Abstract
Microtubule dynamics underlie spindle assembly, yet we do not know how the spindle environment affects these dynamics. We developed methods for measuring two key parameters of microtubule plus-end dynamic instability in Xenopus egg extract spindles. To measure plus-end polymerization rates and localize growing plus ends, we used fluorescence confocal imaging of EB1. This revealed plus-end polymerization throughout the spindle at ∼11 μm/min, similar to astral microtubules, suggesting polymerization velocity is not regionally regulated by the spindle. The ratio of EB1 to microtubule fluorescence revealed an enrichment of polymerizing ends near the spindle middle, indicating enhanced nucleation or rescue there. We measured depolymerization rates by creating a front of synchronized depolymerization in spindles severed with microneedles. This front could be tracked by polarization and fluorescence microscopy as it advanced from each cut edge toward the associated pole. Both imaging modalities revealed rapid depolymerization (∼30 μm/min) superimposed on a subset of microtubules stable to depolymerization. Larger spindle fragments contained a higher percentage of stable microtubules, which we believe were oriented with their minus ends facing the cut. Depolymerization was blocked by the potent microtubule stabilizing agent hexylene glycol, but was unaffected by α-MCAK antibody and AMPPNP, which block catastrophe and kinesin motility, respectively. These measurements move us closer to understanding the complete life history of a spindle microtubule.
INTRODUCTION
To perform their major jobs of aligning and segregating chromosomes, mitotic and meiotic spindles must establish robustly self-organizing, dynamic structures from microtubules. The assembly dynamics of microtubules in the spindle are governed by several factors, most notably the abundance and distribution of microtubule nucleation sites and the dynamic instability of microtubule ends (reviewed in Inoue and Salmon, 1995; Desai and Mitchison, 1997; Karsenti and Vernos, 2001). Dynamic instability is thought to determine the mean microtubule length and the duration of microtubule lifetimes within the spindle. It is described by four parameters: the polymerization rate, depolymerization rate, and the frequencies of transitions between polymerization and depolymerization, known as catastrophe and rescue, at the ends of individual microtubules. Pauses in microtubule polymerization may also make an important contribution to polymerization dynamics. Superimposed on features of microtubule stability, the spatial distribution and polarity of microtubules provide a mechanism for structural organization of the spindle. Microtubule positions within the spindle depend on their sites of nucleation and possibly also on motor-driven movements away from these sites. For example, when coupled to poleward translocation, microtubule plus-end growth and minus-end depolymerization produce the phenomenon of poleward microtubule flux. Poleward flux contributes to microtubule turnover within the metaphase spindle at its steady state-length and to chromosome separation during anaphase. To understand how the spindle assembles and segregates chromosomes, we must determine how these dynamic properties are modified by the spindle environment. How are the dynamic behaviors intrinsic to microtubules modulated to produce this robust structure, and how do factors inside the spindle maintain the dynamics properties of microtubules within it?
Serial section electron microscopy (EM) and time-lapse imaging combined with photobleaching, photoactivation, and fluorescence speckle microscopy (FSM) have provided a partial description of the structure and dynamics of mitotic spindles in yeast and mammalian cells (reviewed in Rieder and Khodjakov, 2003). These techniques have revealed a dramatic turnover of microtubules within a geometrically stable structure. In tissue culture cells, the polymerization rate of plus ends within the spindle has been measured (Tirnauer et al., 2002a). But we still do not know the full life histories of microtubules within the spindle—their sites of nucleation, depolymerization rates, and rescue, catastrophe, and pause frequencies. Meiotic Xenopus egg extracts have become an important experimental system for studying spindle assembly and microtubule dynamics (Desai et al., 1999; Karsenti and Vernos, 2001). Our knowledge of meiosis II spindle structure is less complete than for somatic spindles, and microtubule organization and dynamics may differ significantly from somatic mitotic spindles because of chromatin-induced microtubule nucleation (Waters and Salmon, 1997; Karsenti and Vernos, 2001). We know that microtubule lifetimes are ∼1-2 min and that one key mechanism of their turnover is poleward flux, which occurs at a rate of ∼2 μm/min (Sawin and Mitchison, 1991). Missing information includes the parameters of dynamic instability, including polymerization and depolymerization rates, catastrophe, rescue, and pause frequencies; as well as the sites of nucleation, the distribution of plus and minus ends, and the range of microtubule lengths. The importance of measuring these parameters will increase as we try to come to a deeper understanding through quantitative modeling (Nedelec et al., 2003).
