Abstract
Human cholesteryl ester transfer protein (CETP) mediates the net transfer of cholesteryl ester mass from atheroprotective high-density lipoproteins to atherogenic low-density lipoproteins by an unknown mechanism. Delineating this mechanism would be an important step toward the rational design of new CETP inhibitors for treating cardiovascular diseases. Using EM, single-particle image processing and molecular dynamics simulation, we discovered that CETP bridges a ternary complex with its N-terminal β-barrel domain penetrating into high-density lipoproteins and its C-terminal domain interacting with low-density lipoprotein or very-low-density lipoprotein. In our mechanistic model, the CETP lipoprotein-interacting regions, which are highly mobile, form pores that connect to a hydrophobic central cavity, thereby forming a tunnel for transfer of neutral lipids from donor to acceptor lipoproteins. These new insights into CETP transfer provide a molecular basis for analyzing mechanisms for CETP inhibition.
CETP mediates the transfer of neutral lipids, including cholesteryl esters and triglycerides, between high-density lipoproteins (HDL), low-density lipoproteins (LDL) and very-low-density lipoproteins (VLDL)1. Elevated LDL cholesterol (LDL-C) level and/or low HDL cholesterol (HDL-C) level in human plasma are major risk factors for cardiovascular disease2,3. As increased CETP level can reduce HDL-C concentration4 and CETP deficiency is associated with elevated HDL-C5,6, CETP inhibitors, including torcetrapib, anacetrapib and dalcetrapib, have been investigated in clinical trials for treating cardiovascular disease7–9. Despite the intense clinical interest in CETP inhibition, little is known concerning the molecular mechanisms of CETP-mediated lipid transfer among lipoproteins or even how CETP interacts with lipoproteins.
CETP is a hydrophobic glycoprotein of 476 amino acids (~53 kDa before post-translational modification). Its crystal structure reveals a banana-shaped molecule with N- and C-terminal β-barrel domains, a central β-sheet and an ~60-Å-long hydrophobic central cavity. The cavity, which can accommodate two cholesteryl ester molecules, communicates with two pores near the central β-sheet domain. These pores, which are occupied by two phospholipid molecules, could be gates for the interaction of the central cavity with the aqueous environment or lipoproteins10.
Three CETP neutral lipid transfer hypotheses were proposed in 1980, 1982 and 1993, respectively: (i) a shuttle mechanism that involves CETP collecting cholesteryl esters from one lipoprotein and delivering them through the aqueous phase to another lipoprotein11; (ii) a tunnel mechanism in which CETP bridges two lipoproteins to form a ternary complex, with lipids flowing from the donor to acceptor lipoprotein through the CETP molecule12; and (iii) a modified tunnel mechanism implicating a CETP dimer13.
One difficulty in investigating CETP mechanisms using structural methods is that interaction with CETP can alter the size, shape and composition of lipoproteins, especially HDL14–16. We validated an optimized negative-staining (OpNS) EM protocol14,17 in which flash fixation of lipoprotein particles preserves a near native-state conformation for direct visualization of individual macromolecular particles. We applied this protocol to study the mechanisms by which CETP interacts with spherical HDL, LDL and VLDL. Three-dimensional reconstructions of CETP, free and HDL bound, were obtained by single-particle techniques. In addition, we used inhibitory CETP-specific antibodies to identify the regions of CETP that interact with HDL and LDL. Finally, molecular dynamics simulation was used to assess the molecular mobility of CETP and predict the most likely conformational changes that are associated with lipid transfer.
RESULTS
Structure of CETP using EM
Conventional cryo-EM is often the method of choice for studies of protein structure under physiological conditions because it avoids the potential artifacts induced by fixatives and stains17. Still, cryo-EM studies of CETP are challenging; small molecules (<200 kDa) are difficult to image or reconstruct by the cryo-EM single-particle approach because of low contrast18. Thus, we studied human recombinant CETP by using OpNS14,17 and a cryo–positive-staining EM (cryo-PS-EM) protocol.
Our OpNS protocol, refined from the conventional negative-staining protocol, eliminates rouleaux artifacts of lipoprotein particles and was statistically validated as a way to determine lipoprotein particle shapes and sizes14,17. The cryo-PS-EM protocol was developed by combining our OpNS14,17 and conventional cryo-EM protocols19–21. Instead of air-drying the sample in the last step of the negative-staining protocol, we flash-froze the sample in liquid nitrogen. As the cryo-EM image of a particle has reversed contrast compared to that produced using the reported cryo-NS protocol22 but has image contrast consistent with that of a conventional cryo-EM image, we call it the cryo-PS-EM protocol.
We compared the particle shape and size in images of CETP with that in the CETP crystal structure (Protein Data Bank code 2OBD10). Survey-OpNS-EM micrographs and selected particle views revealed the expected banana-shaped CETP with dimensions of 125 Å × 30 Å (Supplementary Results, Supplementary Fig. 1a,b). When the CETP crystal structure is overlaid on a reference-free class average of OpNS-EM images, a near-perfect match in shape and size is found (Supplementary Fig. 1c, sd), and it is notable that even the concave surface, C-terminal end (more globular) and N-terminal end (more tapered) of CETP are readily distinguished (Supplementary Fig. 1d). These studies validate direct OpNS-EM as a way to visualize the structure of CETP in other settings in which it associates with various lipoproteins.
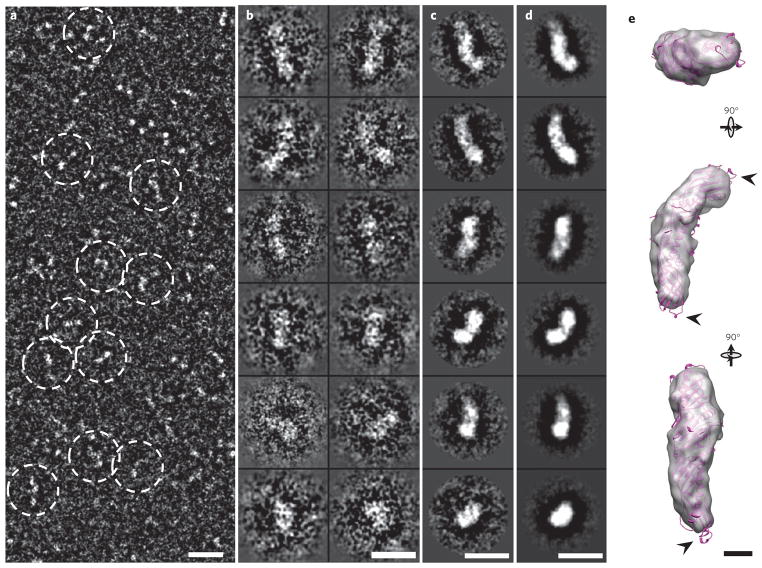
Survey cryo-PS-EM micrographs (Fig. 1a and Supplementary Fig. 2a) and selected particle views (Fig. 1b and Supplementary Fig. 2b) also showed the banana-shaped CETP with a shape and dimensions similar to those observed in micrographs from OpNS-EM (Supplementary Fig. 1). These cryo-PS-EM images of CETP were also used to reconstruct a three-dimensional density map of CETP using the single-particle reconstruction method21. Again, the class averages and the projections of the reconstructed three-dimensional density map show a banana-shaped density (~125 Å × 30 Å) with a shape and size similar to those of the crystal structure (Fig. 1c–e and Supplementary Fig. 2c–g) at ~13-Å resolution (Supplementary Fig. 3); once again, we observed a near-perfect fit of the crystal structure into the envelope of the three-dimensional density map, suggesting that the density map can be used to determine the CETP concave surface orientation (Fig. 1e and Supplementary Fig. 2f,g), with the C-terminal (more globular end) and N-terminal (more tapered end) domains at the ends of the long CETP axis. Some hydrophobic loops were observed outside the CETP envelope at the distal portions of the N- and C-terminal β-barrel domains, suggesting that these regions have structural flexibility (arrows in Fig. 1e).
