild-type from all control patches (wild-type n = 150, median = 4.5 colonies per patch).
Pulsed Field Electrophoresis and In-Gel Hybridization
Chromosome-sized DNA was prepared in agarose plugs (Green et al., 1998). Gels (1%) were run in 0.5× Tris borate-EDTA at 200 V at 14°C for 30 h with continuous ramp switch times of 24–54 s to separate the smaller yeast chromosomes. Gels were stained in 0.5 μg/ml ethidium bromide in dH2O for 1 h, washed in dH2O for 1.5 h, and then photographed. Gels were denatured in 1.5 M NaCl, 0.5 M NaOH for 1.5 h, neutralized in 1.5 M NaCl, 1 M Tris, pH 7.4, for 45 min, washed in dH2O for 30 min, and dried on a vacuum gel-dryer for 1 h at 55°C. Gels were prehybridized in 20 ml of hybridization buffer (4× SSC, 20 mM NaH2PO4, pH 7.4, 0.1% SDS, 5× Denhardt's) at 57°C for at least 30 min. Gels were hybridized with a random hexamer-labeled DNA fragment corresponding to MATALPHA1 gene sequence in 15 ml of hybridization buffer at 57°C overnight. Gels were washed twice for 1 h in 3.5× SSC at 58.5°C, twice for 30 min in 3× SSC, 0.1% SDS at 58.5°C and exposed to Kodak MS film at –70°C overnight. Because of high sequence similarity, the MAT-ALPHA1 probe also hybridizes HMLALPHA and HMRA on the left and right arms of chromosome III, respectively.
Cohesion Assays
ykoΔ::kanMX alleles were amplified by PCR and introduced into strains PS1337 (MATa leu2::LEU2.tetR-GFP BMH1::URA3-TetO array; He et al., 2000), YBL86-74C (MATa leu2::LEU2.tetR-GFP ura3-52::URA3.tetO224 mcd1-1; gift of D. Koshland, Baltimore, MD), or YBS1045 (MATa ade2 trp1 his3 leu2::[LEU2tetR-GFP] ura3::[3 × URA3tetO112] PDS1-13MYC::TRP1; Kenna and Skibbens, 2003) by transformation. All mutants were verified by PCR assay across the novel junction formed by introduction of the drug resistance cassette. To test cohesion fidelity, logarithmically growing cells were synchronized in G1 with alpha factor (5 μg/ml for 2.5 h) or in G2/M with nocodazole (15 μg/ml for 3 h). After fixation in 3.7% formaldehyde, cells were diluted in 1 M sorbitol, 50 mM potassium phosphate buffer, stained with 4,6-diamidino-2-phenylindole (100 ng/ml), resuspended in Fluorsave (Calbiochem, San Diego, CA), and stored at 4°C.
Cells mutant for cohesin function (mcd1-1) were grown in log phase, arrested in nocodazole, and then grown at permissive (25°C) or nonpermissive (37°C) temperature for 3 h before fixation. Cells containing PDS1-13MYC were processed for immunofluorescence to specifically detect preanaphase nuclei by virtue of their high level of PDS1 protein. Fixed cells were mounted on slides pretreated with 10 mg/ml poly-d-lysine, spheroplasted, and permeabilized by a brief incubation in 0.1% Triton X-100 in phosphate-buffered saline (PBS)/1% bovine serum albumin (BSA). The cells were incubated in 1:90 dilution of 9E10 mouse anti-myc antibody (Covance, Princeton, NJ) for 1 h, washed three times in PBS/1% BSA, incubated in 1:800 dilution of goat anti-mouse secondary antibody conjugated to Alexa 594 (Molecular Probes, Eugene, OR) for 1 h, and washed three times in PBS/BSA. Samples were then washed once in 4,6-diamidino-2-phenylindole (100 ng/ml) in PBS/BSA, resuspended in Fluorsave, and stored at 4°C until scoring.
The frequency of cells containing two green fluorescent protein (GFP) spots was determined for three independent transformants per genotype, scoring a minimum of 100 cells per culture. Failure of spindle checkpoint arrest leads to accumulation of an “extrabudded” phenotype indicative of inappropriate entry into the next cell cycle. In all mutants, extrabudded cells (defined as in Warren et al., 2002) occurred at frequencies similar to wild type and were excluded from scoring. For mutants with high a-like faker rates, G1 arrested cells (αfactor) were scored for the frequency of two spots to detect aneuploidy in the cultures. This was <3% in all tested cases.
RESULTS
Microarray-based Synthetic Lethality Screen
Cells lacking CTF4 or CTF18 are able to form colonies but sustain a high level of chromosome loss and exhibit decreased sister chromatid association (Kouprina et al., 1992; Miles and Formosa, 1992; Hanna et al., 2001). ctf4Δ ctf18Δ double mutant cells exhibit synthetic lethality (Miles and Formosa, 1992; Formosa and Nittis, 1999). To identify additional nonessential genes involved in cohesion, we performed a microarray-based synthetic lethal screen. This strategy follows the relative representation of deletion mutation “barcode” tags (as in Giaever et al., 2002) present in a pool of 5916 ykoΔ::kanMX mutants. ctf4Δ::natMX ykoΔ::kanMX double mutants were created en masse by transformation of the pool with a ctf4Δ::natMX DNA fragment to replace CTF4. As a control, replacement of ura3Δ0 with ura3Δ::natMX was performed in parallel. The relative representation of each ykoΔ::kanMX mutant in combination with ura3Δ::natMX or ctf4Δ::natMX was represented as a signal intensity ratio (control:experiment). Analysis of these ratios generated a list of 37 ykoΔ mutations that were underrepresented in combination with a ctf4Δ mutation (see MATERIALS AND METHODS and Supplemental Table 1). To test the microarray-predicted interactions, we individually mated ykoΔ::kanMX and ctf4Δ::natMX mutants to create double deletants. The presence of the haploid selection markers MFA1pr-HIS3 and can1R permitted selection of MATa haploids from each cross by random spore selection (as in Tong et al., 2001). Growth defects were identified by comparison of colony size in single and double mutants (Figure 1). This test confirmed poor or absent growth of ctf4Δ ykoΔ double mutants for 26 of the 37 genes (Table 1, ctf4 interaction and ctf4 SL score columns).
Table 1.
