Abstract
Ctf8p is a component of Ctf18-RFC, an alternative replication factor C-like complex required for efficient sister chromatid cohesion in Saccharomyces cerevisiae. We performed synthetic genetic array (SGA) analysis with a ctf8 deletion strain as a primary screen to identify other nonessential genes required for efficient sister chromatid cohesion. We then assessed proficiency of cohesion at three chromosomal loci in strains containing deletions of the genes identified in the ctf8 SGA screen. Deletion of seven genes (CHL1, CSM3, BIM1, KAR3, TOF1, CTF4, and VIK1) resulted in defective sister chromatid cohesion. Mass spectrometric analysis of immunoprecipitated complexes identified a physical association between Kar3p and Vik1p and an interaction between Csm3p and Tof1p that we confirmed by coimmunoprecipitation from cell extracts. These data indicate that synthetic genetic array analysis coupled with specific secondary screens can effectively identify protein complexes functionally related to a reference gene. Furthermore, we find that genes involved in mitotic spindle integrity and positioning have a previously unrecognized role in sister chromatid cohesion.
INTRODUCTION
The maintenance of proper ploidy during cell division requires both the accurate replication of chromosomes and their faithful segregation during mitosis. The physical association of sister chromatids after DNA replication, or sister chromatid cohesion, is crucial for the proper segregation of sister chromatids at anaphase and is therefore critical for genome stability. In Saccharomyces cerevisiae, cohesion is mediated by a multisubunit protein complex called cohesin that is composed of at least four proteins: Smc1p, Smc3p, Mcd1p/Scc1p, and Irr1p/Scc3p (SA1 or SA2 in mammalian cells) (Guacci et al., 1997; Michaelis et al., 1997; Toth et al., 1999). Pds5p is also required for sister chromatid cohesion and its localization to chromatin requires Mcd1p/Scc1p (Hartman et al., 2000; Panizza et al., 2000). Before the onset of anaphase, Esp1p, a protease required for the separation of sister chromatids, is bound to its inhibitor Pds1p (Ciosk et al., 1998). At the onset of anaphase Pds1p is ubiquitinated and targeted for degradation by the anaphase promoting complex/cyclosome (APC/C) (Cohen-Fix et al., 1996). Degradation of Pds1p releases Esp1p, which then cleaves the cohesin subunit Scc1p resulting in sister chromatid separation (Uhlmann et al., 1999, 2000).
Other proteins required for proper sister chromatid cohesion function during the establishment of cohesion, which takes place during S phase. Scc2p and Scc4p physically interact with each other but are not core components of the cohesin complex (Ciosk et al., 2000). Scc2p and Scc4p are, however, required for the association of cohesin with DNA (Ciosk et al., 2000). Eco1p/Ctf7p is also required for the establishment but not the maintenance of cohesion (Skibbens et al., 1999; Toth et al., 1999). In eco1 mutants, the cohesin complex is able to associate with DNA, but proper sister chromatid cohesion is not established, indicating that the proper establishment of sister chromatid cohesion is likely to be a multistep process requiring both the association of the cohesin complex with DNA as well as the physical pairing of sister chromatids (Toth et al., 1999).
Deletion of SMT4 does not seem to affect the association of the cohesin subunit Mcd1p/Scc1p with chromatin, but smt4 strains are unable to maintain centromeric cohesion (Bachant et al., 2002). Smt4p is capable of reversing Smt3p/SUMO-1 modification (Li and Hochstrasser, 1999, 2000), and several observations suggest that the centromeric cohesion defect of smt4 is partially due to defective Smt3p/SUMO-1 modification of Top2p. Overexpression of TOP2 reduces the degree of defective centromeric cohesion in smt4 strains, and top2-4 mutants exhibit defective centromeric cohesion. Finally, expression of Top2p-SNM, in which the SUMO-1 modification sites of Top2p have been mutated, results in partial suppression of the smt4 cohesion defect at centromeric loci (Bachant et al., 2002). These data suggest that Smt3p/SUMO-1 modification of Top2p may play a role in establishing a modified chromosome structure or localized catenations that are required for efficient centromeric cohesion.
Many other observations have highlighted the important role that the DNA replication machinery plays in sister chromatid cohesion. Overexpression of POL30, which encodes the DNA polymerase processivity factor PCNA, rescues the temperature sensitivity of a ctf7-203 allele (Skibbens et al., 1999). Ctf4p, which is required for efficient sister chromatid cohesion (Hanna et al., 2001), binds specifically to DNA polymerase α (Miles and Formosa, 1992a,b). ctf4 also exhibits genetic interactions with cdc17-1 (which encodes DNA polymerase α) and is synthetically lethal with rfc1/cdc44, a component of the replication factor C (RFC) complex (Formosa and Nittis, 1999; Budd and Campbell, 2000). An additional link between DNA replication and the establishment of sister chromatid cohesion came with the discovery and analysis of Trf4p and Trf5p, redundant homologues that encode the nuclear DNA polymerase σ. Deletion of TRF4 results in a sister chromatid cohesion defect (Wang et al., 2000).
Finally, components of Ctf18-RFC, an alternative RFC complex composed of Rfc2p, Rfc3p, Rfc4p, Rfc5p, Ctf18p, Ctf8p, and Dcc1p, are required for proper sister chromatid cohesion (Hanna et al., 2001; Mayer et al., 2001; Naiki et al., 2001). Because one function of the canonical RFC complex (composed of Rfc1p, Rfc2p, Rfc3p, Rfc4p, and Rfc5p) is to initiate a switch from DNA polymerase α to DNA polymerase δ during lagging strand DNA synthesis (reviewed in Waga and Stillman, 1998), these data suggest a model whereby Ctf18-RFC functions to initiate a polymerase switch to DNA polymerase σ during DNA replication, which would be required for the efficient establishment of sister chromatid cohesion. The human Ctf8, Ctf18, and Dcc1 proteins were recently shown to interact with the p36, p37, p38, and p40 subunits of RFC, indicating that the Ctf18–RFC complex exists in human cells (Bermudez et al., 2003; Merkle et al., 2003). In addition, hCtf18 coimmunoprecipitated PCNA, and the seven-member hCtf18-RFC was shown to be able to load the PCNA clamp onto DNA in an ATP-dependent manner (Bermudez et al., 2003; Merkle et al., 2003).