Mitchison and Kirschner (1985) proposed that selective microtubule stabilization drives spindle assembly. In principle, stabilization could be achieved through increases in microtubule polymerization rate, rescue frequency, or pause duration, or by decreases in depolymerization rate or catastrophe frequency. Selective stabilization was initially proposed for microtubules in direct contact with kinetochores (Mitchison and Kirschner, 1985). In the Xenopus meiotic spindle, chromatin plays a central role in nucleation, and perhaps stabilization, of microtubules (Karsenti and Vernos, 2001). This is proposed to occur via a gradient of Ran-GTP centered on chromatin that promotes biased growth of microtubules (Kalab et al., 2002; Trieselmann and Wilde, 2002). In studies of centrosome-nucleated microtubule asters, Ran primarily affects rescue and catastrophe frequencies (Carazo-Salas et al., 2001; Wilde et al., 2001), but the effects of Ran or other individual proteins on the dynamics of microtubules in the spindle interior are unknown. In addition, spindles are enriched in a number of microtubule regulatory proteins whose effects on dynamic instability have only been measured on individual microtubules in the cytoplasm.
To determine the relative contribution of spindle proteins and chromatin to spindle structure, the dynamic instability parameters of individual microtubules within the spindle must be measured. This has been a challenge because microtubule density in spindles is too great to distinguish individual microtubule ends. To address this, we took two approaches. We used fluorescence confocal microscopy of the microtubule plus-end tracking protein (+tip) EB1 to determine microtubule plus-end polymerization rates. For depolymerization rates, we developed a spindle cutting technique that generated a front of synchronized microtubule depolymerization moving toward the spindle poles. By combining these methods, we can begin to determine a quantitative picture of microtubule dynamic instability in the meiotic spindle.
MATERIALS AND METHODS
Extract Reagents and Spindle Assembly
Crude cytostatic factor (CSF) arrested extracts were prepared from Xenopus laevis eggs as previously described (Desai et al., 1999). Cycled spindle assembly was done with demembranated X. laevis sperm nuclei in the presence of rhodamine-labeled tubulin (Hyman et al., 1991) or Alexa488 or Alexa594-labeled EB1 at 0.01-0.013 mg/ml (Tirnauer et al., 2002b). Perturbations included the addition of 2% DMSO (Pierce Biotechnology, Rockford IL), 150 μg/ml the microtubule depolymerizing kinesin MCAK, or the nonhydrolyzable ATP analog AMPPNP at 5 mM. EB1 imaging was done on cycled spindles except where indicated; cutting experiments were performed on CSF-arrested and cycled spindles, with no differences seen between the two states.
Spindle Cutting Technique
CSF-arrested and cycled spindles were assembled as above. Microneedles were pulled from 100-μl microcapillaries (Drummond scientific cat. no. 1-100-1000) with a long taper over ∼20 mm to a fine point. For some fluorescence experiments, the cutting needles were dipped in rhodamine-labeled bovine serum albumin before cutting to make them more visible. A pair of microneedles were held in micromanipulators (Narashige International USA, Inc. Long Island, NY) mounted on the microscope stage at 180° to each other. Xenopus extract, 5 μl, containing spindles was transferred to a 25-mm round coverslip, mounted with warm VALAP (equal parts Vasoline, lanolin, and paraffin) to the bottom of a steel slide with a 22-mm round cutout, and covered with 250 μl of mineral oil to prevent evaporation.
All observations were made at 20× magnification on an inverted TE300 microscope (Nikon, Tokyo, Japan). For polarization imaging, both Wollaston prisms were removed from the DIC light path, and the polarizer was set a few degrees from extinction. Because of the deSénarmont compensation used in the Nikon DIC system, this resulted in excellent polarization images. Spindles were located using polarization, and paired needles were lowered so that their points pressed down on the glass on opposite sides of the spindle. The two needles touched and crossed over on top of the spindle, resulting in the spindle being trapped and slightly compressed between the two needles. After time-lapse recording was started, the two needles were rapidly drawn apart across the spindle to create a scissoring action that cut the spindle in two.