Candidate list for new cohesion mutants
| Gene | Annotation | ura3:ctf4 ratio (log2) | ctf4 score | ctf18 score | A-like faker |
|---|---|---|---|---|---|
| XRS2 | Double strand break repair | 1.2 | 5 | 4 | 48 |
| KAR3 | Microtubule motor, mitosis | 1.6 | 5 | 5 | 40 |
| DCC1 | Sister chromatid cohesion | 2.8 | 5 | 1 | 18 |
| SRS2 | DNA repair, helicase | 1.7 | 5 | 5 | 12 |
| MRC1 | DNA replication checkpoint | 1.7 | 5 | 4 | 11 |
| CLB2 | G2/M transition of cell cycle | 2.1 | 5 | 5 | |
| RRM3 | DNA replication, helicase | 1.8 | 4 | 3 | 11 |
| TOF1 | DNA topological change | 1.2 | 4 | 5 | 10 |
| CTF19 | Kinetochore protein | 1.6 | 4 | 2 | 8 |
| MDM31 | Mitochondrion organization | 1.2 | 4 | 3 | 6 |
| MCM21 | Kinetochore protein | 2.1 | 4 | 2 | 5 |
| SWM1 | APC/cyclosome | 1.8 | 4 | 3 | |
| RMD7 | Meiotic nuclear division | 2.4 | 4 | 3 | |
| MDM39 | Mitochondrial distribution | 1.4 | 4 | 2 | |
| VPS25 | Protein-vacuolar targeting | 1.1 | 4 | 4 | |
| POL32 | Nucleotide-excision repair | 1.2 | 3 | 3 | 26 |
| SGS1 | DNA helicase | 1.2 | 3 | 3 | 14 |
| CSM3 | Chromosome segregation | 1.7 | 3 | 4 | 12 |
| LTE1 | Cell cycle | 1.7 | 3 | 3 | |
| RAD61 | Radiation sensitivity | 1.3 | 3 | 3 | 4 |
| CHL4 | Chromosome segregation | 1.2 | 3 | 1 | 3 |
| RIM8 | Meiotic regulation | 1.2 | 3 | 3 | |
| MAD2 | Mitotic spindle checkpoint | 1.4 | 3 | 4 | |
| ELM1 | Protein kinase/cell morphology | 1.2 | 3 | 3 | |
| BUB2 | Mitotic exit network | 1.6 | 3 | 3 | |
| MID1 | Calcium ion transport | 1.3 | 3 | 1 |
The gene list that resulted from individual confirmation by random spore analysis is shown. Annotations were abbreviated from information in the Saccharomyces Genome Database (www.yeastgenome.org) or the Yeast Proteome Database (www.incyte.com). Signal intensity ratios from the microarray data (ura3:ctf4 ratio) and ctf4Δ ykoΔ synthetic interaction score (ctf4 score) are listed. Each ctf4Δ-interacting gene was tested in combination with ctf18Δ, and synthetic interaction scores are given (ctf18 score). Finally, the ratio increase of a-like faker colonies (mutant:wild-type) was determined for null mutants of all ctf4Δ interacting genes. a-like faker frequency increase for each mutant exhibiting reproducible values ≥2 are shown. All other mutants were indistinguishing from wild-type.
This analysis was performed on a top segment of the gene list ranked by signal intensity ratio and ended at a ratio chosen to limit false positives (see MATERIALS AND METHODS). It is therefore not expected to be complete. In fact, the 26 genes identified did not include four published nonessential genes (ctf18Δ, rad27Δ, mec1-1, rad52Δ; Kouprina et al., 1992; Miles and Formosa, 1992; Formosa and Nittis, 1999). In contrast, the gene with highest signal intensity ratio was DCC1 (Table 1 and Supplemental Table 1), a subunit of the RFCCTF18 protein complex, which represents a predicted synthetic lethal interaction. For the analyzed segment of the gene list (top 37), the array signal intensity ratio and the synthetic lethality score correlated significantly (Spearman correlation = 0.382, p = 0.0165). Further work to identify and control sources of variation in the method will improve the correlation and the predictive value of signal intensity ratios.
The combined molecular activities of CTF4 and CTF18 are essential for cell viability, suggesting that their distinct activities provide a similar function in parallel. To ask whether the ctf4Δ interactions would be shared, we assayed growth phenotypes of all 26 genes in a ctf18Δ background. Twenty-three of 26 genes that showed a ctf4Δ interaction also exhibited a synthetic lethal or fitness defect with ctf18Δ (Table 1, ctf18 SL score), reflecting functional overlap between Ctf4 and Ctf18 protein complexes. The diverse annotations on genes in the interaction list suggest that several distinct components may be represented. Interestingly, differences in interaction strength also reveal distinctions between CTF4 and CTF18 function. For example, the presence of four mutants previously studied for roles in the kinetochore–microtubule junction (ctf19Δ, mcm21Δ, chl4Δ, and kar3Δ) may reflect a convergent pathway linking DNA replication and cohesion near the centromere. Three of these (ctf19Δ, mcm21Δ, and chl4Δ) exhibit a greater requirement for CTF4 than CTF18, suggesting that primary roles of the replication-associated genes differ near the centromere.
Secondary Screen for Genome Instability
Defects in sister chromatid cohesion cause chromosome missegregation at mitosis. Genetic instability has been tested for many of the 26 interactors with a diverse set of assays (e.g., at www.yeastgenome.org). We used a systematic method for determining the presence of genetic instability by using a convenient marker loss assay that follows the presence or absence of a marker endogenous to ykoΔ haploids (Figure 2). This assay takes advantage of a property of mating type determination in budding yeast. MATα cells that lose their MAT locus information phenocopy MATa status, the default (Strathern et al., 1981). These rare “a-like fakers” are able to mate with tester MATα cells, and their frequency can be scored by selection of prototrophic MATα/[MAT-null] diploids.
Figure 2.
a-like faker assay of ctf4Δ interacting genes. (A) Outline of the method for detecting a-like fakers. (B) Reproducibility of the recovery of a-like faker products after mating is illustrated. Four patches of independent clonal isolates from wild-type (top row) and ctf4Δ (bottom row) are shown. Colony counts from four or more patches per mutant were used to calculate a ratio indicating the increase in frequency over wild type. (C) Representative patches for each ykoΔ mutant with elevated a-like faker frequency. bim1Δ served as a positive control for marker instability. (D) Electrophoretic karyotype and ingel hybridization analysis of 10 wild-type a-like faker mated products are shown. The gel was hybridized with 32P-labeled MATα sequence. The migration positions of yeast chromosomes are indicated on the left. Whole chromosome loss was apparent in mated products containing only the tester chromosome III (lanes 2, 3, 4, 7, 9, and 10). Gross chromosomal rearrangements of chromosome III were observed as appearance of a new band (lanes 5 and 6). Where both chromosomes III are retained, MATα was lost by gene conversion (lanes 1 and 8).
To screen for genome unstable mutants, 1-cm2 patches of MATα his3Δ ykoΔ haploids were replica plated onto a MATα his1 lawn, and prototrophs were then selected by replica plating onto solid media lacking histidine (Figure 2A). Mutants generating an elevated frequency of His+ papillae were noted, and retested using four or more independent colonies. The frequency of His+ papillae was highly reproducible (Figure 2B). A ratio of a-like faker colonies recovered (ykoΔ:wild-type) was calculated as an indicator of marker instability, by using median values from four or more independent clonal lineages per tested genotype (Figure 2B). Among the 26 ctf4Δ interacting mutants, 15 exhibited an a-like faker frequency more than twofold higher than wild type (Table 1, a-like faker column; and Figure 2C).