To identify other genes required for sister chromatid cohesion, we performed synthetic genetic array (SGA) analysis to isolate genes that, when deleted, result in synthetic fitness defects in combination with a deletion of CTF8. We then assessed sister chromatid cohesion in strains carrying a deletion in genes found to be synthetically sick or lethal with ctf8Δ. Here we report that deletion of VIK1, CTF4, CSM3, KAR3, TOF1, CHL1, or BIM1 results in inefficient sister chromatid cohesion. Using mass spectrometric analysis of immunoprecipitated complexes, we find that Vik1p and Kar3p, and Csm3p and Tof1p, physically interact in vivo. These data indicate that SGA analysis of a reference gene can be used to identify functionally relevant protein complexes. Of particular significance, we find that genes involved in mitotic spindle integrity and positioning are required for efficient sister chromatid cohesion. Thus, these pathways make previously uncharacterized contributions to accurate chromosome segregation via their effects on sister chromatid cohesion.
MATERIALS AND METHODS
Synthetic Genetic Array Analysis
SGA analysis was carried out as described previously (Tong et al., 2001). Briefly, a starting strain containing ctf8Δ (MATα ctf8Δ::natR canΔ1::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 met15Δ0) was mated to the yeast genome deletion set (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 with [nonessential orf]Δ::kanMX6). Diploids were sporulated and the resulting haploids were grown on selective plates to identify viable gene deletions that show synthetic genetic interactions with ctf8Δ. Genetic interactions were confirmed by tetrad analysis on YPD.
Assessing Sister Chromatid Cohesion
Tet repressor-GFP/Tet operator repeat strains were as follows: wild-type (YPH1477, MATa ade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2-TetR-GFP ura3::3xURA3-TetO112 PDS1-13Myc:TRP1; Mayer et al., 2001), chl1Δ (YPH1614, YPH1615), vik1Δ (YPH1616, YPH1617), csm3Δ (YPH1618, YPH1619), bim1Δ (YPH1620, YPH1621), tof1Δ (YPH1622, YPH1623), kar3Δ (YPH1624, YPH1625), and top1Δ (YPH1626, YPH1627) strains were grown logarithmically in YPD and diluted to OD600 = 0.150 before arresting in G2/M with 15 μg/ml nocodazole (Sigma-Aldrich, St. Louis, MO) or in G1 with 5 μg/ml alpha factor (Diagnostic Chemicals Limited, Charlottetown, Prince Edward Island, Canada) for 3 h at 25°C. Cells were fixed with an equal volume of fresh 4% paraformaldehyde for 10 min at room temperature, washed once with SK (1M sorbitol, 0.05 M K2PO4), and resuspended in SK for cohesion assessment.
Lac repressor-GFP/Lac operator repeat strains were as follows: wild-type (Y819, MATa ade2-1 trp1-1::LacO-TRP1-LEU2 can1-100 his3-11,15::LacI-GFP-HIS3 leu2–3112 ura3-1; Sanchez et al., 1999), chl1Δ (YPH1628, YPH1629), vik1Δ (YPH1630, YPH1631), csm3Δ (YPH1632, YPH1633), bim1Δ (YPH1634, YPH1635), tof1Δ (YPH1636, YPH1637), and kar3Δ (YPH1638, YPH1639), or wild-type (YPH1444, MATa ade2-101 trp1-1 can1-100 his3-11,15::LacI-GFP(pAFS144, thermostable)-HIS3 leu2-3112 ura3-1 CEN15(1.8kb)-LacO-URA3; Goshima and Yanagida, 2000, and references therein), chl1Δ (YPH1640, YPH1641), vik1Δ (YPH1642, YPH1643), csm3Δ (YPH1644, YPH1645), bim1Δ (YPH1646, YPH1647), tof1Δ (YPH1648, YPH1649), and kar3Δ (YPH1650, YPH1651) strains were grown logarithmically in YPD at 25°C and then collected by centrifugation and resuspended in synthetic complete media lacking histidine and containing 40 mM 3-aminotriazole (Sigma-Aldrich). Cells were grown in this media for 90 min at 25°C to induce the Lac repressor-GFP fusion protein that was expressed from the HIS3 promoter. These cells were then collected by centrifugation, resuspended in YPD, and arrested in G2/M with nocodazole or in G1 with alpha factor for3hat25°C. Cells were processed as described above, and the number of GFP signals in each cell was scored.
Epitope Tagging and Immunoprecipitations
C-terminal epitope tagging of proteins at their endogenous loci was performed as described previously (Longtine et al., 1998). All tagged strains are derived from YPH1652 (MATa ade2-101 trp1Δ63 hi3Δ200 leu2Δ1 ura3-52 lys2-801). For mass spectrometric analysis of immunoprecipitated complexes, immunoprecipitations of YPH1653 (MATa CHL1–13Myc:TRP1), YPH1654 (MATa CSM3–13Myc:TRP1), YPH1655 (MATa VIK1-13Myc:TRP1) and YPH1652 (untagged control) were performed as described previously (Lamb et al., 1994) from 500 to 700 mg of protein extract. For coimmunoprecipitation experiments, immunoprecipitations of YPH1656 (MATa TOF1-3HA:kanMX6), YPH1657 (MATa TOF1-3HA:kanMX6 CSM3-13Myc:TRP1), YPH1658 (MATa TOP1-3HA:kanMX6), YPH1659 (MATa TOP1-3HA:kanMX6 CSM3-13Myc: TRP1), YPH1654 (MATa CSM3-13Myc:TRP1), YPH1660 (MATa TOF1-3HA: kanMX6 TOP1-13Myc:HIS3MX6), YPH1661 (MATa TOP1-13Myc:HIS3MX6), and YPH1652 (untagged control) were performed as described previously (Tyers et al., 1992; Measday et al., 2002). Briefly, cells were lysed in 50 mM Tris-HCl pH 7.5, 250 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, and 0.1% NP-40 buffer containing protease inhibitors, and equal amounts of protein (at least 4 mg) were immunoprecipitated using anti-hemagglutinin (HA) or anti-Myc conjugated beads (Covance, Princeton, NJ). Forty micrograms of total lysate and 15% of the immunoprecipitated fractions were loaded on SDS-PAGE gels, and tagged proteins were detected on Western blots with anti-HA (12CA5) or anti-Myc (9E10) antibodies (Roche Diagnostics, Indianapolis, IN).
Pds1 Assay
Green fluorescent protein (GFP) and Pds1-13Myc were detected by indirect immunofluorescence in nocodazole arrested, paraformaldehyde fixed cells by using anti-GFP (AbCam) and anti-Myc (Roche Diagnostics) antibodies, respectively. The presence or absence of Pds1-13Myc signal in cells containing two separated GFP dots was assessed. At least 20 two-dot cells were examined for each strain.