Image Acquisition and Analysis
Images of EB1 and tubulin fluorescence in cycled spindles were acquired with a 60-mW argon/krypton laser for illumination on a Nikon TE300 inverted microscope with a 100× plan 1.4 NA lens (Nikon) through a CS10 Spinning Disk Confocal Head (Perkin Elmer-Cetus Life Sciences, Boston, MA) and acquired digitally at 5-s intervals onto an ORCA ER cooled CCD camera (Hamamatsu Photonics, Bridgewater, NJ) using MetaMorph software (Universal Imaging Corp., West Chester, PA). Unaltered image data was analyzed using the kymograph feature of MetaMorph software and rate calculations were performed with Excel (Microsoft Corp., Redmond, WA).
Spindle cutting was done using polarization or wide field fluorescence optics to acquire time lapse images at 2- or 2.5-s intervals onto an Orca ER camera (Hamamatsu Photonics) using MetaMorph software. Kymographs and rate calculations were done as above.
RESULTS
Distribution of Polymerizing Microtubule Plus Ends and Polymerization Rate of Spindle Microtubules
We added fluorescently labeled EB1, which associates preferentially with polymerizing microtubule plus ends (Tirnauer et al., 2002b), to cycled Xenopus egg extracts before DNA replication, to identify polymerizing microtubule plus ends. Although adding labeled EB1 increases the relative amount of time microtubules spend polymerizing, it does not appreciably affect the rate of polymerization or spindle morphology, and it is thus a useful tool for tracking polymerization rates (Tirnauer et al., 2002b). We used spinning disk confocal microscopy to acquire images of EB1 alone or in the presence of rhodamine-labeled tubulin. These images revealed the distribution of EB1-labeled microtubule plus ends as well as their polymerization rates. Polymerizing plus ends were located within the spindle, extending outward from the spindle (analogous to astral fibers), just distal to the kinetochores (marked by α-CenpA; Maddox et al., 2003; unpublished data), and at spindle poles (Figure 1A).
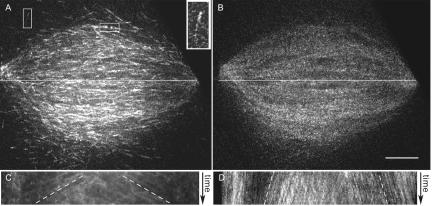
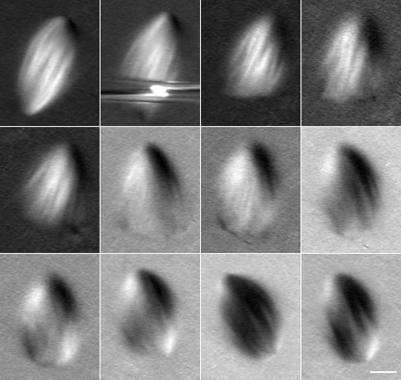
Figure 1.
Microtubule polymerization rates measured using fluorescent EB1. Spindles were assembled in Xenopus egg extracts after DNA replication in the presence of Alexa488-EB1 with or without rhodaminetubulin. (A) EB1 forms comets on microtubule plus ends when they polymerize. Within the spindle, EB1 localizes to spindle poles, spindle microtubules, astral microtubules (vertical box), and just distal to kinetochores (horizontal box). Inset shows a higher magnification example of a comet on a polymerizing microtubule plus end. Tracking polymerizing spindle microtubule ends over time allowed measurement of their polymerization rates. White line, region used to construct kymograph. (B) Corresponding image of rhodamine tubulin fluorescence in the same spindle. (C) Kymograph along the line in A. The obvious lines on the kymographs (white dashed lines) were used to determine the mean microtubule polymerization rate of 10.9 μm/min. (D) Kymograph along the line in B. The slower rate of microtubule movement is due to poleward microtubule flux. Bar, 10 μm.
To obtain plus-end polymerization velocities, we measured the movement of EB1 by time-lapse microscopy (Movie 1A). Kymographs were constructed from lines drawn along the long spindle axis, to show the movements of polymerizing plus ends over time. Visual inspection of these kymographs suggested that the majority of plus ends were growing away from the spindle equator rather than toward it, suggesting that a large percentage of microtubules extended beyond the chromatin region (Figure 1A). We used the slope of the prominent EB1 lines in the kymographs to determine spindle microtubule polymerization rates (Figure 1C). These rates were relatively uniform regardless of position along the spindle, at 10.9 ± 3.1 μm/min (n = 61 measurements in 9 spindles, Figure 1B). When visualized simultaneously, speckle-level tubulin fluorescence within microtubules moved poleward at the rate of microtubule flux (Movie 1B; Figure 1D). Microtubule polymerization rates were similar in spindles formed without DNA replication (11.7 ± 2.8 μm/min; n = 26 kymographs in 3 spindles) and to the rates of astral microtubules growing away from the spindle in the same set of time-lapse series (12.3 ± 4.9 μm/min; n = 12) as well as to astral microtubule polymerization rates previously measured using centrosome-nucleated microtubules (Belmont and Mitchison, 1996; Tournebize et al., 2000; Wilde et al., 2001; Tirnauer et al., 2002b). Thus, we found that plus-end polymerization rates are relatively unaffected by the internal spindle environment.