Figure 1. Three-dimensional reconstruction of CETP by cryo-PS-EM.
(a) Survey view (reversed contrast) of CETP molecule alone (dashed circles). (b–d) Twelve representative views of selected and windowed raw particles of CETP (b) and their class averages (c) are compared with corresponding views of the projections of the three-dimensional reconstruction (d). (e) The crystal structure of CETP (purple) inserted into the three-dimensional EM density map (~13-Å resolution) demonstrates a near-perfect match to the density map in shape and size, except for the distal portions of the N- and C-terminal β-barrel domains, segments of which protrude (arrowheads) from the density envelope. Scale bars: a–d, 100 Å; e, 20 Å.
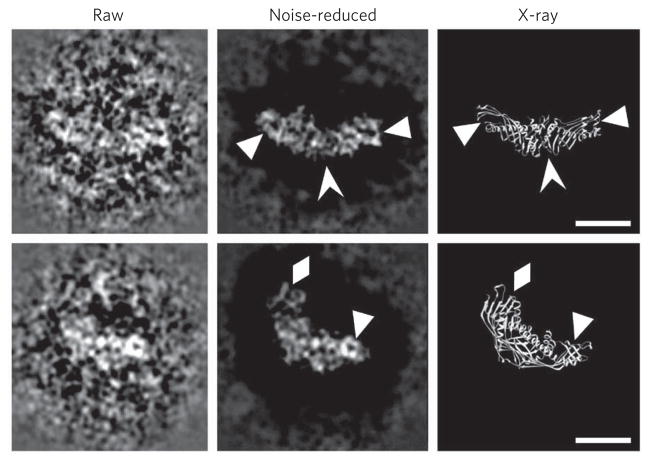
Notably, some raw particle images (~50%) show substantial structural detail (Fig. 2 and Supplementary Fig. 4). For example, at specific viewing orientations, a manual noise-decreasing procedure performed on the raw particle images near the particle boundary (Fig. 2 and Supplementary Fig. 4) revealed that the particle outer features are remarkably similar to the crystal structure (Fig. 2 and Supplementary Fig. 4). Thus, the particle internal features show parallel fringes that are well matched to the β-sheet strands within the C-terminal β-barrel domain of the crystal structure as well as to end loops and holes (Fig. 2 and Supplementary Fig. 4). Imaging of internal structural features was achieved by using EM imaging conditions of near-Scherzer focus (~0.1 μm) and higher doses (~60–70 e− Å−2) rather than conventional cryo-EM conditions such as high defocus (~2–3 μm) and low doses (~20 e− Å−2). These high-resolution images show that CETP can be visualized directly by this cryo-PS-EM protocol, confirming our findings above using OpNS-EM. Moreover, the cryo-PS-EM images show that the staining reagent, uranyl formate, penetrates the molecular surface, challenging the conventional view that staining can visualize only the outer surface structure23. The mechanism of how uranyl formate penetrates the molecular surface is unknown. One possibility is that the uranyl cation binds available protein carboxyl groups, and thus the surrounding vitreous ice is of lower density than the staining of the protein and uranyl groups, thereby acting as a positive.
Figure 2. High-resolution CETP images by cryo-PS-EM.
Two selected high-resolution and raw particle images of CETP (contrast reversed, left column) were manually noise reduced around the raw particle images’ edges for easy visualization (middle column). The latter images can be compared with those of the X-ray crystal structure’s main chain at a similar orientation (right column). The high-resolution raw image areas show detailed structural features of CETP. These particles have very similar features to the crystal structure; for example, the small circular holes near both distal ends can be visualized in the noise-reduced images (triangles in both rows); further, the striations in the C-terminal domains of the images match the β-sheets of the crystal structure (arrowheads in top row), and loops protruding from the C-terminal end of the CETP images are similar to the crystal structure (diamonds in bottom row). Scale bars, 50 Å.
Even though we show that unexpectedly detailed secondary structural elements can be observed with the cryo-PS-EM method in individual images, the final three-dimensional reconstruction did not show the high-resolution details shown in our cryo-PS-EM two-dimensional images. This may be due to (i) the flexibility of the entire CETP molecule, (ii) variation of two-dimensional image quality in that only ~50% of these images contain more or less high-resolution structural details (the loss of detail may be due to the variation of the coated stains, particle orientation, ice thickness or a combination of the three) and/or (iii) the limited capability of the two-dimensional class-average program used for three-dimensional reconstructions. Therefore, the high-resolution details were quickly smeared after the class-average process. Because the three-dimensional reconstructions from cryo-PS-EM images provided no additional insights beyond those resulting from our OpNS-EM images and the cryo-PS-EM is a time-consuming procedure, the simpler, OpNS-EM protocol was used for the remainder of experiments (described below).
Structure of the HDL–CETP complex by electron microscopy
To determine how CETP interacts with HDL, we incubated CETP with HDL at CETP:HDL molar ratios between 0.5:1 and 5:1 and examined the products using the OpNS-EM protocol. We observed spherical HDL particles (diameter ~85–110 Å) with a single protruding feature that was not seen in the absence of CETP, and we concluded that the protrusion is CETP that is part of a binary complex (Fig. 3a,b and Supplementary Figs. 5 and 6); less than ~10% of HDL–CETP complexes have two or more protruding CETP molecules that often share the same half of the HDL surface (Supplementary Fig. 7). No CETP molecule that was configured with its concave surface adjacent to the convex surface of HDL or that bridged two HDL particles to form a ternary complex was observed, suggesting that CETP has HDL binding directionality. As the total number of CETP molecules bound to HDL can be greater than the total number of apolipoprotein A-I (apoA-I) molecules in HDL, apoA-I is not likely to be involved in CETP binding, supporting the previous model based on protein-free vesicles known as liposomes24,25.