Mass Spectrometry Analysis
The proteins contained in silver-stained bands were analyzed by capillary chromatography, electrospray ionization tandem mass spectrometry (LC-MS/MS) essentially as described previously (Gygi et al., 2000). Silver stained bands were excised and the proteins therein were trypsinized in situ. The generated peptide fragments were extracted and analyzed by LC-MS/MS by using an LCQ Classic ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) equipped with an in-house microelectrospray ionization source. Needle voltage was set at 2 kV. Ion signals above a predetermined threshold automatically triggered the instrument to switch from MS to MS/MS mode for generating collision-induced dissociated spectra (data-dependent MS/MS). The generated collision-induced dissociated spectra were searched against the whole genome yeast sequence database using the computer algorithm SEQUEST (Eng et al., 1994).
RESULTS
Genome-wide Synthetic Genetic Interaction Screen with ctf8Δ
Previously, we have shown that a deletion of CTF8 is synthetically lethal with mutations in genes required for sister chromatid cohesion including smc3-42, scc1-73, and scc2-4 (Mayer et al., 2001). In contrast, deletion of CTF8 is not synthetically lethal with deletions of other nonessential components of the Ctf18-RFC complex including ctf18Δ and dcc1Δ. These data suggested that CTF8 becomes essential when genes that function in cohesion pathways distinct from that of the Ctf18–RFC complex are impaired. They also suggested that a synthetic lethal screen could be used to identify other nonessential genes required for efficient sister chromatid cohesion. We performed a synthetic genetic interaction screen using an array containing the ∼4600 viable haploid yeast open reading frame deletion mutants, as described previously (SGA analysis) (Tong et al., 2001). The SGA screen was repeated three times and 55 deletion mutants were identified that displayed synthetic growth defects with ctf8Δ in at least two of the three screens (Table 1). Because CTF4 is the only known nonessential gene required for efficient cohesion other than those in the Ctf18–RFC complex, isolation of ctf4Δ in the ctf8Δ SGA screen indicated that this approach was successful in identifying other genes required for efficient sister chromatid cohesion.
Table 1.
Synthetic genetic interactions with ctf8Δ and initial sister chromatid cohesion analysis
| Systematic name | Gene name | Genetic interaction on YPD | % Two GFP dots: initial analysis |
|---|---|---|---|
| YDR332W | No | 9 | |
| YGL168W | Sick | 18.5 | |
| YML095C-A | Sick | 9.8 | |
| YMR166C | No | 14.4 | |
| YDR375C | BCS1 | No | 11.8 |
| YER016W | BIM1 | Lethal | 16.9 |
| YER014C-A | BUD25 | No | Not done |
| YGL003C | CDH1 | No germination | 19 |
| YPL008W | CHL1 | Lethal | 27.4 |
| YMR198W | CIK1 | No | 7.3 |
| YNL298W | CLA4 | Sick | 8.1 |
| YPR119W | CLB2 | Sick | 9.2 |
| YMR048W | CSM3 | Lethal | 21.1 |
| YPR135W | CTF4 | Lethal | 31.7 |
| YPL194W | DDC1 | No | 11 |
| YAL026C | DRS2 | No | 10 |
| YDR518W | EUG1 | No | 10.5 |
| YFL023W | FYV11 | No | 8.6 |
| YNL153C | GIM3 | Sick | Not done |
| YEL003W | GIM4 | Sick | 9.9 |
| YDR225W | HTA1 | Sick | 10 |
| YNL106C | INP52 | No | 8.7 |
| YPR141C | KAR3 | Lethal | 16.5 |
| YGL173C | KEM1 | Sick | 14.2 |
| YLR260W | LCB5 | No | 8 |
| YAL024C | LTE1 | No | 8 |
| YGL086W | MAD1 | Sick | Not done |
| YJL030W | MAD2 | Sick | Not done |
| YIR021W | MRS1 | No | 10.2 |
| YJR073C | OPI3 | No | 12 |
| YGR078C | PAC10 | Sick | 12.6 |
| YNL264C | PDR17 | No | 9.7 |
| YGL153W | PEX14 | No | 9.1 |
| YDR329C | PEX3 | No | 7.8 |
| YNL329C | PEX6 | No | 10.2 |
| YGL167C | PMR1 | Sick | 12.3 |
| YKL113C | RAD27 | No | 14.7 |
| YLR039C | RIC1 | No | 12.3 |
| YMR142C | RPL13B | No germination | 31 |
| YGL244W | RTF1 | Sick | 6.8 |
| YHR154W | RTT107 | No | 11.6 |
| YMR272C | SCS7 | No | 10 |
| YLR268W | SEC22 | No | 13.6 |
| YGL066W | SGF73 | Sick | 11.7 |
| YBL031W | SHE1 | No | 12 |
| YOR195W | SLK19 | Sick | 10.2 |
| YGR229C | SMI1 | Sick | Not done |
| YLR452C | SST2 | No | 9 |
| YPL057C | SUR1 | No | 8.3 |
| YJL176C | SWI3 | No | Not done |
| YNL273W | TOF1 | Lethal | 15.1 |
| YFR010W | UBP6 | No | 6.3 |
| YBR173C | UMP1 | Sick | 9.4 |
| YLR373C | VID22 | No | 8.6 |
| YPL253C | VIK1 | Sick | 19.3 |
Assessing Sister Chromatid Cohesion in Mutants Synthetically Lethal with ctf8Δ
We first analyzed sister chromatid cohesion directly in the 55 mutants identified in the ctf8Δ SGA screen (Table 1). To identify those genes required for efficient sister chromatid cohesion, we mated these mutants to a strain containing both a Tet repressor-GFP fusion and a Tet operator repeat located 35 kb from the centromere of chromosome V. The resulting diploids were sporulated and dissected, and two independent haploids containing the deletion of interest, as well as the Tet repressor-GFP fusion and the Tet operator repeats, were assayed for defective cohesion. Cells were grown logarithmically, arrested in G2/M with nocodazole for 3 h, fixed, and then scored to determine the number of GFP dots in each cell. MAD1 and MAD2 were excluded from this analysis because they encode mitotic checkpoint proteins required for proper G2/M arrest in response to nocodazole (Li and Murray, 1991). Four other genes (BUD25, SMI1, GIM3, and SWI3) were excluded from further analysis due to abnormal budding phenotypes or special growth requirements. The deletion of 10 genes (ygl168wΔ, bim1Δ, cdh1Δ, chl1Δ, csm3Δ, ctf4Δ, kar3Δ, rpl13bΔ, tof1Δ, and vik1Δ) increased the percentage of nocodazole-arrested cells with two GFP dots to at least 15% (wild type was 10.7%), indicative of either defective sister chromatid cohesion or polyploidy in these cells (Table 1). Further analysis of rpl13bΔ cells indicated that the high percentage of cells with two GFP signals in these strains was due to high levels of polyploidy, not defective sister chromatid cohesion. Because CTF4 had previously been reported to be required for efficient sister chromatid cohesion (Hanna et al., 2001), we did not investigate the role of this gene in cohesion any further. We also did not pursue CDH1 because it is a substrate specificity factor for the APC/C (Schwab et al., 1997; Visintin et al., 1997). The increased percentage of cdh1Δ cells with two GFP dots could be indicative of abrogated APC/C activity or high Clb2p levels in the cell, rather than a direct role of Cdh1p in cohesion. Finally, we did not pursue ygl168wΔ because, upon further analysis, this mutant did not consistently exhibit at least 15% of cells with two GFP signals in the cohesion assay (our unpublished data). Therefore, this initial analysis identified six genes (VIK1, CSM3, TOF1, BIM1, KAR3, and CHL1) with previously undescribed roles in sister chromatid cohesion.