We next measured the density of plus ends relative to microtubules along the spindle pole-to-pole axis. When compared with the distribution of microtubule fluorescence in the same sample, EB1 fluorescence was enriched in a wide band in the middle of the spindle. The ratio of EB1 fluorescence to microtubule fluorescence in this zone, compared with the area just inside the poles, was 2.5 ± 0.6 (n = 12 spindles). The higher equatorial ratio of EB1 to microtubule fluorescence indicates a higher relative steady state concentration of polymerizing microtubule ends near the middle of the spindle, compared with the regions around the poles.
Synchronization of Microtubule Depolymerization by Cutting
Currently there are no specific chemical or protein markers for imaging depolymerizing microtubule ends, making it difficult to track individual depolymerizing microtubules within the spindle. We therefore induced synchronous microtubule depolymerization in a large population of microtubules by mechanical severing, as was done by Nicklas et al. (1989), except that we used paired needles to slice rather than press the spindle in half (Figure 2A). Extract (with spindles) was spread underneath a layer of mineral oil in a small open chamber, and paired glass microneedles were used to slice spindles along or across their long axis, severing them into two spindle fragments that could be observed by time-lapse imaging before, during, and after cutting. We first imaged the spindles by polarization microscopy in order to avoid photobleaching and photodamage during imaging (Figure 2B). Spindles were oriented with their axes at 45° to the polarizer direction for maximum contrast from spindle microtubule birefringent retardation. Only spindle fragments that remained at approximately this orientation after cutting were analyzed. When cuts were made along the spindle axis, two longitudinal bipolar spindles were formed with half the width of the original. There was no evidence of microtubule depolymerization (unpublished data). Cuts across the interpolar spindle axis severed the spindle microtubules, and a fraction of the severed ends underwent rapid depolymerization toward their associated pole (Figure 3A and Movie 3A). The intensity difference between the front of reduced signal and the stable fraction of microtubules decreased as the zone moved poleward; typically we could track it for 5 to 10 μm before it became obscured.
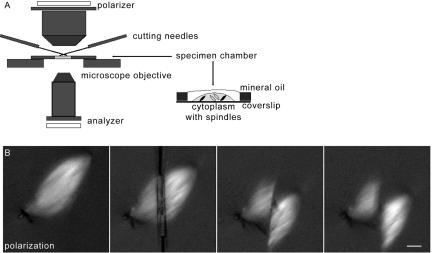
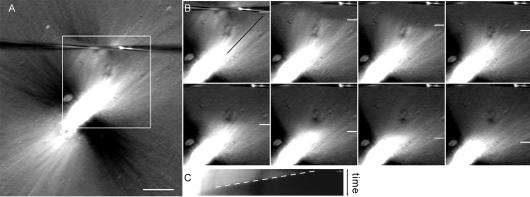
Figure 2.
Experimental setup for spindle cutting. (A) Xenopus egg extract containing spindles was placed in open chambers, covered with a thin layer of mineral oil, and cut (sliced) across the microtubules using paired microneedles. (A) Cartoon showing the cutting setup and specimen chamber. (B) Spindle microtubule depolymerization was observed in samples imaged by polarization microscopy (shown) or fluorescence microscopy. Note that spindles appear brightest when microtubules are at a 45° angle to the polarizer. A series of images obtained before, during, and immediately after the cut is shown. Bar, 10 μm.