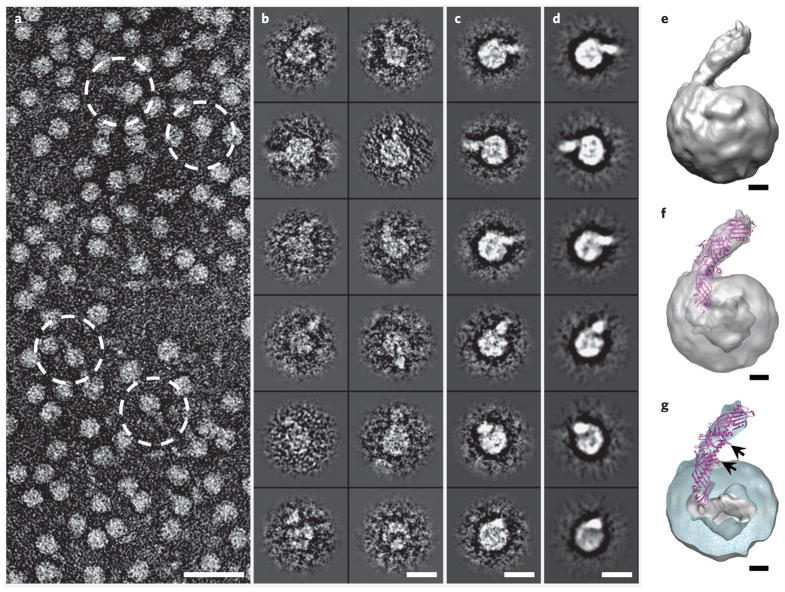
Figure 3. Three-dimensional reconstruction of HDL–CETP complex by OpNS-EM.
(a) Survey view of the negative-stained EM structure of HDL–CETP complexes (dashed circles). (b–d) Twelve representative views of selected and windowed individual raw particles of HDL–CETP complexes (b) and their corresponding class averages (c) are compared with the same views of the reconstructed projections (d). (e) The three-dimensional density map (~14-Å resolution, reconstructed from 6,607 homogenous particle images) shows a binary complex in which the banana-shaped CETP is attached to the HDL surface at a ~45° angle. (f,g) Inserting the CETP crystal structure into the EM density map of an HDL–CETP complex shows the N-terminal β-barrel domain of CETP penetrating the outer shell of HDL and its distal end in the low-density core in transparency (f) and cut-away surface view (g). The cut-away surface view (cyan) in g shows the HDL portion containing a high-density outer shell (~18–27 Å thick) and a low-density cavity (~55 Å in diameter). The black arrows in g point to two phospholipid-binding pores, one of which is adjacent (~15 Å) to the HDL surface, whereas the other is further away, indicating that the phospholipid-binding pores of CETP do not interact directly with the HDL core. Scale bars: a, 300 Å; b–d, 100 Å; e–g, 20 Å.
In binary complexes, the CETP N-terminal domain (more tapered end) appears to interact with HDL. Measurement of more than 100 binary complexes revealed a CETP width of ~30 Å, whereas the length protruding from the HDL surface was ~77 ± 10 Å (Supplementary Fig. 5b), which is notably shorter than the length of CETP alone (~125 Å). This observation suggests that the remainder of CETP (~48 ± 10 Å) penetrates through the HDL surface or, alternatively, partitions tangentially into the HDL lipid surface and that the free-end portion of CETP seems more likely to be the globular end of CETP (C-terminal domain, Supplementary Fig. 5c). This measurement provoked the hypothesis that the distal CETP N-terminal end penetrates into or through the phospholipid portion of the HDL surface under physiological conditions. Preliminary studies of native human HDL, prepared by immunoabsorption, and liposomes show similar CETP-macromolecule relationships, as shown in Figure 3a,b and Supplementary Figures 5, 6a,b and 7.
CETP penetration of HDL revealed by 3D reconstructions
To study the CETP interaction with HDL in more detail, we generated a three-dimensional density map of the HDL–CETP complex from OpNS-EM images (Fig. 3a–e and Supplementary Fig. 6a–f) with the single-particle reconstruction protocol used for LDL three-dimensional reconstructions21. To obtain higher-resolution reconstruction of the HDL–CETP complex, we computationally selected a homogenous population of ~6,600 particle images from an original pool of ~20,000 images using the same batch program that was used for LDL reconstruction21. The convergent reconstruction from this subpopulation of particle images (Fig. 3b,c and Supplementary Fig. 6b,c) provides a statistically defined and robust density map showing the most prominent and reliable structural features of the HDL–CETP complex (Fig. 3d and Supplementary Fig. 6d,e).
A 14-Å-resolution (Supplementary Fig. 3) three-dimensional map reveals a quasispherical HDL (~100 Å × 110 Å × 115 Å) attached to the end of a banana-shaped CETP (~80-Å free end × 45-Å penetrating end × 25-Å width) at a contour level corresponding to the molecular volume of the complex (Fig. 3e and Supplementary Fig. 6f). This spherical HDL contains an 18- to 27-Å high-density outer shell and a low-density core (diameter ~55 Å; Supplementary Fig. 6g), whereas the banana-shaped CETP portion has a concave surface protruding ~45° from the HDL surface. The curvature of the banana-shaped portion is similar to that observed after EM reconstruction of CETP alone (Fig. 1e).
To determine the CETP orientation, we inserted the CETP crystal structure into the banana-shaped portion of the binary HDL–CETP complex density envelope (Fig. 3f) using two structural analysis methods in Chimera26. Using the atom-in-map method26, we fit the CETP C-terminal domain of the crystal structure into the free-end portion of the HDL–CETP density map, producing an average map value fit of 5.373. In contrast, inserting the N-terminal domain into the free end gave a lower value, 5.232. Using the map-in-map method26, a 14-Å-resolution density map generated from the crystal structure was inserted into the free end to calculate the correlation values, which were r = 0.9480 and r = 0.8972 for the C- and N-terminal CETP domains, respectively. Both analyses confirm the interaction of the N-terminal domain with HDL. Inserting the CETP crystal structure into the HDL–CETP envelope shows that 48 Å of the tapered N-terminal end of CETP penetrates the HDL surface (Fig. 3f), reaching the cholesteryl ester–rich, low-density core (Fig. 3g).
Inserting the CETP crystal structure into the binary complex also shows that the phospholipid-binding pores of CETP, which are thought to be involved in lipid transfer, do not interact directly with the cholesteryl ester–rich HDL core. One phospholipid-binding pore is adjacent (~15 Å) to the HDL surface, whereas the other is further away (arrows in Fig. 3g), suggesting that the pore adjacent to the HDL surface may be a neutral lipid pore for cholesteryl ester transfer through the HDL surface. Alternatively, the portion of the N-terminal domain that penetrates into the cholesteryl ester core may generate another, perhaps more efficient, pore for cholesteryl ester transfer.