Confirmation of Genetic Interactions by Tetrad Analysis
The synthetic genetic interactions between ctf8Δ and the gene deletions identified in the SGA screen were confirmed by tetrad dissection. A ctf8Δ strain was mated to a deletion of each gene identified in the SGA screen and the resulting diploid was sporulated and dissected on rich medium (YPD) (Figure 1B). Of the 55 mutants in the initial set, six mutants were lethal when combined with ctf8Δ on YPD, and 18 mutants were sick when combined with ctf8Δ on YPD (Table 1 and Figure 1A). Thus, 44% of the genetic interactions identified in the SGA screen were confirmed by tetrad analysis. The genetic interactions identified show an enrichment for genes involved in the spindle checkpoint, microtubule dynamics, chromosome segregation, and chromatin structure, consistent with the role of CTF8 in sister chromatid cohesion. Of particular significance, the mutants that were synthetically lethal with ctf8Δ, namely, chl1Δ, csm3Δ, tof1Δ, ctf4Δ, bim1Δ, and kar3Δ, include six of the seven mutants that displayed defects in sister chromatid cohesion (Figure 1A). Thus, the severity of the fitness defect in the ctf8Δ double mutants served as an excellent predictor of biological function. These data further suggest that cumulative defects in sister chromatid cohesion pathways are likely to result in lethality.
Figure 1.
CTF8 genetic interactions. (A) Results from SGA analysis with ctf8Δ presented as a genetic interaction map. Lines represent a confirmed synthetic sick interaction between the connected genes. Broken lines represent synthetic lethal interactions. Gene functions are indicated. Genes required for efficient sister chromatid cohesion are indicated in bold. (B) Examples of tetrad analyses for a synthetic lethal interaction (ctf8Δ bim1Δ) and a synthetic sick interaction (ctf8Δ clb2Δ). Double mutants are indicated by the arrowheads.
Analysis of Sister Chromatid Cohesion at a Locus 35 kb from CEN5
We next proceeded with a detailed analysis of sister chromatid cohesion defects in the six novel cohesion mutants. Each gene was deleted in YPH1477, which contains a Tet repressor-GFP fusion as well as Tet operator repeats located 35 kb from the centromere of chromosome V, and the effect on sister chromatid cohesion at this arm locus was assayed. Strains containing each of the single gene deletions were grown logarithmically and arrested either in G2/M using nocodazole or in G1 using alpha mating factor for 3 h at 25°C. These cells were then fixed, and the number of GFP dots per cell was scored. Deletion of VIK1, CSM3, TOF1, BIM1, KAR3, or CHL1 resulted in defective sister chromatid cohesion at this arm locus as evidenced by an increased percentage of G2/M-arrested cells with two GFP dots compared with the wild-type strain (Figure 2). The increased number of GFP dots could not be attributed to an increase in polyploidy because when these strains were arrested in G1 with alpha factor, the cells had either no increase or only a slight increase in the number of cells with more than one GFP dot compared with a wild-type strain arrested in G1. Furthermore, 80–90% of the nocodazole-arrested cells with separated sister chromatids had high levels of Pds1p, indicating that these cells had not broken through the nocodazole arrest and had not initiated anaphase (Figure 2B). Finally, we observed a range of cohesion defects, with chl1Δ displaying the greatest defect, followed by kar3Δ, tof1Δ, csm3Δ, bim1Δ, and then vik1Δ. The magnitudes of the cohesion defects in the chl1Δ and kar3Δ strains were similar to that observed in ctf8Δ mutants (Mayer et al., 2001). In contrast, the vik1Δ mutant exhibited a less pronounced cohesion defect than the other mutants (Figures 2, 3, 4), consistent with the weaker genetic interaction observed between vik1Δ and ctf8Δ in comparison to that of other mutants.
Figure 2.
(A) Assessing cohesion at an arm locus 35 kb from CEN5. CHL1, VIK1, CSM3, BIM1, TOF1, KAR3, and TOP1 were deleted in strain YPH1477, which expresses a Tet repressor-GFP fusion protein and Pds1-13Myc, and contains a Tet operator repeat integrated 35 kb from the centromere of chromosome V. Two independent strains for each deletion were scored in two independent experiments for sister chromatid cohesion defects. At least 200 cells were scored for each strain. (B) Sister chromatid cohesion defects occur in the presence of Pds1p. The average percentage of nocodazole-arrested cells with two GFP signals from two strains in two independent experiments is given along with the SD. The presence or absence of Pds1-13Myc was also scored in nocodazole-arrested cells with two GFP signals. At least 20 two-dot cells were scored for Pds1p in each strain.
Figure 3.
(A) Assessing cohesion at an arm locus 12 kb from CEN4. CHL1, VIK1, CSM3, BIM1, TOF1, and KAR3 were deleted in strain Y819, which expresses a Lac repressor-GFP fusion protein and contains a Lac operator repeat integrated 12 kb from the centromere of chromosome IV. The number of GFP signals in each cell was scored for two independent strains for each deletion in two independent experiments. At least 200 cells were scored for each strain. (B) Sister chromatid cohesion quantified. The average percentage of nocodazole-arrested cells with two GFP signals from two strains in two independent experiments is given along with the SD.