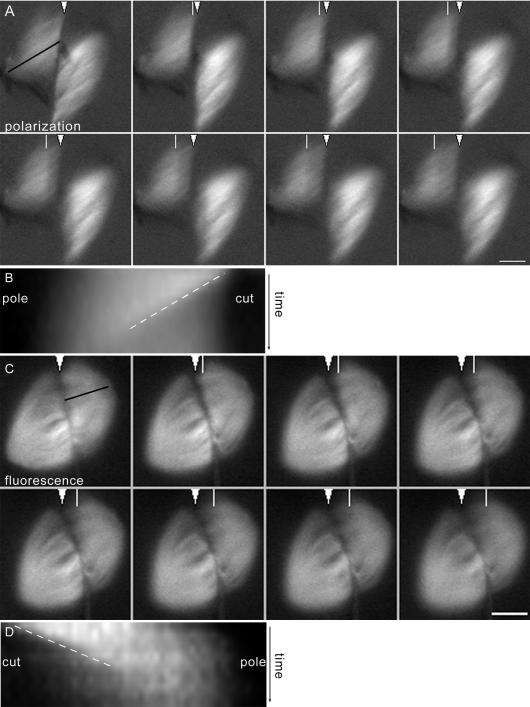
Figure 3.
Spindle microtubules depolymerize rapidly after cutting. (A) Frames from a time-lapse image of a cut spindle imaged by polarization microscopy. In the first frame, the cut is marked with a white arrowhead, and the line used for making the kymograph is shown with a black line along the fibers. In subsequent frames, a white line shows the position of the depolymerization front. See Supplemental Movie 3A. (B) Kymograph of the spindle in A showing the depolymerization front as it moves from the cut edge toward the associated spindle pole. The slope of the line of the kymograph (white dashed line, position vs. time) was used to calculate depolymerization rates. (C) Frames from a time-lapse image of a cut spindle imaged by fluorescence microscopy, labeled as in A. See Supplemental Movie 3C. (D) Kymograph of the spindle in C showing the depolymerization front, labeled as above. Bar, 10 μm.
We hypothesized that the front of reduced birefringence induced by spindle cutting was due to synchronized depolymerization of a subset of spindle microtubules. To confirm this, we directly visualized microtubules in cut spindles by assembling spindles in the presence of rhodamine-labeled tubulin. To reduce photobleaching, the spindles were manipulated under polarization optics before cutting and imaged by fluorescence microscopy only during and after cutting. Over this brief time period, photobleaching in control spindles was <5% (unpublished data). With fluorescence imaging, we also observed a front of reduced fluorescence moving toward the associated spindle pole and a fraction of microtubule fluorescence remaining (Figure 3C and Movie 3C). We could again track the front's movement for 5 to 10 μm before it became obscured near the pole. This lead us to conclude that the front of reduced birefringence and the front of reduced fluorescence represent the same population of depolymerizing microtubules and that the bulk of the remaining signal in both cases corresponds to a subset of microtubules stable to depolymerization induced by the cut.
Spindle Microtubule Depolymerization Rates Are Rapid
The uniform movement of the front of reduced signal (birefringent retardation or fluorescence) produced by spindle cutting allowed us to infer individual microtubule depolymerization rates from bulk measurements. These rates of microtubule depolymerization were determined by analyzing kymographs constructed from lines along the long spindle axis, parallel to the microtubules (Figure 3, B and D). Mean depolymerization rates were 32 and 28 μm/min for polarization and fluorescence images, respectively (Table 1). The rate of depolymerization was constant as the front moved toward the pole. This result suggests that microtubules depolymerize rapidly and for long distances poleward within the spindle without undergoing rescue events or changes in depolymerization rates after cutting. We could not directly compare catastrophe-induced (physiological) and cut-induced depolymerization, but we suspect the two processes are the same, because the rate of microtubule depolymerization in vitro was similar whether it occurred after UV microbeam cutting or spontaneous catastrophe (Walker et al., 1989).
Table 1.
Microtubule depolymerization rates after spindle cutting
| Treatment | Polarization microscopy | Fluorescence microscopy |
|---|---|---|
| Spindle cut alone | 32.3 ± 5.1 (39) | 28.1 ± 6.5 (19) |
| Spindle cut + hexylene glycol | No depolymerization (n = 6) | |
| Spindle cut + DMSO | 24.1 ± 3.3 (10) | |
| Spindle cut + α-MCAK | 32.0 ± 6.4 (21) | |
| Spindle cut + AMPPNP | 30.1 ± 4.3 (9) | |
| Aster cut + α-MCAK | 39.5 ± 5.3 (15) |
CSF (cytostatic factor) arrested Xenopus egg extract spindles or asters were cut with paired microneedles and imaged by time-lapse polarization or fluorescence microscopy. Where indicated, extracts were preincubated with antibody to MCAK at a final concentration of 150 μg/ml, DMSO at a final dilution to 2%, AMPPNP at a final concentration of 5 mM, or hexylene glycol at a final dilution to 2%. Rates are given ± SD. The number of cut half spindles or asters measured is in parentheses.