CETP domains interact with HDL and LDL or VLDL
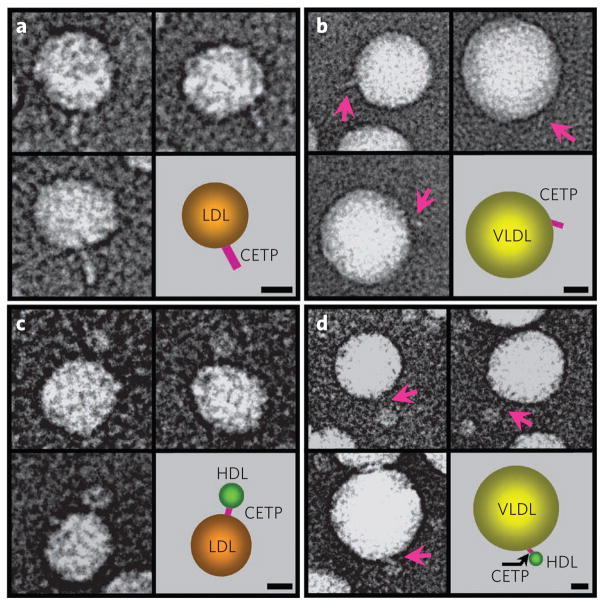
CETP was incubated separately with LDL and VLDL and was examined by the OpNS-EM protocol. Spherical LDL ( diameter ~200–270 Å) and VLDL (diameter ~370–570 Å) particles were observed with a single CETP protruding from their surfaces as part of a binary complex (Fig. 4a,b and Supplementary Fig. 8). Although we observed neither LDL–CETP complexes with two protruding CETP molecules nor two LDLs or VLDLs bridged by one CETP, we did observe occasional VLDL particles with more than one attached CETP molecule. The ability of multiple CETP to attach to one VLDL particle may be due to different surface properties induced by differing apolipoprotein compositions and lipid surface curvatures and areas. Measurements of >100 of these binary complexes revealed that the free-end width of CETP is ~30 Å, similar to that observed for the HDL–CETP complexes. The free-end lengths on LDL and VLDL are ~100 ± 10 Å and ~105 ± 10 Å, respectively, and are shorter than the length of CETP alone (~125 Å), suggesting that the hidden portions of CETP penetrating the LDL and VLDL surfaces are ~25 ± 10 Å and ~20 ± 10 Å, respectively (Fig. 4a,b and Supplementary Fig. 8).
Figure 4. Structural conformations of CETP bound to LDL or VLDL and CETP interactions between lipoproteins by OpNS-EM.
(a) Linear- or banana-shaped CETPs (~100 ± 10 Å long) associated with the surfaces of LDL (diameter ~200–270 Å) particles. (b) Linear-shaped CETPs (~105 ± 10 Å long, pink arrows) protruding from the surfaces of VLDL particles (diameter ~370–570 Å). (c) Linear-shaped CETPs (~25–55 Å long) bridge HDL particles (diameter ~85–110 Å) to LDL particles, forming ternary complexes. (d) VLDL particles connected to HDL particles via linear-shaped CETPs (~35–65 Å long, pink arrows), forming ternary complexes similar to those formed with LDL. The schematic on the bottom right of each panel represents the corresponding complex on the bottom left of each panel. Scale bars, 100 Å.
An antibody can be used as a landmark to define the orientation of protein binding by EM27,28. A domain-specific polyclonal antibody to CETP, H300, whose epitopes are within a region containing the entire C-terminal β-barrel domain and part of the central β-sheet (residues 194–493), was used to identify the parts of CETP that penetrate lipoproteins. The OpNS-EM micrographs of the HDL–CETP complex show a characteristic ‘Y’-shaped density, usually near the free end of CETP (Supplementary Fig. 9). In contrast, LDL–CETP–H300 complexes were rarely seen in the micrographs obtained from the incubations of H300 with LDL and CETP (Supplementary Fig. 10), suggesting that the CETP epitopes to H300 are buried within the LDL particle. Moreover, the percentage of binary complexes (LDL–CETP) formed in the presence of H300 was also lower than that without H300 (<15% versus ~38%), suggesting that H300 inhibits CETP-LDL interaction. These experiments are consistent with the N-terminal domain of CETP interacting with HDL while the C-terminal domain preferentially interacts with LDL. These data are also consistent with the fitting of the crystal structure into the three-dimensional density map and two-dimensional images of the HDL–CETP complex and with the dimensions calculated from the CETP pores to the HDL surface.
CETP bridges HDL to LDL or VLDL to form ternary complexes
After co-incubation of CETP, HDL and LDL, the OpNS-EM micrographs showed ~25% of LDL particles connecting to HDL particles by a ~25- to 55-Å-long CETP bridge (Fig. 4c and Supplementary Fig. 11a–c). The length of the bridge was much shorter than the length of CETP alone (~125 Å), indicating that CETP penetrates into one or both lipoprotein surfaces or cores to form the ternary complex. When co-incubation was repeated with VLDL, ~30% of the VLDL particles were connected to HDL particles by CETP bridges (length ~35–65 Å; Fig. 4d and Supplementary Fig. 11d–f). Unlike LDL, ~30% of the VLDL complexes were bound to more than one HDL–CETP complex, most likely because they contain more protein, which provides more ‘binding sites’; bridges between lipoproteins of the same class were not observed, further supporting the hypothesis that HDL and LDL or VLDL bind different CETP domains. The coexistence of ternary complexes of HDL–CETP–LDL and HDL–CETP–VLDL and lipid transfer (shown in Fig. 5 and Supplementary Fig. 12) are consistent with the mechanistic model of cholesteryl ester transfer through a tunnel within CETP. These observations do not totally exclude the shuttle mechanism, as the ternary complexes coexisted with the binary complexes HDL–CETP and LDL–CETP. However, the tunnel mechanism seems more plausible because it is directly supported by the observation that CETP bridges HDL to LDL or VLDL to form a ternary complex.
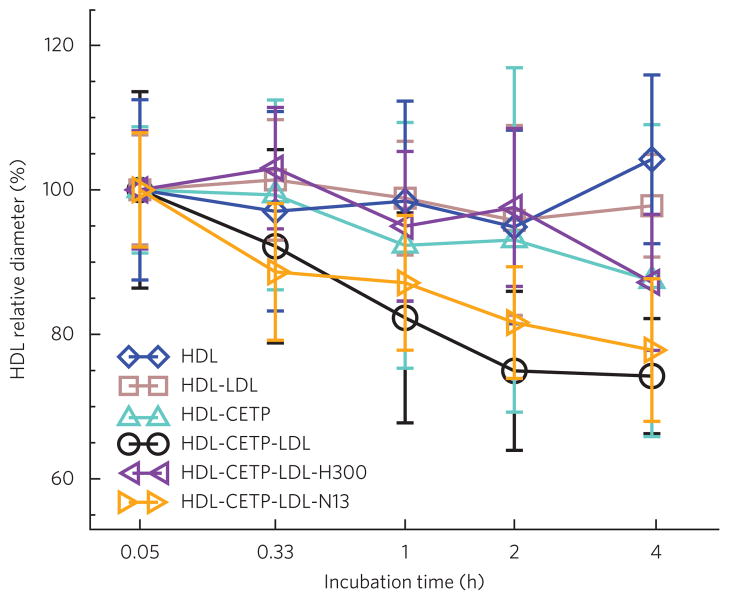
Figure 5. Analysis of HDL size change during incubation with CETP.