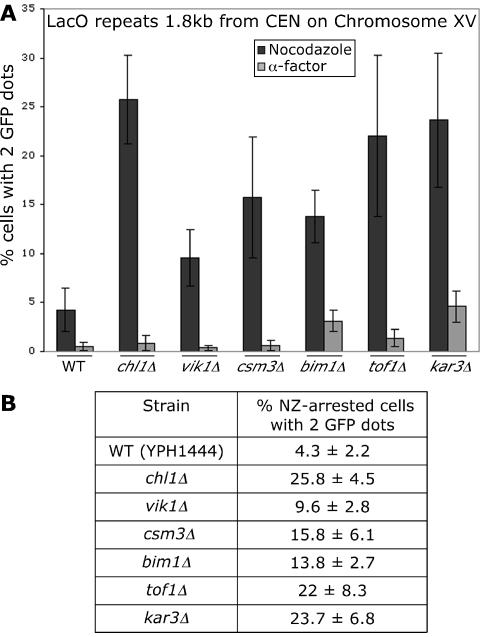
Figure 4.
(A) Assessing cohesion at a CEN locus 1.8 kb from CEN15: CHL1, VIK1, CSM3, BIM1, TOF1, and KAR3 were deleted in strain YPH1444, which expresses a Lac repressor-GFP fusion protein and contains a Lac operator repeat integrated 1.8 kb from the centromere of chromosome XV. The number of GFP signals in each cell was scored for two independent strains for each deletion in two independent experiments. At least 200 cells were scored for each strain. (B) Sister chromatid cohesion quantified. The average percentage of nocodazole-arrested cells with two GFP signals from two strains in two independent experiments is given along with the SD.
Finally, we measured cohesion in a top1Δ mutant, because Tof1p is reported to physically interact with Top1p (Park and Sternglanz, 1999) and because topoisomerase II has been implicated in sister chromatid cohesion (Bachant et al., 2002). We found no detectable defect in cohesion at this arm site in the top1Δ mutant (Figure 2).
Analysis of Sister Chromatid Cohesion at a Locus 12 kb from CEN4
We assessed sister chromatid cohesion at a second arm locus in chl1Δ, kar3Δ, vik1Δ, bim1Δ, tof1Δ, and csm3Δ strains. The genes were individually deleted in strain Y819, which contains Lac operator repeats integrated 12 kb from the centromere of chromosome IV and expresses a Lac repressor-GFP fusion protein. As with the chromosome V locus, deletion of each of these genes resulted in a significant increase in defective sister chromatid cohesion at this locus (Figure 3). Although 9.6% of wild-type (WT) cells arrested in nocodazole had two GFP dots, 29% of chl1Δ cells had two GFP signals (Figure 3B). This increase in the percentage of cells with two GFP signals could be attributed to defective sister chromatid cohesion and not to high levels of polyploidy because chl1Δ cells arrested in G1 had about the same percentage of cells with two GFP signals as did the WT strain (3.7% in chl1Δ and 2.5% in WT) (Figure 3A). Similarly, deletion of CSM3, VIK1, TOF1, BIM1, or KAR3 resulted in an increased percentage of G2/M-arrested cells with two GFP dots (15.1, 14.6, 25.1, 19.5, and 25.6% respectively), and this increase was not due to excessive polyploidy in these cells (Figure 3).
Characterization of Sister Chromatid Cohesion at a CEN Locus
Centromeres in S. cerevisiae have previously been found to have unique features with respect to sister chromatid cohesion. Mcd1p/Scc1p and Smc1p have been shown by chromatin immunoprecipitation experiments to bind to defined regions along the chromosome with increased abundance at and around centromeres (Blat and Kleckner, 1999; Megee and Koshland, 1999; Tanaka et al., 1999; Laloraya et al., 2000). Centromere DNA has also been shown to nucleate binding of the cohesin subunit Mcd1p/Scc1p, resulting in the association of Mcd1p/Scc1p with at least 2 kb of DNA flanking the centromere sequence (Megee and Koshland, 1999; Tanaka et al., 1999). To assess sister chromatid cohesion at a CEN locus, KAR3, BIM1, CHL1, CSM3, TOF1, and VIK1 were individually deleted in YPH1444, which has a Lac operator repeat integrated 1.8 kb from the centromere of chromosome XV and expresses a Lac repressor-GFP fusion protein. Deletion of CHL1 resulted in 25.8% of nocodazole-arrested chl1Δ cells having two GFP dots compared with only 4.3% in WT cells (Figure 4). When chl1Δ cells were arrested in G1, 0.83% of cells had two GFP dots, whereas 0.5% of WT cells arrested in G1 had two GFP dots, indicating that the high percentage of separated signals in nocodazole-arrested chl1Δ cells is not due to increased polyploidy. Thus, CHL1 is required for efficient cohesion at a centromeric locus on chromosome XV. Likewise, deletion of CSM3, VIK1, TOF1, BIM1, or KAR3 increased the percentage of nocodazole-arrested cells with two GFP dots (15.8, 9.6, 22, 13.8, and 23.7%, respectively), indicating that these genes are also important for efficient cohesion at this centromere proximal locus (Figure 4). Collectively, these data indicate that KAR3, CHL1, CSM3, TOF1, BIM1, and VIK1 are required for efficient sister chromatid cohesion at both CEN and arm loci.
Identification of Protein Complexes Important for Cohesion
Kar3p has been reported to interact with two accessory proteins, Vik1p and Cik1p (Page et al., 1994; Manning et al., 1999). Whereas kar3Δ, cik1Δ, and vik1Δ were identified in our initial screen as having synthetic genetic interactions with ctf8Δ, only the interactions with kar3Δ and vik1Δ could be confirmed by tetrad analysis, and only kar3Δ and vik1Δ were found to have mitotic cohesion defects. The reported physical association between Kar3p and Vik1p, two of six proteins found to have roles in sister chromatid cohesion in the course of this work, encouraged us to look for protein interactions with the six genes found to have a role in sister chromatid cohesion. We tagged Vik1p, Chl1p, and Csm3p with 13Myc epitopes at their C termini by integrating sequences encoding the tag into their respective chromosomal loci. Vik1-13Myc, Chl1-13Myc, and Csm3-13Myc were immunoprecipitated from cell extracts and the immunoprecipitates were run on a 10% polyacrylamide gel that was then silver stained (Figure 5A). Two specific proteins were evident in the Vik1-13Myc immunoprecipitate; one was Vik1–13Myc, whereas the other protein was identified as Kar3p using mass spectrometric analysis (Figure 5A). We were unable to identify any prominent specific bands in the Chl1-13Myc immunoprecipitate, other than Chl1-13Myc itself (Figure 5A). We identified two specific proteins in the Csm3-13Myc immunoprecipitate, one of which corresponded to Csm3-13Myc. Mass spectrometry was used to identify the other protein as Tof1p, one of the other gene products we found to be required for efficient sister chromatid cohesion (Figure 5A).