Loss of MATα can occur by local deletion, gene conversion from the silent mating type loci, or whole chromosome loss (Strathern et al., 1981). To distinguish between these possibilities, 40 independent a-like faker mated products from the wild-type parental strain were analyzed by pulsed field gel analysis. A MATα his5 tester strain containing a chromosome III of distinct size was used to distinguish among the three possibilities. Chromosome III bands were detected using a radiolabeled probe derived from the MAT locus. This probe will hybridize at three dispersed chromosome III positions (HML, MAT, and HMR) and should detect virtually all rearranged forms of the chromosome. Figure 2D shows separated chromosomes from 10 His+ papillae as examples. Among 40 independent colonies analyzed, whole chromosome loss was the predominant mode observed comprising 68% (27/40) of total. Rearranged chromosome III was observed in 20% (8/40). No rearrangement or loss was observed in 12% (5) due to conversion of the MAT locus from alpha to a (our unpublished data).
Identification of Sister Chromatid Cohesion Defects
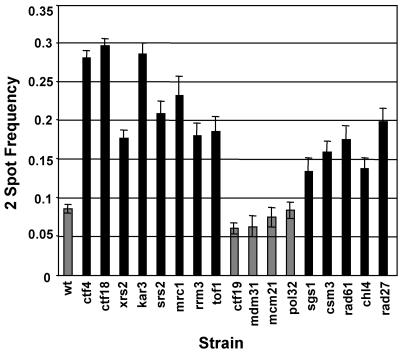
The 15 ctf4Δ interacting genes that exhibited genome instability were evaluated for sister chromatid cohesion. The DCC1 gene encoded a subunit of RFCCTF18 and was already known to contribute to sister chromatid cohesion (Mayer et al., 2001; our unpublished data). The 14 additional ctf4Δ interacting mutants were characterized using a cytological assay for sister chromatid association. A tandem array of tetracycline repressor binding sites on distal chromosome V was detected with a repressor-GFP fusion protein (He et al., 2000), and the frequency of cells containing separated spots was determined. More than 90% of wild-type large-budded cells contained single GFP spots, indicative of tight association between sisters. Ten strains (xrs2Δ, kar3Δ, srs2Δ, mrc1Δ, rrm3Δ, tof1Δ, sgs1Δ, csm3Δ, rad61Δ, and chl4Δ) exhibited a significant defect (p < 0.01; Figure 3, black bars). We also tested rad27Δ, a previously known ctf4Δ interacting gene (Formosa and Nittis, 1999), encoding a FLAP endonuclease (Paques and Haber, 1999). The rad27Δ::kanMX deletion mutant exhibited high a-like faker frequency (our unpublished data), and also a cohesion defect (Figure 3). Eight of the 11 newly defined cohesion-defective genes shown in Figure 3 are annotated for roles in DNA metabolism. These findings strongly support the view that replication- or repair-associated roles of these proteins, as well as Ctf4 and Ctf18, contribute to sister chromatid cohesion.
Figure 3.
Sister chromatid cohesion test of candidate genes. Each ykoΔ strain was tested for sister association in nocodazole-induced metaphase arrest. The frequency of separation between GFP-marked tet operator loci on sister chromatids was determined from three or more independent transformants per mutant at ≥100 cells per sample. Independent scorings were combined to generate a single two-spot proportion (cells with separated spots divided by total), and error bars indicate proportion ±SE. Black bars indicate rates greater than wild type, at significance p < 0.05 calculated as chi square for proportions under a null hypothesis for identity (Fleiss, 1981). All comparisons are pairwise, with 1 degree of freedom. p values for significant samples are ctf4Δ = 0, ctf18Δ = 0, xrs2Δ = 0, kar3Δ = 0, srs2Δ = 0, mrc1Δa = 0, mrc1Δb = 0, tof1Δ = 0, rrm3Δ = 0, sgs1Δ = 0.0016, csm3Δ = 0, rad61Δ = 0, chl4Δ = 0.0001, and rad27Δ = 0.
For KAR3 (encoding a kinesin-like protein; Meluh and Rose, 1990) and CHL4 (encoding a kinetochore protein; Mythreye and Bloom, 2003; Pot et al., 2003), elevated sister separation might be the result of spindle checkpoint failure. This seemed unlikely for two reasons. First, kar3 and chl4 mutants require the spindle checkpoint for viability or normal growth kinetics (Gardner et al., 2001; Lee and Spencer, unpublished data), indicating that the spindle checkpoint functions in many cells. Second, cells exhibiting supernumerary buds indicative of cell cycle progression were excluded from scoring (see MATERIALS AND METHODS). However, for these two genes, checkpoint arrest was directly assessed in cells containing epitope-tagged Pds1. Both null mutants again exhibited elevated frequency of separated sisters (see Supplemental Figure 1). This indicates a surprising new role for KAR3 and CHL4 in promoting sister chromatid association of a locus near the telomere of chromosome V.
The MRX Complex Contributes to Cohesion
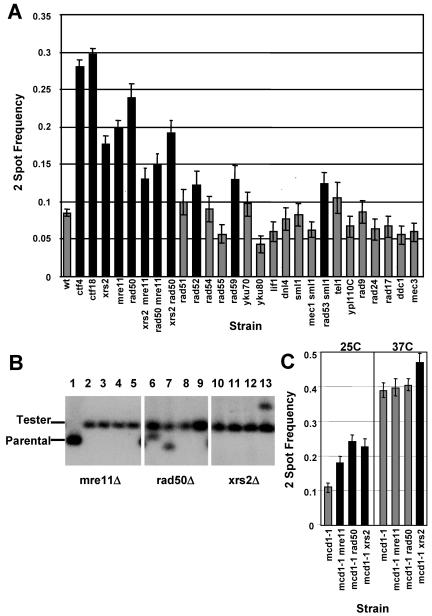
XRS2, a member of the MRX complex, was found to be important for sister chromatid cohesion (Figure 3). To further investigate the role for the MRX complex, rad50Δ and mre11Δ were tested for synthetic interactions with ctf4Δ and ctf18Δ (Table 2) and for cohesion defects (Figure 4A). Like xrs2Δ, rad50Δ and mre11Δ individually exhibited decreased growth in ctf4Δ and ctf18Δ double mutant cells, and cohesion defective phenotypes, indicating that MRX proteins may contribute to cohesion together in a complex. This hypothesis is supported by the phenotype of double mutants, which did not exhibit a higher proportion of failed cohesion than single mutants (Figure 4A). This epistasis among MRX complex members is consistent with the relationships seen for mating type switching, damage sensitivity, trinucleotide repeat expansion, and S-phase checkpoint regulation in earlier studies (Ivanov et al., 1994; Johzuka and Ogawa, 1995; Richard et al., 2000; D'Amours and Jackson, 2001). MRX subunits are also equally required in vitro for negative supercoiling of circular DNA and formation of DNA chains (Trujillo et al., 2003). We note that loss of multiple MRX subunits often exhibited cohesion defective phenotypes that differed from the single mutants. In theory, MRX proteins present in pairs or singly in cells can be imagined to be null, partially functional, or even aberrantly regulated. In this view, the idea that MRX complex contributes to cohesion solely as a single functional unit may be overly simplistic. However, it is clear that the double mutants do not exhibit an additive phenotype.