Samples were collected and viewed by OpNS-EM. A total of ~500 HDL particles for each incubation condition were selected from EM micrographs for quantitative size analysis. The geometric sizes of particles were calculated (Supplementary Methods) and expressed as the mean ± s.d. Quantifying the size of HDL particles for HDL alone (blue) and for HDL-LDL (brown) and HDL-CETP (cyan) mixtures shows no substantial size changes during the incubation time except for in the ternary HDL-CETP-LDL mixture (black). The diameters of HDL particles following incubations with LDL and CETP in the presence of antibodies H300 (purple) and N13 (gold) are also shown.
CETP reaction mechanism
CETP with HDL, LDL or both was incubated with or without the CETP-specific polyclonal antibodies H300 and N13 at physiological temperatures for up to 48 h, during which HDL size was measured using the OpNS-EM. These experiments (Fig. 5, Supplementary Fig. 12 and Supplementary Table 1; details shown in Supplementary Results) do not support the shuttle mechanism in which CETP dissociates from the HDL surface after it has removed the maximum amount of cholesteryl ester from the HDL core.
Although the results do not favor the shuttle mechanism for CETP-mediated transfer of neutral lipids between HDL and LDL, they do not exclude the possibility that CETP shuttles neutral lipids between HDL particles themselves (Supplementary Results).
CETP hydrophobic central cavity
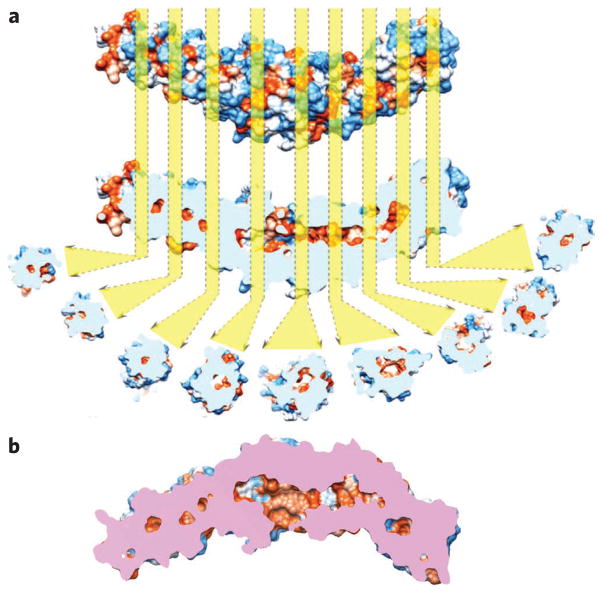
A series of hydrophobic cavities exist in the crystal structure of CETP. The occurrence of a hydrophobic pore within the N-terminal distal portion of CETP and a hydrophobic tunnel connecting the distal portions of the N- and C-terminal regions would support the tunnel mechanism. Therefore, the van der Waals surface of the CETP crystal structure was visualized as a colored surface according to its hydrophobicity (Fig. 6a). Longitudinal cross-sections of the surface density of CETP from the N-terminal β-barrel domain to the C-terminal β-barrel domain reveal a 60-Å-long hydrophobic central cavity traversing the central β-sheet domain, as reported previously10. To our surprise, a series of additional small, hollow, isolated hydrophobic cavities (diameter >5 Å) are present along the central axis of both β-barrel domains, lateral to the central β-sheet and the previously reported central cavity (Fig. 6b). The occurrence of unconnected cavities aligned linearly over ~105 Å, including both barrel domains and the ~60-Å central cavities, suggests that the crystal structure of CETP might show a ‘closed’ state with respect to cholesteryl ester transfer. Connecting the central cavity with these isolated lateral cavities could, if the protein is in an ‘activated’ state, connect all of the cavities to form a continuous tunnel between the distal portions of the N- and C-terminal domains for cholesteryl ester transfer. At this point, the continuous hydrophobic tunnel is hypothetical, and CETP’s activated state is speculative and requires confirmation from X-ray crystallography.
Figure 6. Analyses of the crystal structure of CETP.
(a) The CETP crystal structure is shown according to its van der Waals surface and is colored by its hydrophobicity (ranging from royal blue for the most hydrophilic to orange-red for the most hydrophobic). Longitudinal cut-away cross-sections show hydrophobic cavities (orange) from the center of the N-terminal β-barrel domain to the center of the C-terminal β-barrel domain. The distal portions of both β-barrels of CETP appear to be sealed. The cavities were aligned along the center of each domain, potentially reflecting the cholesteryl ester transfer pathway. The longest dimension of the pathway is about ~105 Å. (b) The cut-away surface view of CETP. The side openings near the center of the molecule are the phospholipid-binding pores. Pink regions show the cross surface between the cutting plane and the CETP molecule.
Molecular dynamics simulations predict a continuous central tunnel within CETP. Interaction of CETP with the surface monolayer and the core of a lipoprotein would have to trigger a conformational change at the distal portions of the β-barrel domains to open the continuous tunnel that accommodates entry of a cholesteryl ester into the central cavity and its exit from the opposite end. This would require high flexibility within the distal portions of the β-barrel domains. Because the N- and C-terminal β-barrel domains (i) penetrate the HDL, LDL and VLDL surfaces; (ii) have distal ends rich in hydrophobic amino acids; (iii) are in close proximity to the lipoprotein core; and (iv) have structurally flexible regions as suggested by the hydrophobic loops outside the CETP EM envelope at both distal portions of the β-barrel domains, we hypothesized that the ends of both the N- and C-terminal β-barrel domains undergo conformational changes that initiate cholesteryl ester transfer. To test this hypothesis, we analyzed the regional mobility of CETP and predicted the molecular motions within CETP using two molecular dynamics simulation methods. The nanoscale molecular dynamics method29 showed that the residues containing the top 30% of fluctuations are mostly on both ends of the CETP β-barrel domains and on the α-helix X (Supplementary Fig. 13a–c). The greatest structural fluctuation occurs near the distal end of the N-terminal β-barrel domain, which is consistent with the distal portions outside the EM envelope depicted in Figure 1e. Similar events most likely occur at the C terminus as well.