Figure 5.
(A) Identification of proteins that physically interact with Chl1p, Csm3p, and Vik1p. Chl1p, Csm3p, and Vik1p were epitope tagged with 13Myc epitopes at their C termini under their endogenous promoter. Cellular extracts were prepared from these strains and an isogenic wild-type strain. Immunoprecipitations were performed from equal amounts of protein (500–700 mg), and the immunoprecipitates were run on a 10% SDS-PAGE gel, which was subsequently silver stained. The identity of unique protein bands was determined by mass spectrometry. (B) Confirmation of protein interactions. Tof1p, Csm3p, and Top1p were tagged at their C termini with either 13Myc or 3HA epitopes at the endogenous loci. Immunoprecipitations were performed from equal amounts of protein using anti-HA conjugated beads. Western blot analysis of the immunoprecipitate was performed using either anti-HA or anti-Myc monoclonal antibodies as indicated.
To confirm the physical interaction between Tof1p and Csm3p suggested by mass spectrometric analysis, we tagged Tof1p with 3HA epitopes at its C terminus. Tof1–3HA was expressed from its endogenous promoter. Immunoprecipitation of Tof1–3HA with anti-HA conjugated beads specifically coimmunoprecipitated Csm3–13Myc from extracts prepared from Tof1-3HA Csm3-13Myc cells (Figure 5B). The association of Tof1-3HA with Csm3-13Myc was also observed when the immunoprecipitation was carried out with anti-Myc–conjugated beads (our unpublished data). We conclude that Csm3p and Tof1p physically interact in vivo.
Tof1p was first identified as a topoisomerase-interacting protein by two-hybrid analysis using the central domain (not containing the active site) of topoisomerase I as bait (Park and Sternglanz, 1999). These data prompted us to investigate whether we could confirm a physical interaction between topoisomerase I (encoded by the TOP1 gene) and Csm3p or Tof1p by coimmunoprecipitation. We tagged Top1p with either 13Myc or 3HA epitopes at its C terminus. We found no physical association between Top1-3HA and Csm3-13Myc or between Tof1-3HA and Top1-13Myc under the same experimental conditions used to coimmunoprecipitate Csm3-13myc and Tof1-3HA (Figure 5B).
DISCUSSION
Because ctf8 is synthetically lethal with mutations in essential genes required for cohesion, we hypothesized that a synthetic lethality screen using a ctf8Δ strain as a bait may identify other genes required for sister chromatid cohesion. The only other nonessential gene previously shown to be required for sister chromatid cohesion, other than the nonessential genes in the Ctf18–RFC complex, is CTF4. Isolation of ctf4Δ in the ctf8Δ SGA analysis indicated that this screen was successful in identifying other nonessential genes required for sister chromatid cohesion. In addition to CTF4, six other genes isolated in the ctf8Δ SGA screen were found to be required for efficient sister chromatid cohesion: BIM1, CSM3, TOF1, VIK1, KAR3, and CHL1. Warren et al. (2004; this issue) describe a similar screen, by using a chip-based method, to isolate genes that are synthetically lethal with ctf4Δ. From the list of interactors, they define 17 deletion mutants that exhibit cohesion defects, three of which were also found in our screen: kar3Δ, tof1Δ, and csm3Δ. Together, these results provide a list of 20 new nonessential genes in addition to CTF4 that define pathways involved in proper sister chromatid cohesion. Moreover, our study shows that the proteins encoded by four of the six identified genes in our screen form two distinct complexes: Kar3p and Vik1p, which had been reported previously (Manning et al., 1999), and Csm3p and Tof1p. Thus, our data demonstrate that a synthetic genetic array screen followed by cohesion assays as a secondary screen was a successful approach to identify novel protein complexes involved in sister chromatid cohesion.
DNA Topology and Sister Chromatid Cohesion
In all of our cohesion assays, chl1 mutants consistently had cohesion defects of similar magnitude to that observed in ctf8, dcc1, and ctf18 mutants; ∼30% of nocodazole arrested chl1 cells had 2 GFP signals. CHL1 was previously identified in two screens designed to isolate mutations in genes required for high fidelity chromosome segregation (Spencer et al., 1990; Kouprina et al., 1993). Sequence analysis reveals that Chl1p is an evolutionarily conserved DEAH family helicase (Shiratori et al., 1999) and human Chl1 (hChlR1) has been shown to possess both ATPase and DNA helicase activities (Hirota and Lahti, 2000). Consistent with CHL1 having a role in sister chromatid cohesion, we find that deletion of CHL1 is synthetically lethal with scc1-73, ctf7-203, smc1-259, scc2-4, and smc3-42 (our unpublished data). Furthermore, deletion of CHL1 results in an accumulation of cells with G2/M DNA content that is dependent on the mitotic spindle checkpoint gene MAD2 (Li and Murray, 1991; our unpublished data). Just as Smt3p/SUMO-1 modification of Top2p may play a role in establishing a modified chromosome structure or localized catenations that are required for efficient centromeric cohesion (Bachant et al., 2002), Chl1p's strong sequence homology to helicases suggests that Chl1p may function to alter DNA topology during the establishment or maintenance of sister chromatid cohesion. The observation that Chl1p is required for efficient sister chromatid cohesion adds to a growing body of evidence suggesting that DNA topology is critical to proper cohesion.
Identification of Protein Complexes Required for Efficient Sister Chromatid Cohesion
Csm3p was previously found to be required for high-fidelity chromosome segregation in meiosis (Rabitsch et al., 2001). Consistent with the role of Csm3p in chromosome transmission, we find that CSM3 is required for efficient sister chromatid cohesion in mitosis. Furthermore, we find that Csm3p physically interacts with Tof1p. A physical interaction between Csm3p and Tof1p had previously been suggested by two-hybrid analysis (Ito et al., 2001). We also isolated tof1Δ in the ctf8Δ SGA screen and found that TOF1 is required for efficient cohesion. Tof1p was previously reported to interact in a two-hybrid assay with the central domain of topoisomerase I (Park and Sternglanz, 1999). Tof1-GST expressed in Escherichia coli has also been shown to physically associate with the central domain of topoisomerase I in an in vitro binding assay (Park and Sternglanz, 1999). Given the effect of defective Smt3p modification of Top2p on centromeric cohesion (Bachant et al., 2002), we tested whether Top1p physically interacted with either Csm3p or Tof1p. We were unable to identify any physical interaction between Csm3-13Myc and Top1-3HA or between Top1-13Myc and Tof1-3HA by coimmunoprecipitation experiments. It is therefore unclear whether full-length Top1p physically interacts with Tof1p or Csm3p in vivo. Deletion of TOP1 had no significant effect on sister chromatid cohesion at an arm locus 35 kb from CEN5. Thus, although the Csm3p–Tof1p protein complex seems to be required for efficient sister chromatid cohesion, we were unable to find any interaction between this complex and Top1p, and deletion of TOP1 seemed to have no significant effect on cohesion.