Table 2.
Specificity of the genetic interactions with ctf4Δ and ctf18Δ
| Gene | DNA repair category | ctf4 score | ctf18 score | A-like faker |
|---|---|---|---|---|
| RAD51 | Homologous recombination | 1 | 1 | 7 |
| RAD52 | 3 | 3 | 22 | |
| RAD54 | 1 | 1 | 23 | |
| RAD55 | 1 | 1 | 18 | |
| RAD59 | 1 | 1 | ||
| YKU70 | Nonhomologous end joining | 1 | 1 | |
| YKU80 | 1 | 1 | ||
| LIF1 | 1 | 1 | ||
| DNL4 | 1 | 1 | ||
| SML1 | DNA damage signaling | 1 | 1 | |
| MEC1 SML1a | 1 | 1 | nd | |
| RAD53 SMLa | 3 | 3 | nd | |
| TEL1 | 1 | 1 | ||
| YPL110C | 1 | 1 | ||
| RAD9 | DNA damage adaptor | 1 | 3 | 10 |
| RAD24 | DNA damage sensor | 1 | 1 | 6 |
| RAD17 | 1 | 1 | 10 | |
| DDC1 | 1 | 1 | 10 | |
| MEC3 | 1 | 1 | 11 | |
| XRS2 | Signal modifier | 5 | 4 | 48 |
| MRE11 | 5 | 4 | 30 | |
| RAD50 | 5 | 4 | 36 |
nd, not done.
Null alleles of the DNA repair genes listed were tested for genetic interaction with ctf4Δ or ctf18Δ. Strength of the genetic interaction (SL score) is shown, as defined in Figure 1B. Each null mutant was also tested for a-like faker frequency, and mutant:wild-type ratio is shown. Data for mutants showing ratios of less than two-fold in a primary screen is omitted.
MEC1 and RAD53 were each tested in sml1Δ mutant backgrounds to support viability of the deletion alleles for these genes (Zhao et al., 1998). Our observations fail to confirm a previously reported growth defect in ctf4 mec1-1 double mutants (Miles and Formosa, 1992), an observation made before the contribution of spontaneous sml1 mutants was appreciated. In our hands, the mec1Δ ctf4Δ triple mutant exhibited a growth phenotype similar to mec1Δ sml1Δ and ctf4Δ sml1Δ double mutant strains, which were both slow growing
Figure 4.
A subset of genes that act in DNA repair is important for cohesion. (A) Additional repair mutants are tested for sister association in metaphase arrest as in Figure 2B. Left, MRX complex components function together in cohesion. Sister chromatid association was tested as in Figure 2B for a null allele of each component and in double mutant combinations. mre11Δ and rad50Δ were significantly elevated above wild type (p < 0.05). Double mutant samples showed significant elevation of two-spot frequency in comparison with wild type, with the following p values (calculated as in Figure 2B): xrs2Δ mre11Δ = 0.001, mre11Δ rad50Δ = 0, and xrs2Δ rad50Δ = 0. Although all double mutants clearly fail to show additive frequencies, we observed statistically significant differences among the mrx single and double mutant two-spot frequencies in pairwise comparisons among them. Specifically, the null hypothesis of equal frequency is rejected (p > 0.05) for the following five comparisons: xrs2 versus xrs2 rad50, xrs2 versus mre11 rad50, mre11 versus xrs2 rad50, xrs2 rad50 versus mre11 rad50, and xrs2 mre11 versus mre11 rad50. Right, p values for significantly (p < 0.05) increased samples are rad52Δ = 0.0291, rad59Δ = 0.0088, and rad53Δ sml1Δ = 0.0082. Controls (wt, ctf4, and ctf18) and xrs2 data are those from Figure 3, shown again for clarity. (B) Pulsed field gel-separated chromosomes were hybridized with 32P-labeled MATα sequence as in Figure 2D. Tester, tester chromosome III; Parental, parental chromosome III. Lane 1, mre11Δ haploid parental strain; lanes 2–5, mre11Δ a-like faker mated products; lanes 6–9, rad50Δ a-like faker mated products; and lanes 10–13, xrs2Δ a-like faker mated products. Whole chromosome loss is evident in lanes 2, 3, 4, 5, 8, 9, 10, 11, and 12. Rearranged chromosome III is evident in lanes 6, 7, and 13. (C) Left, two-spot frequency is evaluated for null mutants lacking MRX components in nocodazole arrested mcd1-1 cells at permissive temperature (25°C). Black bars indicate significant difference from the control strain mcd1-1 (alone), calculated as in Figure 3. As expected, all strains lacking MRX genes are significantly elevated above mcd1-1 alone, indicated by the black bars. The p values were mcd1-1 mre11Δ = 0.0016, mcd1-1 rad50Δ = 0, and mcd1-1 xrs2Δ = 0. Right, two-spot frequency is evaluated for the same strains after arrest in nocodazole at nonpermissive temperature (37°C). Double mutants combining loss of MRX and Cohesin (mcd1-1) exhibit epistasis for cohesion defective phenotypes in mre11Δ mcd1–1 and rad50Δ mcd1–1 cells. However, xrs2Δ mcd1-1 cells exhibit a significant increase over the mcd1-1 control (p = 0.0191) in sister separation, indicated by the black bar.
To ascertain whether whole chromosome loss was associated with increased a-like faker production in cells lacking MRX proteins, we examined electrophoretic karyotypes of a-like faker mated products from MATα xrs2Δ, rad50Δ, and mre11Δ mutants (Figure 4B). The majority of a-like faker mated products recovered from each of these mutants was associated with complete loss of chromosome III, suggestive of a nondisjunction mechanism as would be expected for a cohesion defect. Gross chromosomal rearrangements were also observed, as would be expected for aberrant repair of DNA lesions. To confirm that the elevated chromosome separation observed in the two spot assay was not due to spindle checkpoint failure, xrs2Δ, rad50Δ, and mre11Δ mutants were examined in a strain where only Pds1-containing nuclei were scored. In this stringent assay, all three mutants exhibited a significant sister separation frequency (see Supplemental Figure 1).