To investigate whether the flexibility of the domains contributes to the formation of a continuous tunnel between unconnected hydrophobic cavities, we analyzed the vibrational motions of CETP under 20 interrupting modes using the anisotropic network model, which predicts large-scale fluctuations30,31. This analysis showed that the ends of the β-barrel domains are flexible (Supplementary Fig. 13d). Remarkably, a ~15° twist along the long axis of CETP (Supplementary Fig. 13d) is associated with a ~10° tilt of the β-barrel strands (Supplementary Fig. 13e). Torsional analysis shows slight widening of the tunnel (~5 Å); thus, the central cavity connects with all of the small hydrophobic cavities to form a ~120-Å tunnel that is ~5–15 Å wide (Supplementary Fig. 13f–h). These analyses are consistent with the hypothesis that the ends of CETP β-barrel domains are subject to low-energy flexing that may gate cholesteryl ester entry into the central tunnel, which must open at both ends for cholesteryl ester transfer. The tunnel is hydrophobic and connects the distal ends of the β-barrel domains to form a continuous central tunnel that mediates cholesteryl ester transfer.
DISCUSSION
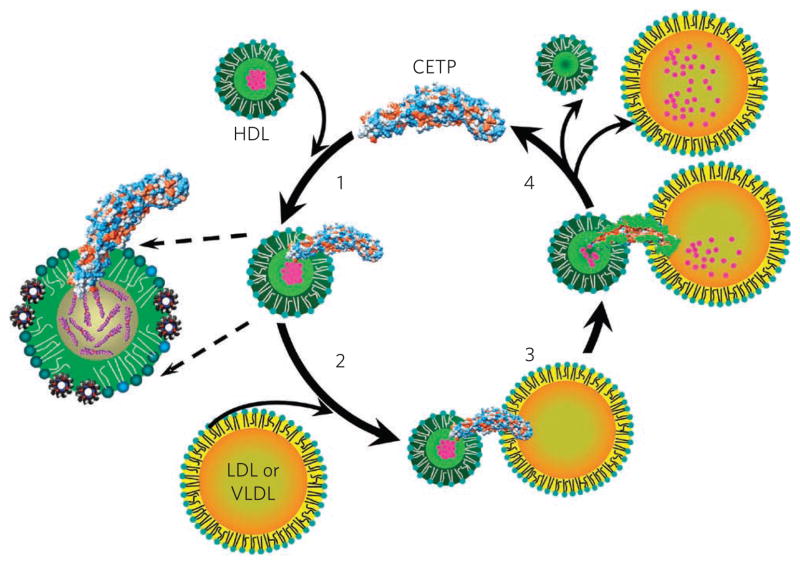
On the basis of the present findings, we propose a model (Fig. 7) in which both CETP terminal ends, including the N-terminal domain’s hydrophobic mini helix and strands and the C-terminal domain’s hydrophobic Ω2 strand, penetrate surface sites on lipoproteins to access the surface, central core or both (Fig. 7), similar to that for the association of exchangeable apolipoproteins to lipoprotein surfaces32,33. These interactions induce torsion of the two distal domains of CETP, which is the key to opening the tunnel for neutral lipid transfer. We also propose that cholesteryl ester migrates from HDL, which resides in a high-energy kinetic trap34,35, to LDL or VLDL through a hydrophobic tunnel structure (Fig. 7). The thermodynamic forces that drive cholesteryl ester transfer from HDL are (i) differential cholesteryl ester concentrations in donor and acceptor particles, (ii) changes in hydrophobicity along the central cavity favoring cholesteryl ester N- to C-terminus transfer and (iii) different internal densities, for example, LDL (less dense cholesteryl ester regions)21 or VLDL (triglyceride-rich environment)35. We further speculate that phospholipid pores could permit phospholipid movement out of the surface layer of HDL to accommodate an increase in surface curvature and into the surface of the acceptor lipoprotein (for example, LDL and VLDL) to accommodate a decrease in surface curvature. In step 4 in Figure 7, the ternary complex dissociates. Our model is consistent with mutagenesis data36 showing that substitution of polar amino acids for hydrophobic ones is associated with less neutral lipid transfer, that mutation of the N-terminal phospholipid pore decreases neutral lipid transfer by interfering with tunnel hydrophobicity and that a six-amino-acid deletion mutation of the small amphipathic α-helix X of C-terminal origin decreases maximal neutral lipid transport but not initial CETP binding to HDL, suggesting that α-helix X is important for function.
Figure 7. Proposed mechanism for cholesteryl ester transfer by CETP.
The N-terminal β-barrel domain of CETP initially penetrates the HDL surface, and the distal end of the domain interacts with the cholesteryl ester core of HDL (1). Upon interaction of the C-terminal β-barrel domain with LDL or VLDL (2), molecular forces introduced by the lipoproteins at either end of CETP cause twisting of the β-barrel domains and pore formation at the distal ends of the N- and C-terminal domains. These pores connect with the hydrophobic, continuous and central cavities of CETP to form a tunnel (3) that serves as a conduit, resulting in a transfer of cholesteryl esters to LDL or VLDL and a reduction in HDL size (4).
In summary, CETP was directly visualized as a banana-shaped asymmetric molecule with a tapered N-terminal end and a globular C-terminal end; according to our observations, the N-terminal end penetrates into HDL. There are near-perfect fits of overlays and insertion of the crystal structure into two- and three-dimensional structures. The dimensions of the portions of CETP that penetrate HDL and LDL or VLDL are distinct, and their subtraction from the total length of CETP approximates the length of the bridge observed between HDL and LDL or VLDL. Additionally, the C terminus–specific H300 antibody associates with the free end of CETP protruding from HDL, and H300 blocks formation of a LDL–CETP binary complex but not that of a HDL–CETP complex. The appearance of HDL–CETP–LDL ternary complexes strongly supports the tunnel mechanism for neutral cholesteryl ester transfer. We observed CETP terminal ends bridging lipoproteins; cholesteryl ester transfer occurs in the presence of the ternary complexes; the H300 antibody inhibits ternary complex formation and transfer activity; even after long incubations, the percentage of HDL–CETP complexes shows no observed decrease over time; and molecular simulations suggest that a hydrophobic tunnel spans the length of CETP. Our proposed CETP structure reveals the portions that are involved in mediating cholesteryl ester transfer and could be targeted by new classes of CETP inhibitors.
METHODS
Protein isolation and purification
Recombinant human CETP (~53 kDa before post-translational modifications) and lipoprotein were purified as described in detail in Supplementary Methods.
CETP–lipoprotein complex formation
CETP–HDL, HDL–CETP–LDL and HDL–CETP–VLDL complexes were prepared as described in detail in Supplementary Methods.