Tof1p also functions in the checkpoints that respond to DNA damage or replication fork stalling during S phase and is important for activation of the Rad53 checkpoint protein kinase (Foss, 2001). Tof1p was recently shown to interact with Cdc45p, a component of the DNA replication machinery, and to move with the replication machinery as DNA synthesis proceeds during S phase. This suggested that Tof1p functions during checkpoint response in a replication-pausing complex that ensures that the replication complex does not uncouple from sites of DNA synthesis (Katou et al., 2003). The role of Tof1p in sister chromatid cohesion suggests a functional link between checkpoint pathways, DNA replication, and cohesion. It is currently unclear whether efficient cohesion requires intact checkpoint pathways, or whether Tof1p performs two separable functions. However, it is becoming increasingly evident that S phase checkpoint pathways are critical for replication fork integrity (Lopes et al., 2001; Tercero and Diffley, 2001; Sogo et al., 2002), and given the links between DNA replication and the establishment of sister chromatid cohesion, it is possible that checkpoint pathways contribute directly to efficient cohesion. Interestingly, Tof1p shows limited homology to a C. elegans protein of the TIMELESS family, TIM-1, that is essential for proper chromosome segregation in mitosis and meiosis and was recently shown to interact with cohesin, and hypothesized to facilitate cohesin loading or stability (Chan et al., 2003).
KAR3 encodes a microtubule motor protein with minus end directionality (Endow et al., 1994). Kar3p functions in a range of cellular processes, including spindle integrity, nuclear fusion during mating (Meluh and Rose, 1990), and mitotic spindle positioning (Cottingham et al., 1999). Kar3p physically associates with two homologous accessory factors, Cik1p and Vik1p (Page et al., 1994; Manning et al., 1999). The Kar3p–Cik1p and the Kar3p–Vik1p complexes seem to be functionally distinct. For example, Kar3p complexed with Cik1p, but not Vik1p, has been implicated in spindle positioning (Cottingham et al., 1999) and karyogamy (Page et al., 1994; Manning et al., 1999). Although we identified kar3Δ, vik1Δ, and cik1Δ in the ctf8Δ SGA screen, we found that only KAR3 and VIK1 were required for efficient sister chromatid cohesion. It is therefore likely that the role of Kar3p in cohesion is mediated through its interaction with Vik1p rather than Cik1p. Interestingly, Kar3p has previously been reported to interact with the cohesin protein Smc1p in a two-hybrid assay (Newman et al., 2000).
BIM1 and Sister Chromatid Cohesion
Bim1p is a microtubule binding protein that interacts with the plus end of microtubules and with the spindle itself (Schwartz et al., 1997; Tirnauer et al., 1999). Bim1p is a critical element of the Kip3p pathway that ensures correct orientation of the mitotic spindle (Tirnauer et al., 1999; Lee et al., 2000), and as such is important for polarized cell growth. SGA analysis with bim1Δ identified genetic interactions with spindle assembly checkpoint genes and genes encoding kinetochore components, suggesting that Bim1p might also be involved in the attachment of microtubules to the kinetochore (Tong et al., 2001). Although a role for Bim1p in sister chromatid cohesion had not previously been hypothesized, there are several links between Bim1p function and accurate chromosome segregation. The bim1Δ SGA identified genetic interactions with the Ctf18-RFC genes DCC1 and CTF8 (Tong et al., 2001), suggesting a functional relationship between BIM1 and Ctf18-RFC. bim1Δ is also synthetically sick with csm3Δ (Tong et al., 2001), which we find is important for sister chromatid cohesion. Genetic interactions with the spindle checkpoint genes (Tong et al., 2001) could be due to defective cohesion in bim1Δ, rather than to defective microtubule attachment or spindle positioning. Finally, mutations in the S. pombe homologue of BIM1, mal3+, cause a 400-fold increase in chromosome loss (Beinhauer et al., 1997). We find that deletion of BIM1, in addition to being synthetically lethal with ctf8Δ, confers a two- to threefold increase in premature separation of sister chromatids. This suggests that efficient sister chromatid cohesion might be important for establishing or maintaining the position of the mitotic spindle. Alternatively, the cohesion defect in bim1Δ might reflect a separate function for Bim1p, distinct from its role in spindle orientation. This additional function of Bim1p is of particular interest given the connections between the human Bim1p homologue EB1, the adenomatous polyposis coli tumor suppressor, and genetic instability in human cancers (Su et al., 1995).
Acknowledgments
We thank Tushara Weerasooriya for assistance with the SGA analysis. This work was supported by grants from the Canadian Institutes of Health Research (to P.H., G.W.B., and C.B.), the National Cancer Institute of Canada (to C.B.), and the National Institutes of Health (to P.H.). I.P. was supported by a National Cancer Institute of Canada Research Studentship.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–08–0619. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–08–0619.