To test the effect of lattice stabilization on cutting-induced depolymerization, we added the strong microtubule lattice stabilizing agent 2-methyl-2,4-pentanediol (“hexylene glycol”; Forer et al., 1976; Harris and Clason, 1992) to spindles before cutting. This agent blocked propagation of the depolymerization front, confirming that the front depends on an unstable microtubule lattice (Table 1). The weaker lattice stabilizing agent DMSO did not block front propagation, but slowed it significantly. We also tested the effect of inhibition of the major catastrophe factor, MCAK (also called XKCM1; Kinoshita et al., 2001). Inhibiting MCAK resulted in massive polymerization of microtubule plus ends out of the spindle, as blocking catastrophes would be expected to do (analyzed in Mitchison et al., unpublished results). However, cuts still induced a depolymerization front, showing that MCAK is required for catastrophes, but not for the propagation of depolymerization. Finally, we tested the effect of inducing rigor binding of spindle kinesins using the nonhydrolysable ATP analog AMPPNP. Again, there was no effect on the propagation of the depolymerization front. The microtubule depolymerization rates measured from these perturbations are summarized in Table 1.
A Subpopulation of Spindle Microtubules Is Stable to Cutting
As noted above, time-lapse polarization and fluorescence microscopy revealed that cut spindle fragments contain a subset of microtubules stable to cutting-induced depolymerization (Figure 3). We estimated the fraction of microtubules that remained stable by measuring the difference in signal at the same location before and after cutting, using the kymographs. Precise measurement was difficult, especially on polarization images, due to changes in spindle orientation and microtubule organization. We estimated that on average 39% of the signal was lost in the polarization sequences (n = 26) and 52% in the fluorescence (n = 22). Thus, after cutting, approximately half of the microtubules depolymerized, and half were stable.
We hypothesized that one factor governing microtubule stability after cutting was microtubule polarity relative to the cut edge, with plus ends undergoing depolymerization and minus ends remaining stable. If this were the case, the position of the cut should influence the fraction of microtubules that depolymerize. Each spindle fragment contains microtubules from its associated pole as well as microtubules of opposite polarity originating from the middle of the spindle and/or the opposite pole. When cut, these microtubules of opposite polarity would expose their minus ends at the cut edge of the fragment. Smaller spindle fragments are expected to have relatively fewer of these minus ends and relatively more exposed plus ends at the cut edge and therefore to have a larger percentage of microtubules that depolymerize upon cutting. Larger fragments would have the converse: a larger fraction of stable microtubules. To test this hypothesis, we determined the correlation between spindle fragment length (as a percent of total spindle length before cutting) and percentage of microtubules that depolymerized. In several sequences where the spindle was cut markedly off center, we observed a larger stable fraction in the larger spindle fragment (Figure 4, A and B) and a smaller stable fraction in the smaller fragment. When we made the measurement over all quantifiable fluorescence experiments, we found a weak correlation between the size of the spindle fragment and the percent of spindle microtubules stable to depolymerization (Figure 4C). For individual pairs of fragments, the larger fragment had a greater fraction of stable microtubules in 8 of 10 cases (Figure 4D). As discussed below, this finding supports the conclusion that microtubule polarity influences stability to depolymerization in the spindle, although it does not rule out other mechanisms of selective stabilization.
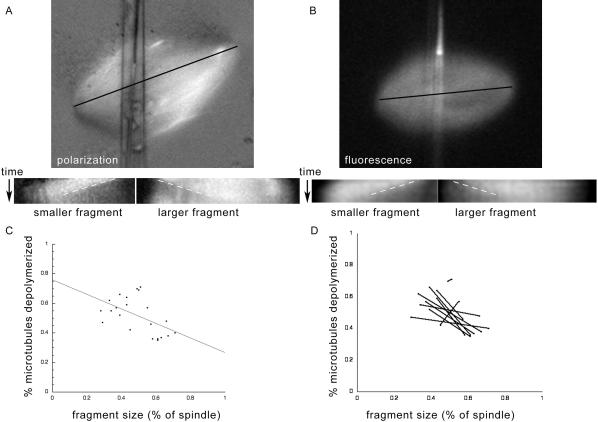
Figure 4.