Negative-staining EM specimen preparation by the OpNS protocol
Specimens were prepared for EM by the OpNS protocol as described14,17. In brief, CETP (final concentration 0.93 mg ml−1 (17.5 μM)) and HDL–CETP (final concentration 0.93 mg ml−1) complexes were diluted to 0.005 mg ml−1 with Dulbecco’s phosphate-buffered saline (DPBS: 2.7 mM KCl, 1.46 mM KH2PO4, 136.9 mM NaCl and 8.1 mM Na2HPO4; Invitrogen) buffer. An aliquot (~3 μl) was placed on a thin, carbon- coated 300-mesh copper grid (Cu-300CN, Pacific Grid-Tech) that had been glow discharged. After ~1 min, excess solution was blotted with filter paper. The grid was washed by briefly touching the surface of the grid with a drop (~30 μl) of distilled water on parafilm and blotting dry with filter paper. This touching and blotting step was performed three times, each time with a clean drop of distilled water. Three drops of 1% (w/v) uranyl formate negative stain (Pacific Grid-Tech) on parafilm were then applied successively, and excess stain was removed by blotting in the same fashion. The grid was allowed to remain in contact with the last drop of stain with the sample side down for 1–3 min in the dark before removal of excess stain and was air dried at ~22 °C (refs. 14,17). Because uranyl formate solutions are light sensitive and unstable, the newly prepared solution was aliquoted and stored in the dark at −80 °C. Just before use, each aliquot was thawed in a water bath in the dark and then filtered (0.02-μm filter). The filter syringe was wrapped with aluminum foil to protect the uranyl formate solution from light. The same protocol was used to prepare other binary and ternary complexes. The lipoprotein particle sizes and shapes obtained from our OpNS protocol have <5% difference from those obtained from the frozen-hydrated native state by cryo-EM of apolipoprotein E4 (apoE4) HDL particles14,17. In the conventional negative-staining protocol17, lipoprotein particles result in the generation of stain-induced structural artifacts, including rouleaux, whereas with our OpNS protocol, rouleaux artifacts are reduced by using uranyl formate instead of phosphotungstic acid at low salt conditions14,17.
Cryo-PS-EM specimen preparation and EM data collection
CETP specimens were also prepared by a cryo-PS-EM method. In detail, CETP (final concentration 0.93 mg ml−1) was diluted to 0.005 mg ml−1 with DPBS buffer, and an aliquot (~3 μl) was applied to a glow-discharged holey thin-carbon-film–coated 300-mesh copper grid (Cu-300HN, Pacific Grid-Tech) for 1 min. The grid was washed with deionized water droplets, and 1% (w/v) uranyl formate (Pacific Grid-Tech) was applied as in the OpNS protocol. Instead of air-drying in the last step, the samples were flash-frozen in liquid ethane under conditions of 100% humidity and 4 °C with a FEI Vitrobot rapid-plunging device after being blotted with filter paper (#595, Schleicher & Schuell) from both sides for 2 s. The frozen-hydrated specimens were transferred to liquid nitrogen for storage. Cryo-PS-EM micrographs were acquired at −180 °C on a Gatan UltraScan 4K × 4K CCD camera by a FEI Tecnai 20 transmission electron microscope (Philips Electron Optics–FEI) or a Zeiss Libra 120 Plus transmission electron microscope (Carl Zeiss NTS) with 20-eV in-column energy filter operating at 200 kV or 120 kV at ×80,000 magnification and low-electron-dose conditions under Scherzer focus, whereas NS-EM micrographs were acquired at ~20 °C with the same CCD camera and magnification but under a defocus of 0.1–0.7 μm. Each pixel of the micrographs corresponded to 1.41 Å or 1.48 Å in the specimens.
The contrast of the image obtained from this cryo-PS method is inverted compared to that from the cryo-NS method reported previously22. In the prior cryo-NS method, an exceptionally high concentration of negative stain, 16% (w/v) ammonium molybdate, was used to maintain the negative image contrast from cryo-NS specimens22, whereas 1% uranyl formate was used in this paper.
Three-dimensional reconstruction of the HDL–CETP complex
Images were processed with FREALIGN, EMAN and SPIDER software packages37–39. The defocus and stigmatism of each micrograph were determined by fitting the contrast transfer function (CTF) parameters with its power spectrum by using ctffind3 in the FREALIGN software package37. Micrographs with poor correlation of phase residuals, large stigmatism (>0.1 μm) or distinguishable drift were excluded. The phase of each micrograph was corrected by a Wiener filter with the SPIDER software package38. Only isolated particles from the NS-EM images were initially selected and windowed as 256 × 256 pixel images (~360 Å × 360 Å at the specimen) using the boxer program in EMAN39. For three-dimensional reconstruction and refinement, we used the same program as that used for further selecting lipoprotein particles with a homogeneous size, which we term a ‘computational size-exclusion gel-filtration’ algorithm21. Using this method, ~38% of the particles in total were used for three-dimensional reconstruction, from which ~317 class averages were generated by reference-free class averages computed using refind2d.py in EMAN39. To prevent bias from a starting model, we used a featureless, smooth, solid cylinder (length ~75 Å, diameter ~35 Å) perpendicularly attached to a featureless, solid Gaussian globule (diameter 120 Å × 100 Å × 80 Å) as an initial starting model. This model was generated on the basis of typical features in reference class averages40. For the first four rounds of refinement, only very-low-resolution particle information was used (below the first CTF 0 in reciprocal space). Iterative refinement proceeded to convergence. Then, CTF amplitude and phase corrections, finer angular sampling and solvent flattening via masking were performed for higher-resolution refinement. This process was iterated to convergence. According to the 0.5 Fourier shell correlation criterion41, the final resolution of the asymmetric reconstruction of HDL–CETP complex was 14 Å (Supplementary Fig. 3).
Three-dimensional reconstruction of CETP
The image processing is described in detail in Supplementary Methods.
Supplementary Material
Acknowledgments
We thank D.A. Agard, I. Bahar and K. Dill for valuable discussions and A. Cheng and J. Song for helpful comments. This work was supported by Basic Energy Sciences–US Department of Energy (DE-AC02-05CH11231) and the W. M. Keck Foundation (no. 011808); the Keygrant Project of the Chinese Ministry of Education no. 708082 (S.Z.); US National Institutes of Health grant NIH-HL077268 and Tobacco-Related Disease Research Program of California grant 16FT-0163 (M.O.).
Footnotes
Accession codes. Electron Microscopy Data Bank: the cryo-PS-EM density map of CETP and the NS-EM map of HDL–CETP are deposited under ID codes 5346 and 5347, respectively. Protein Data Bank: the previously published crystal structure for CETP is deposited under accession code 2OBD.