The MRX complex exhibits structural similarity to Cohesin and has been proposed to support sister chromatid association during double-strand break repair (Wyman and Kanaar, 2002). Here, we have observed a role for MRX complex proteins in sister chromatid cohesion fidelity in unchallenged cell cycles. MRX complex could act independently in a complementary nonessential pathway that enhances cohesion fidelity or may promote cohesion through maintenance of Cohesin function. We considered the possibility of an independent role because temperature conditional mutants of Cohesin typically do not result in 100% sister dissociation in either two spot or fluorescence in situ assays, but rather exhibit failure in 50–70% of metaphases (Guacci et al., 1997; Michaelis et al., 1997). To test the independent pathway hypothesis, we characterized double mutants combining null alleles of MRX components and a conditional allele of an essential Cohesin subunit (mcd1-1; Guacci et al., 1997). If the MRX complex comprised an alternative supporting pathway, these double mutants would be expected to exhibit additive frequencies of cohesion failure. mre11Δ, rad50Δ, and xrs2Δ null alleles were introduced into an mcd1-1 strain containing an array of tetracycline repressor binding sites at ura3 and a tet-repressor-GFP fusion gene. Cells were treated with nocodazole at permissive temperature to achieve a metaphase arrest. Arrest was maintained as the cells were further cultured at permissive (25°C) or nonpermissive (37°C) temperature for 3 h. At permissive temperature, each of the strains exhibited levels of cohesion failure consistent with loss of an MRX protein. At nonpermissive temperature, the frequency of separated spots in mcd1-1 mre11Δ or mcd1-1 rad50Δ double mutants was not significantly different from mcd1-1 alone (p > 0.05; Figure 4C). These results indicated that the MRX complex did not contribute to cohesion fidelity independent of Cohesin. Interestingly, the mcd1-1 xrs2Δ double mutant did exhibit a marginally significant (0.01 < p < 0.05) difference (Figure 4C). This finding suggests that Xrs2p functions independently from Rad50 and Mre11 to contribute to cohesion under conditions where Cohesin is severely compromised.
A Sister Chromatid Cohesion Role Is Specific to a Subset of DNA Replication/Repair Genes
From the synthetic lethal screen, eight of the 11 new cohesion defective mutants had known roles in DNA repair. To determine whether a cohesion defect is a general characteristic of cells lacking DNA repair pathways (reviewed in Donaldson and Blow, 2001; Melo and Toczyski, 2002; Nyberg et al., 2002; Symington, 2002), we tested 22 additional DNA repair mutants. We assayed genetic interaction with ctf4Δ and ctf18Δ, determined a-like faker frequency, and tested for cohesion defects (Table 2 and Figure 4A). Many of the repair mutants exhibited elevated a-like faker frequencies. However, only three of them showed a synthetic growth defect or an increase in two spot frequency in the cohesion assay. This indicated that the presence of unrepaired damage intrinsic to unchallenged cell cycles does not in general contribute to cohesion failure.
RAD52, RAD59, and RAD53 each showed a marginally significant cohesion phenotype (0.01 < p < 0.05). RAD52 and RAD59 encode related proteins with strand-annealing functions important for double-strand break repair (reviewed in Paques and Haber, 1999; Symington, 2002). RAD53 encodes a protein kinase important for S-phase checkpoint signaling (reviewed in Donaldson and Blow, 2001; Melo and Toczyski, 2002; Nyberg et al., 2002). Curiously, we did not find genetic interactions or a cohesion defect for RAD24. RAD24 encodes one of three nonessential RFC1-like proteins (together with CTF18 and ELG1) that may provide partially overlapping complementary functions (Naiki et al., 2001; Bellaoui et al., 2003; Ben-Aroya et al., 2003; Kanellis et al., 2003; Kenna and Skibbens, 2003), particularly in response to induced DNA damage. Effects on sister chromatid cohesion after intentionally induced DNA damage was not tested here and may reveal participation of additional genes such as these.
Biological Pathways Implicated by the Interaction List
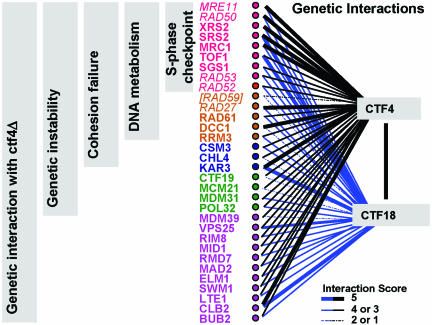
The ctf4Δ interacting gene set detected in this study and the observed phenotypes of null mutants are summarized in Figure 5. Combining the results from all assays, 31 genes with decreased viability in double mutant combination with ctf4Δ were identified and studied. 29 of these constitute new observations. Of the 31, 21 showed genetic instability in null mutants in an a-like faker assay. Sixteen of the 21 showed a significant increase in sister separation by a two-spot assay. To this we add Rad59 (which joined the cohesion-defective list as an outlier showing neither genetic interaction nor a-like faker phenotype), bringing the total to 17 with increased sister separation.
Figure 5.
Analysis of ctf4Δ synthetic lethal genes: a summary. The list of 33 genes with ctf4Δ genetic interaction or newly described sister separation phenotype is shown. This list includes results from the initial microarray-based screen and subsequent analyses. The strength of genetic interaction with ctf4Δ is indicated by the width of the blue bars as indicated. All genes were also tested for genetic interaction with ctf18Δ, and the strength of interaction is indicated by the black bars. Genes that enter through analyses after the array-based synthetic lethal list was established are indicated in italics (MRE11, RAD50, RAD53, RAD52, RAD59, and RAD27). Null mutants for all 33 genes exhibited a decrease in viability with ctf4Δ, except RAD59. (RAD59 is included solely because of its sister chromatid separation phenotype. It does not show synthetic lethality with ctf4Δ or ctf18Δ, nor does it have an increased a-like faker frequency. All other genes shown conform to the indicated categories.) Of the 32 ctf4Δ interacting genes, 21 showed marker instability as assayed by a-like faker frequency. Seventeen of 21 exhibited a newly identified sister separation phenotype. Nine of the 17 have been previously described as having an S-phase checkpoint defect.
Two of the 17 genes are known for their functions at the microtubule–kinetochore interface. The KAR3 gene encodes a microtubule-based motor protein (Meluh and Rose, 1990; Middleton and Carbon, 1994). Its effect on sister chromatid association of two genomic locations distant from kinetochores (35 kb and 393 kb away on chromosome V, respectively) indicates an additional role in chromosome structure. Chl4 has been characterized as a kinetochore protein that may be required for maturation of newly synthesized daughter kinetochores (Mythreye and Bloom, 2003; Pot et al., 2003). Its effect on sites far from chromosomal centromeres likewise indicates an additional function. Future work will be required to understand the involvement of these kinetochore-associated proteins in cohesion on chromosome arms.