References
- Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N., and Elledge, S.J. (2002). The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9, 1169-1182. [DOI] [PubMed] [Google Scholar]
- Beinhauer, J.D., Hagan, I.M., Hegemann, J.H., and Fleig, U. (1997). Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139, 717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, V.P., Maniwa, Y., Tappin, I., Ozato, K., Yokomori, K., and Hurwitz, J. (2003). The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc. Natl. Acad. Sci. USA 100, 10237-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., and Kleckner, N. (1999). Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98, 249-259. [DOI] [PubMed] [Google Scholar]
- Chan, R.C., Chan, A., Jeon, M., Wu, T.F., Pasqualone, D., Rougvie, A.E., and Meyer, B.J. (2003). Chromosome cohesion is regulated by a cloch gene paralogue TiM-1. Nature 423, 1002-1009. [DOI] [PubMed] [Google Scholar]
- Budd, M.E., and Campbell, J.L. (2000). The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459, 173-186. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Shirayama, M., Shevchenko, A., Tanaka, T., Toth, A., and Nasmyth, K. (2000). Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243-254. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., and Nasmyth, K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., Peters, J.M., Kirschner, M.W., and Koshland, D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081-3093. [DOI] [PubMed] [Google Scholar]
- Cottingham, F.R., Gheber, L., Miller, D.L., and Hoyt, M.A. (1999). Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147, 335-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S.A., Kang, S.J., Satterwhite, L.L., Rose, M.D., Skeen, V.P., and Salmon, E.D. (1994). Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 13, 2708-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J., McCormack, A.L., and Yates, J.R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976-989. [DOI] [PubMed] [Google Scholar]
- Formosa, T., and Nittis, T. (1999). Dna2 mutants reveal interactions with DnNA polymerase alpha and Ctf4, a Pol alpha accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics 151, 1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss, E.J. (2001). Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157, 567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619-633. [DOI] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi, S.P., Corthals, G.L., Zhang, Y., Rochon, Y., and Aebersold, R. (2000). Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA 97, 9390-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, J.S., Kroll, E.S., Lundblad, V., and Spencer, F.A. (2001). Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell Biol. 21, 3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, T., Stead, K., Koshland, D., and Guacci, V. (2000). Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151, 613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, Y., and Lahti, J.M. (2000). Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 28, 917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou, Y., Kanoh, Y., Bando, M., Noguchi, H., Tanaka, H., Ashikari, T., Sugimoto, K., and Shirahige, K. (2003). S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078-1083. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., Tsouladze, A., Koryabin, M., Hieter, P., Spencer, F., and Larionov, V. (1993). Identification and genetic mapping of CHL genes controlling mitotic chromosome transmission in yeast. Yeast 9, 11-19. [DOI] [PubMed] [Google Scholar]
- Laloraya, S., Guacci, V., and Koshland, D. (2000). Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151, 1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.R., Michaud, W.A., Sikorski, R.S., and Hieter, P.A. (1994). Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 13, 4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L., Tirnauer, J.S., Li, J., Schuyler, S.C., Liu, J.Y., and Pellman, D. (2000). Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287, 2260-2262. [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray, A.W. (1991). Feedback control of mitosis in budding yeast. Cell 66, 519-531. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398, 246-251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell Biol. 20, 2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lopes, M., Cotta-Ramusino, C., Pellicioli, A., Liberi, G., Plevani, P., Muzi-Falconi, M., Newlon, C.S., and Foiani, M. (2001). The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557-561. [DOI] [PubMed] [Google Scholar]
- Manning, B.D., Barrett, J.G., Wallace, J.A., Granok, H., and Snyder, M. (1999). Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 144, 1219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M.L., Gygi, S.P., Abersold, R., and Hieter, P. (2001). Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell 7, 959-970. [DOI] [PubMed] [Google Scholar]
- Measday, V., Hailey, D.W., Pot, I., Givan, S.A., Hyland, K.M., Cagney, G., Fields, S., Davis, T.N., and Hieter, P. (2002). Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16, 101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P.C., and Koshland, D. (1999). A functional assay for centromere-associated sister chromatid cohesion. Science 285, 254-257. [DOI] [PubMed] [Google Scholar]
- Meluh, P., and Rose, M.D. (1990). KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60, 1029-1041. [DOI] [PubMed] [Google Scholar]
- Merkle, C.J., Karnitz, L.M., Henry-Sanchez, J.T., and Chen, J. (2003). Cloning and characterization of hCTF18, hCTF8, and hDCC 1, Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J. Biol. Chem. 278, 30051-30056. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- Miles, J., and Formosa, T. (1992a). Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol. Cell Biol. 12, 5724-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, J., and Formosa, T. (1992b). Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc. Natl. Acad. Sci. USA 89, 1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki, T., Kondo, T., Nakada, D., Matsumoto, K., and Sugimoto, K. (2001). Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell Biol. 21, 5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, J.R., Wolf, E., and Kim, P.S. (2000). A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 13203-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, B.D., Satterwhite, L.L., Rose, M.D., and Snyder, M. (1994). Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J. Cell Biol. 124, 507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza, S., Tanaka, T., Hochwagen, A., Eisenhaber, F., and Nasmyth, K. (2000). Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10, 1557-1564. [DOI] [PubMed] [Google Scholar]
- Park, H., and Sternglanz, R. (1999). Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast 15, 35-41. [DOI] [PubMed] [Google Scholar]
- Rabitsch, K.P., et al. (2001). A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11, 1001-1009. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M., and Elledge, S.J. (1999). Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286, 1166-1171. [DOI] [PubMed] [Google Scholar]
- Schwab, M., Lutum, A.S., and Seufert, W. (1997). Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90, 683-693. [DOI] [PubMed] [Google Scholar]
- Schwartz, K., Richards, K., and Botstein, D. (1997). BIM1 encodes a microtubule-binding protein in yeast. Mol. Biol. Cell 8, 2677-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori, A., Shibata, T., Arisawa, M., Hanaoka, F., Murakami, Y., and Eki, T. (1999). Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and northern analysis. Yeast 15, 219-253. [DOI] [PubMed] [Google Scholar]
- Skibbens, R.V., Corson, L.B., Koshland, D., and Hieter, P. (1999). Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599-602. [DOI] [PubMed] [Google Scholar]
- Spencer, F., Gerring, S.L., Connelly, C., and Hieter, P. (1990). Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124, 237-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L.K., Burrell, M., Hill, D.E., Gyuris, J., Brent, R., Wiltshire, R., Trent, J., Vogelstein, B., and Kinzler, K.W. (1995). APC binds to the novel protein EB1. Cancer Res. 55, 2972-2977. [PubMed] [Google Scholar]
- Tanaka, T., Cosma, M.P., Wirth, K., and Nasmyth, K. (1999). Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98, 847-858. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., and Diffley, J.F. (2001). Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553-557. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., O'Toole, E., Berrueta, L., Bierer, B.E., and Pellman, D. (1999). Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145, 993-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers, M., Tokiwa, G., Nash, R., and Futcher, B. (1992). The Cln3 Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 11, 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann, F., Lottspeich, F., and Nasmyth, K. (1999). Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37-42. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., Wernic, D., Poupart, M.A., Koonin, E.V., and Nasmyth, K. (2000). Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375-386. [DOI] [PubMed] [Google Scholar]
- Visintin, R., Prinz, S., and Amon, A. (1997). CDC20 and CDH 1, a family of substrate-specific activators of APC-dependent proteolysis. Science 278, 460-463. [DOI] [PubMed] [Google Scholar]
- Waga, S., and Stillman, B. (1998). The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721-751. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., De Las Penas, A., Adams, C., and Christman, M.F. (2000). Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science 289, 774-779. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., Eckley, D.M., Lee, M.S., Hanna, J.S., Hughes, A., Peyser, B., Jie, C., Irizarry, R., and Spencer, F.A. (2004) S-phase checkpoint genes safeguard high fidelity sister chromatid cohesion. Mol. Biol. Cell 15, 1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]