Author contributions
G.R., K.H.W. and X.Q. initiated and designed the project; X.Q. provided the soluble CETP; H.J.P. and R.M.K. contributed the LDL and VLDL; K.-A.R., M.O. and G.C. provided HDL; L.Z., F.Y. and G.R. collected the data; S.Z., D.L. and G.R. conducted the molecular simulation; G.R. solved the structure; G.R., L.Z., M.A.C. and X.Q. analyzed and interpreted the data; G.R. and S.Z. drafted the initial manuscript; M.A.C., H.J.P., K.H.W., K.-A.R., R.M.K., X.Q. and L.Z. discussed and revised the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available online at http://www.nature.comnaturechemicalbiology/.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Barter PJ, et al. Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23:160–167. doi: 10.1161/01.atv.0000054658.91146.64. [DOI] [PubMed] [Google Scholar]
- 2.Camejo G, Waich S, Quintero G, Berrizbeitia ML, Lalaguna F. The affinity of low density lipoproteins for an arterial macromolecular complex. A study in ischemic heart disease and controls. Atherosclerosis. 1976;24:341–354. doi: 10.1016/0021-9150(76)90126-x. [DOI] [PubMed] [Google Scholar]
- 3.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 4.Hayek T, et al. Hypertriglyceridemia and cholesteryl ester transfer protein interact to dramatically alter high density lipoprotein levels, particle sizes, and metabolism. Studies in transgenic mice. J Clin Invest. 1993;92:1143–1152. doi: 10.1172/JCI116683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown ML, et al. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature. 1989;342:448–451. doi: 10.1038/342448a0. [DOI] [PubMed] [Google Scholar]
- 6.Inazu A, et al. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N Engl J Med. 1990;323:1234–1238. doi: 10.1056/NEJM199011013231803. [DOI] [PubMed] [Google Scholar]
- 7.Niesor EJ. Different effects of compounds decreasing cholesteryl ester transfer protein activity on lipoprotein metabolism. Curr Opin Lipidol. 2011;22:288–295. doi: 10.1097/MOL.0b013e3283475e00. [DOI] [PubMed] [Google Scholar]
- 8.Miyares MA. Anacetrapib and dalcetrapib: two novel cholesteryl ester transfer protein inhibitors. Ann Pharmacother. 2011;45:84–94. doi: 10.1345/aph.1P446. [DOI] [PubMed] [Google Scholar]
- 9.Kappelle PJ, van Tol A, Wolffenbuttel BH, Dullaart RP. Cholesteryl ester transfer protein inhibition in cardiovascular risk management: ongoing trials will end the confusion. Cardiovasc Ther. 2010;29:e89–e99. doi: 10.1111/j.1755-5922.2010.00201.x. [DOI] [PubMed] [Google Scholar]
- 10.Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 11.Barter PJ, Jones ME. Kinetic studies of the transfer of esterified cholesterol between human plasma low and high density lipoproteins. J Lipid Res. 1980;21:238–249. [PubMed] [Google Scholar]
- 12.Ihm J, Quinn DM, Busch SJ, Chataing B, Harmony JA. Kinetics of plasma protein-catalyzed exchange of phosphatidylcholine and cholesteryl ester between plasma lipoproteins. J Lipid Res. 1982;23:1328–1341. [PubMed] [Google Scholar]
- 13.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res. 1993;34:1255–1274. [PubMed] [Google Scholar]
- 14.Zhang L, et al. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J Lipid Res. 2011;52:175–184. doi: 10.1194/jlr.D010959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, et al. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein Sci. 2009;18:921–935. doi: 10.1002/pro.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva RA, et al. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc Natl Acad Sci USA. 2008;105:12176–12181. doi: 10.1073/pnas.0803626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, et al. An optimized negative-staining protocol of electron microscopy for apoE4·POPC lipoprotein. J Lipid Res. 2010;51:1228–1236. doi: 10.1194/jlr.D002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren G, Reddy VS, Cheng A, Melnyk P, Mitra AK. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proc Natl Acad Sci USA. 2001;98:1398–1403. doi: 10.1073/pnas.041489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren G, Cheng A, Reddy V, Melnyk P, Mitra AK. Three-dimensional fold of the human AQP1 water channel determined at 4 Å resolution by electron crystallography of two-dimensional crystals embedded in ice. J Mol Biol. 2000;301:369–387. doi: 10.1006/jmbi.2000.3949. [DOI] [PubMed] [Google Scholar]
- 21.Ren G, et al. Model of human low-density lipoprotein and bound receptor based on CryoEM. Proc Natl Acad Sci USA. 2010;107:1059–1064. doi: 10.1073/pnas.0908004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adrian M, Dubochet J, Fuller SD, Harris JR. Cryo-negative staining. Micron. 1998;29:145–160. doi: 10.1016/s0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- 23.Hayat MA, Miller SE. Negative Staining. McGraw-Hill; 1990. [Google Scholar]
- 24.Morton RE, Greene DJ. The surface cholesteryl ester content of donor and acceptor particles regulates CETP: a liposome-based approach to assess the substrate properties of lipoproteins. J Lipid Res. 2003;44:1364–1372. doi: 10.1194/jlr.M300063-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Swenson TL, Brocia RW, Tall AR. Plasma cholesteryl ester transfer protein has binding sites for neutral lipids and phospholipids. J Biol Chem. 1988;263:5150–5157. [PubMed] [Google Scholar]
- 26.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemming D, Thierbach K, Stelter P, Böttcher B, Hurt E. Precise mapping of subunits in multiprotein complexes by a versatile electron microscopy label. Nat Struct Mol Biol. 2010;17:775–778. doi: 10.1038/nsmb.1811. [DOI] [PubMed] [Google Scholar]
- 29.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atilgan AR, et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doruker P, Atilgan AR, Bahar I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: application to α-amylase inhibitor. Proteins. 2000;40:512–524. [PubMed] [Google Scholar]
- 32.Weers PM, Ryan RO. Apolipophorin III: role model apolipoprotein. Insect Biochem Mol Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Saito H, et al. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J Biol Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 34.Mehta R, Gantz DL, Gursky O. Human plasma high-density lipoproteins are stabilized by kinetic factors. J Mol Biol. 2003;328:183–192. doi: 10.1016/s0022-2836(03)00155-4. [DOI] [PubMed] [Google Scholar]
- 35.Han M, et al. Disruption of human plasma high-density lipoproteins by streptococcal serum opacity factor requires labile apolipoprotein A-I. Biochemistry. 2009;48:1481–1487. doi: 10.1021/bi802287q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S, Kussie P, Deng L, Tall A. Defective binding of neutral lipids by a carboxyl-terminal deletion mutant of cholesteryl ester transfer protein. Evidence for a carboxyl-terminal cholesteryl ester binding site essential for neutral lipid transfer activity. J Biol Chem. 1995;270:612–618. doi: 10.1074/jbc.270.2.612. [DOI] [PubMed] [Google Scholar]
- 37.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 39.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 40.Galkin VE, Orlova A, Cherepanova O, Lebart MC, Egelman EH. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc Natl Acad Sci USA. 2008;105:1494–1498. doi: 10.1073/pnas.0708667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.