Fourteen of the 17 are annotated in the Saccharomyces Genome Database (www.yeastgenome.org) as having known roles in DNA metabolism. None have been previously described as having a role in sister chromatid cohesion. Here, we note that among these 14 genes with newly identified roles in cohesion fidelity, nine are known to be required for a fully functional S-phase checkpoint. In particular, a clearly defined role for the Rad53 checkpoint kinase in the stabilization of stalled replication forks in S phase has recently been shown (Tercero et al., 2003). The MRX complex is proposed to act as a signal modifier required for full activation of Rad53 in the S-phase checkpoint (Usui et al., 2001; D'Amours and Jackson, 2002). MRC1 and TOF1 genes are required for full activation of Rad53, for slowed replication progression in response to damage within S phase, and for preservation of replication fork integrity during arrest (Alcasabas et al., 2001; Foss, 2001; Tanaka and Russell, 2001; Katou et al., 2003). The helicases SRS2 and SGS1, and double-strand break repair protein RAD52, are also required for full activation of Rad53 and for recovery from S-phase check-point arrest (Liberi et al., 2000; Vaze et al., 2002; Cobb et al., 2003). Other genome unstable ctf4Δ interactors with known or potential S-phase roles (RAD27, RAD59, RAD61, DCC1, and RRM3) have not yet been characterized specifically for S-phase checkpoint function to our knowledge. In addition to the 14, Csm3 protein has been identified as a binding partner for Tof1 in a high-throughput two-hybrid study (Ito et al., 2001) and after immunoprecipitation (Mayer et al., 2004). Csm3 may therefore represent another protein acting at the replication fork to promote sister chromatid cohesion.
Absence of increased sister separation in rad9Δ (Figure 4) further emphasizes the specificity implied by the recurrent S-phase checkpoint theme. Rad9p functions within G1 and G2 DNA damage checkpoints, where it acts as a “mediator” or “adaptor” protein that concentrates Rad53p at sites of damage and promotes damage signal generation (reviewed in Melo and Toczyski, 2002; Nyberg et al., 2002). Mrc1p is proposed to function similarly to Rad9p, but specifically within the S-phase checkpoint (Alcasabas et al., 2001; Tanaka and Russell, 2001). In contrast to rad9Δ, mrc1Δ cells exhibit a significant cohesion defect (Figure 3). Thus, the S-phase–specific adaptor protein MRC1 also acts to ensure high-fidelity sister chromatid cohesion.
DISCUSSION
Seventeen previously studied yeast genes exhibited a sister chromatid cohesion defect. This new role was identified through characterization of a ctf4Δ genetic interaction list that was subjected to a series of sequential phenotype tests. This approach has provided more evidence for an intimate connection between DNA replication and cohesion and has identified starting points for further study. The identification of several genes with roles in S phase, comprising a specific subset among DNA checkpoint/repair proteins, suggests a parallel role for Ctf4 at the replication fork. This is consistent with the established association of Ctf4 with DNA polymerase alpha. Our screen also identified a cohesion role for microtubule-kinetochore–associated proteins Kar3 and Chl4. This observation is independently confirmed by a synthetic lethal study with ctf8Δ as query, where Kar3, its regulatory subunit Vik1, and the microtubule binding protein Bim1 were found to contribute to sister chromatid cohesion (Mayer et al., 2004). In that study, CTF4, CSM3, TOF1, and CHL1 were also identified as ctf8Δ interacting genes with roles in cohesion. The efficient application of synthetic lethal screening to deletion collections can clearly reveal novel and important functional intersections among genes previously studied for other properties.
Among newly revealed cohesion contributors in budding yeast, we have identified a prominent subset of DNA repair proteins with roles in the S-phase checkpoint. This is consistent with an S-phase contribution to robust cohesion by governance of proper functioning of replication forks. The assays we have used so far do not distinguish between mechanisms at work in cohesion establishment during DNA replication or its maintenance until anaphase. These are not mutually exclusive, and the genes we have identified may act at specific cell cycle phases or may be important across a broad portion of each cycle. We propose that S-phase check-point proteins acting in concert with the replication fork promote the establishment of chromatin configurations permissive for Cohesin activity. Moreover, we propose that the S-phase checkpoint safeguards the integrity of both DNA and chromatin during replication and repair.
Although our goal was to identify cohesion defective mutants, our screen identified additional functionally important interactions. In particular, we note there are six ctf4Δ interacting genes that have roles in the regulation of progression through mitosis (Lte1, Clb2, Bub2, Mad2, Elm1 and Swm1; see e.g., www.yeastgenome.org). These genes do not contribute strongly to genetic stability on their own, and for that reason, they were not studied further in this work. However, their interactions with ctf4Δ suggest that disruptions in the governance of mitotic progression may lead to impaired chromosome structure and subsequent defects in DNA replication or sister chromatid cohesion.
The phenotypes followed in this work differ from most S-phase checkpoint studies in that no DNA damage has been intentionally induced. However, the consequences of loss of a single protein could affect cohesion through checkpoint function loss, through formation of aberrant DNA structures (i.e., mutation-induced DNA damage), or both. A full understanding of the S-phase checkpoint has been made challenging by the recognition that the replication fork serves as both activator and target of the checkpoint pathway. Thus, structural components of the fork may contribute sensor, signaling, repair, or a combination of these functions, as can checkpoint proteins. Is defective checkpoint signaling or compromised replication fork structure responsible for decreased sister association? We find that loss of Rad53, a protein kinase essential for S-phase checkpoint signaling, causes a decrease in sister chromatid cohesion fidelity, even in untreated cells (Figure 4A). This may indicate that absence of S-phase checkpoint signaling can lead to a cohesion decrease. However, other mutants with less severely compromised S-phase checkpoint signaling cause larger deficiencies in cohesion, suggesting that structural damage may also be important. For example, Srs2 protein is a DNA helicase that controls spontaneous homologous recombination frequency (Klein, 2001). After DNA damage, Srs2 shows MEC1-dependent phosphorylation contributes to full Rad53 phosphorylation and is required for checkpoint arrest recovery after DNA strand repair is complete (Liberi et al., 2000; Vaze et al., 2002). Recent studies have indicated that the Srs2 helicase can remove Rad51 from DNA (Krejci et al., 2003; Veaute et al., 2003), providing a molecular mechanism consistent with its role limiting recombination. Jointly, these facts suggest that Srs2's molecular activities restore proper chromatin configuration after repair. Our data suggest this activity of Srs2 may be required to restore chromatin to a configuration that supports cohesion. In this scenario, the combined structural and checkpoint defects in srs2Δ may lead to a sister chromatid cohesion defect greater than that observed for rad53Δ.
Based on the structural similarity between Cohesin and MRX complex, an intriguing model with a sister chromatid association role for MRX has been proposed as a mechanism for supporting double strand break repair after DNA damage (Hartsuiker et al., 2001; Wyman and Kanaar, 2002). On finding an increase in sister separation in mutants lacking MRX subunits, we tested a simple hypothesis wherein the MRX complex provided a nonessential cohesion pathway complementary to Cohesin. When all the data are considered, a role for the MRX complex in support of Cohesin is observed, with the possibility of an additional contribution by Xrs2 alone. The conservation of function observed for the MRX complex in many organisms (reviewed in D'Amours and Jackson, 2002) strongly suggests that the human MRN complex may play a similar role contributing to cohesion. Several biochemical and functional properties of human MRN support this idea. Coimmunoprecipitation between human Rad50 and Smc1 has been observed in extracts from human fibroblast and HeLa cell lines, even without induced DNA damage (Kim et al., 2002a). Furthermore, human MRN complex is observed in PCNA-containing foci during replication in untreated cells, an association that can be enhanced by induction of replication fork stalling (Maser et al., 2001; Mirzoeva and Petrini, 2003). Interestingly, the concentration of human MRN complex at sites of replication fork stalling depends on Blm1 (Franchitto and Pichierri, 2002). Blm1 is the Bloom's Syndrome helicase homologous to budding yeast Sgs1, a ctf4Δ interacting gene that also contributes to cohesion.
Several of the DNA replication and repair genes with sister chromatid cohesion roles we identify in this work have human orthologues known to be associated with disease conditions. Loss of function mutations in members of the MRX complex are associated with Nijmegen Breakage Syndrome (Carney et al., 1998; Varon et al., 1998) and Ataxia Telangiectasia Like Disease (Stewart et al., 1999). In addition, Werner's Syndrome and Bloom's Syndrome are two cancerprone premature ageing diseases caused by mutations in human orthologues of budding yeast SGS1 (reviewed in Mohaghegh and Hickson, 2001). DNA replication and repair defects in these human disease states lead to genome instability. For Werner's Syndrome, this includes a defect in recovery after DNA replication arrest (Pichierri et al., 2001), similar to the defect seen in budding yeast sgs1 and srs2 mutants (Cobb et al., 2002, 2003; Vaze et al., 2002). The yeast cohesion-defective phenotypes suggest that loss of sister association may be an important factor in the human phenotypes observed in these diseases. Other yeast genes required for high fidelity cohesion identified in this study are candidate orthologues for human genome instability loci that contribute to disease.
Supplementary Material
Acknowledgments
We thank S. Ooi, X. Pan, D. Yuan, and J. Boeke for key reagents and discussions relevant to synthetic lethal screening on arrays. We are grateful to O. Chen, C. Boone, J. Hackett, X. He, D. Koshland, and R. Skibbens for yeast strains and methods and to J. Boeke and C. Greider for critical reading of the manuscript. This work was supported by National Human Genome Research Institute and National Institute of General Medical Sciences (to F.S.), and a National Heart, Lung, and Blood Institute training grant (to D.M.E.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–09–0637. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-09-0637.
Online version of this article contains supplementary material. Online version is available at www.molbiolcell.org.
References
- Alcasabas, A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. (2001). Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Chang, M., Ou, J., Xu, H., Boone, C., and Brown, G.W. (2003). Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 22, 4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya, S., Koren, A., Liefshitz, B., Steinlauf, R., and Kupiec, M. (2003). ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100, 9906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, V.P., Maniwa, Y., Tappin, I., Ozato, K., Yokomori, K., and Hurwitz, J. (2003). The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci USA 100, 10237-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, J.M., and Jackson, S.P. (2003). The complex matter of DNA double-strand break detection. Biochem. Soc. Trans. 31, 40-44. [DOI] [PubMed] [Google Scholar]
- Carney, J.P., Maser, R.S., Olivares, H., Davis, E.M., Le Beau, M., Yates, J.R., 3rd, Hays, L., Morgan, W.F., and Petrini, J.H. (1998). The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477-486. [DOI] [PubMed] [Google Scholar]
- Celeste, A., et al. (2002). Genomic instability in mice lacking histone H2A.X. Science 296, 922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb, J.A., Bjergbaek, L., and Gasser, S.M. (2002). RecQ helicases: at the heart of genetic stability. FEBS Lett. 529, 43-48. [DOI] [PubMed] [Google Scholar]
- Cobb, J.A., Bjergbaek, L., Shimada, K., Frei, C., and Gasser, S.M. (2003). DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22, 4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D., and Gautier, J. (2001). Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8, 137-147. [DOI] [PubMed] [Google Scholar]
- Cutler, D.J., et al. (2001). High-throughput variation detection and genotyping using microarrays. Genome Res. 11, 1913-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and Jackson, S.P. (2001). The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15, 2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and Jackson, S.P. (2002). The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3, 317-327. [DOI] [PubMed] [Google Scholar]
- Donaldson, A.D., and Blow, J.J. (2001). DNA replication: stable driving prevents fatal smashes. Curr. Biol. 11, R979-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S., Li, C.M., Levy, D.L., Brown, J., Snow, P.M., and Campbell, J.L. (2003). Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol. Cell. Biol. 23, 2733-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss, J. (1981). Statistical Methods for Rates and Proportions, New York: John Wiley & Sons, Inc.
- Formosa, T., and Nittis, T. (1999). Dna2 mutants reveal interactions with DNA polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics 151, 1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss, E.J. (2001). Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto, A., and Pichierri, P. (2002). Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol. 157, 19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R.D., Poddar, A., Yellman, C., Tavormina, P.A., Monteagudo, M.C., and Burke, D.J. (2001). The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics 157, 1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., et al. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- Goldberg, M., Stucki, M., Falck, J., D'Amours, D., Rahman, D., Pappin, D., Bartek, J., and Jackson, S.P. (2003). MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421, 952-956. [DOI] [PubMed] [Google Scholar]
- Green, E., Hieter, P., and Spencer, F. (1998). Yeast artificial chromosomes. In: Genome Analysis, vol. 3, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 297-565. [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, J.S., Kroll, E.S., Lundblad, V., and Spencer, F.A. (2001). Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21, 3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker, E., Vaessen, E., Carr, A.M., and Kohli, J. (2001). Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20, 6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Asthana, S., and Sorger, P.K. (2000). Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101, 763-775. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, D., Schleiffer, A., Eisenhaber, F., Mechtler, K., Haering, C.H., and Nasmyth, K. (2002). Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol. 12, 323-328. [DOI] [PubMed] [Google Scholar]
- Ivanov, E.L., Sugawara, N., White, C.I., Fabre, F., and Haber, J.E. (1994). Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger, R. (2002). The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell. Biol. 3, 767-778. [DOI] [PubMed] [Google Scholar]
- Johzuka, K., and Ogawa, H. (1995). Interaction of Mre11 and Rad 50, two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139, 1521-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis, P., Agyei, R., and Durocher, D. (2003). Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr. Biol. 13, 1583-1595. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078-1083. [DOI] [PubMed] [Google Scholar]
- Kenna, M.A., and Skibbens, R.V. (2003). Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23, 2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S., Krasieva, T.B., LaMorte, V., Taylor, A.M., and Yokomori, K. (2002a). Specific recruitment of human cohesin to laser-induced DNA damage. J. Biol. Chem. 277, 45149-45153. [DOI] [PubMed] [Google Scholar]
- Kim, S.T., Xu, B., and Kastan, M.B. (2002b). Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, H.L. (2001). Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157, 557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, J., Tauchi, H., Sakamoto, S., Nakamura, A., Morishima, K., Matsuura, S., Kobayashi, T., Tamai, K., Tanimoto, K., and Komatsu, K. (2002). NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12, 1846-1851. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., et al. (1992). CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 5736-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci, L., Van Komen, S., Li, Y., Villemain, J., Reddy, M.S., Klein, H., Ellen-berger, T., and Sung, P. (2003). DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305-309. [DOI] [PubMed] [Google Scholar]
- Liberi, G., Chiolo, I., Pellicioli, A., Lopes, M., Plevani, P., Muzi-Falconi, M., and Foiani, M. (2000). Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19, 5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby, M., Rothstein, R., and Mortensen, U.H. (2001). Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98, 8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R.S., Mirzoeva, O.K., Wells, J., Olivares, H., Williams, B.R., Zinkel, R.A., Farnham, P.J., and Petrini, J.H. (2001). Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 21, 6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M.L., Gygi, S.P., Aebersold, R., and Hieter, P. (2001). Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7, 959-970. [DOI] [PubMed] [Google Scholar]
- Mayer, M.L., Pot, I., Chang, M., Xu, H., Aneliunas, V., Kwok, T., Newitt, R., Aebersold, R., Boone, C., Brown, G.W., and Hieter, P. (2004). Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Cell. Biol. 15, 1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, D. (2002). A unified view of the DNA-damage check-point. Curr. Opin. Cell Biol. 14, 237-245. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., and Rose, M.D. (1990). KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60, 1029-1041. [DOI] [PubMed] [Google Scholar]
- Merkle, C.J., Karnitz, L.M., Henry-Sanchez, J.T., and Chen, J. (2003). Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J. Biol. Chem. 278, 30051-30056. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- Middleton, K., and Carbon, J. (1994). KAR3-encoded kinesin is a minus-end-directed motor that functions with centromere binding proteins (CBF3) on an in vitro yeast kinetochore. Proc. Natl. Acad. Sci. USA 91, 7212-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, J., and Formosa, T. (1992). Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol. Cell. Biol. 12, 5724-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovich, M., and Koshland, D.E. (2003). Molecular biology. SMC complexes–wrapped up in controversy. Science 300, 1101-1102. [DOI] [PubMed] [Google Scholar]
- Mirzoeva, O.K., and Petrini, J.H. (2003). DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1, 207-218. [PubMed] [Google Scholar]
- Mohaghegh, P., and Hickson, I.D. (2001). DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10, 741-746. [DOI] [PubMed] [Google Scholar]
- Mythreye, K., and Bloom, K.S. (2003). Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160, 833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., Datta, A., and Kolodner, R.D. (2001). Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104, 397-408. [DOI] [PubMed] [Google Scholar]
- Naiki, T., Kondo, T., Nakada, D., Matsumoto, K., and Sugimoto, K. (2001). Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21, 5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, K., Taniguchi, T., Ranganathan, V., New, H.V., Moreau, L.A., Stotsky, M., Mathew, C.G., Kastan, M.B., Weaver, D.T., and D'Andrea, A.D. (2002). Interaction of FANCD2 and NBS1 in the DNA damage response. Nat. Cell Biol. 4, 913-920. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673-745. [DOI] [PubMed] [Google Scholar]
- Nyberg, K.A., Michelson, R.J., Putnam, C.W., and Weinert, T.A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617-656. [DOI] [PubMed] [Google Scholar]
- Ooi, S.L., Shoemaker, D.D., and Boeke, J.D. (2003). DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35, 277-286. [DOI] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri, P., Franchitto, A., Mosesso, P., and Palitti, F. (2001). Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12, 2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot, I., Measday, V., Snydsman, B., Cagney, G., Fields, S., Davis, T.N., Muller, E.G., and Hieter, P. (2003). Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14, 460-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M., and Norbury, C.J. (2002). Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99, 12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, G.F., Goellner, G.M., McMurray, C.T., and Haber, J.E. (2000). Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11-RAD50-XRS2 complex. EMBO J. 19, 2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., and Russell, P. (2002). Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109, 563-573. [DOI] [PubMed] [Google Scholar]
- Skibbens, R.V., Corson, L.B., Koshland, D., and Hieter, P. (1999). Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, G.S., Maser, R.S., Stankovic, T., Bressan, D.A., Kaplan, M.I., Jaspers, N.G., Raams, A., Byrd, P.J., Petrini, J.H., and Taylor, A.M. (1999). The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577-587. [DOI] [PubMed] [Google Scholar]
- Stewart, G.S., Wang, B., Bignell, C.R., Taylor, A.M., and Elledge, S.J. (2003). MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961-966. [DOI] [PubMed] [Google Scholar]
- Strathern, J., Hicks, J., and Herskowitz, I. (1981). Control of cell type in yeast by the mating type locus. The alpha 1-alpha 2 hypothesis. J. Mol. Biol. 147, 357-372. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630-670, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., and Russell, P. (2001). Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3, 966-972. [DOI] [PubMed] [Google Scholar]
- Tauchi, H., Matsuura, S., Kobayashi, J., Sakamoto, S., and Komatsu, K. (2002). Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene 21, 8967-8980. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., Longhese, M.P., and Diffley, J.F. (2003). A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11, 1323-1336. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo, K.M., Roh, D.H., Chen, L., Van Komen, S., Tomkinson, A., and Sung, P. (2003). Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J. Biol. Chem. 278, 48957-48964. [DOI] [PubMed] [Google Scholar]
- Usui, T., Ogawa, H., and Petrini, J.H. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255-1266. [DOI] [PubMed] [Google Scholar]
- Varon, R., et al. (1998). Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93, 467-476. [DOI] [PubMed] [Google Scholar]
- Vaze, M.B., Pellicioli, A., Lee, S.E., Ira, G., Liberi, G., Arbel-Eden, A., Foiani, M., and Haber, J.E. (2002). Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373-385. [DOI] [PubMed] [Google Scholar]
- Veaute, X., Jeusset, J., Soustelle, C., Kowalczykowski, S.C., Le Cam, E., and Fabre, F. (2003). The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309-312. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Cortez, D., Yazdi, P., Neff, N., Elledge, S.J., and Qin, J. (2000a). BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14, 927-939. [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., De Las Penas, A., Adams, C., and Christman, M.F. (2000b). Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289, 774-779. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., Brady, D.M., Johnston, R.C., Hanna, J.S., Hardwick, K.G., and Spencer, F.A. (2002). Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13, 3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- Wittmeyer, J., and Formosa, T. (1997). The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17, 4178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, C., and Kanaar, R. (2002). Chromosome organization: reaching out to embrace new models. Curr. Biol. 12, R446-R448. [DOI] [PubMed] [Google Scholar]
- Yazdi, P.T., Wang, Y., Zhao, S., Patel, N., Lee, E.Y., and Qin, J. (2002). SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16, 571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S., Renthal, W., and Lee, E.Y. (2002). Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 30, 4815-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.