Spindle fragment size correlates inversely with the percentage of depolymerizing microtubules. (A) Example of a spindle cut asymmetrically and imaged by polarization microscopy, with corresponding kymographs of both resulting fragments. (B) Example of a spindle cut asymmetrically and imaged by fluorescence microscopy, with corresponding kymographs. For both A and B, the lines used to make the kymographs were drawn parallel to the spindle, as illustrated by the black line. In each case, the short fragment is on the left, and a white dashed line shows the angle of the depolymerization front (position vs. time). Kymographs show that the shorter fragment has a greater contrast difference, representing a higher percentage of depolymerized microtubules. (C) Scatter plot of spindle fragment size (expressed as percent of the total spindle) vs. percent of spindle depolymerized (calculated from the associated kymograph), for fluorescence images. Each spindle fragment is represented by a black dot. The best linear fit for these data has a slope of -0.49 and R value of 0.53. (D) The same data as in C, with each pair fragments from the same spindle graphed together. These pairs show that in all cases except the two most symmetrical cuts (producing the most equal-sized fragments), the larger fragment has less depolymerization and the smaller fragment greater depolymerization.
To determine whether spindles could recover their bipolar structure after cutting, we also followed some cut spindles for longer time periods by polarization microscopy. We found that if the two cut halves were within ∼20 μm of each other, they often made a microtubule connection and drew together to reform a spindle within ∼10 min (unpublished data). If the halves were farther apart, individual halves gradually reestablished bipolarity and normal spindle morphology over ∼20 min (unpublished data). When the cuts were highly asymmetric, leaving one side with all of the chromatin and the other side with none, the side with chromatin recovered to form a bipolar spindle over ∼10 min (Figure 5), and the side without chromatin disappeared (as evidenced by disappearance of birefringence) over ∼10 min. These observations suggest that spindle fragments containing chromatin possess the structural and dynamic information necessary for reestablishing bipolarity.
Figure 5.
Large spindle fragments are able to reform complete spindles after cutting. Spindles were cut as above and imaged by polarization microscopy for 10-20 min. Frames from a time-lapse image are shown; the interval between frames was 80 s. By ∼10 min, a recognizable spindle pole has formed where the cut edge was originally produced. This spindle rotated counterclockwise during the second part of the time-lapse image. The smaller spindle fragment was brushed away by the lower cutting needle. Bar, 10 μm.
Astral Microtubule Depolymerization Rates Measured after Inhibition of MCAK
The depolymerization rates we report here for spindle microtubules are significantly higher than the rates measured for spontaneous depolymerization of centrosomally nucleated microtubules in Xenopus egg extracts. To better understand the reasons for this, we performed cutting experiments on astral microtubules, using the same extract preparations, cutting needles, and polarization optics as above to allow for direct comparison. Astral microtubules associated with unmanipulated spindles were not visible at 20× magnification, so we took advantage of α-MCAK antibody addition, which did not appreciably affect the microtubule depolymerization rate in spindles. In these MCAK-inhibited preparations, astral microtubules extended from the spindle poles for >100 μm into the extract, allowing for efficient cutting and extended observation periods. The long microtubules extending throughout the extract were apparently under tension, because cutting frequently led to retraction of both free and spindle-associated microtubules away from the cut. Cuts of astral microtubules produced rapid plus-end depolymerization toward the spindle that could be analyzed by kymographs (Figure 6 and Movie 6), with the rates corrected for retraction of the microtubules by subtracting movement of the spindle itself. After this correction, the plus-end depolymerization rate calculated from lines on the kymographs was 39.5 ± 5.3 μm/min (n = 15 cuts), slightly faster than the rates we observed for microtubules within the spindle. Thus, depolymerization rates were similar between spindle microtubules and astral microtubules in extracts treated with α-MCAK antibody.
Figure 6.
Astral microtubules depolymerize rapidly after cutting. Astral microtubules were induced to polymerize by the addition of α-MCAK antibody, in order to allow their visualization at 20× magnification. (A) Spindle treated with α-MCAK antibody, imaged by polarization microscopy. Contrast was maximized to emphasize astral microtubules. While microtubules oriented at 45° angles relative to the polarizer are most evident (white and black), the spindle is encircled by astral microtubules at all angles. The cutting needles are visible near the top of the frame. Bar, 20 μm. (B) Frames from a time-lapse image showing the boxed region of the spindle in A. The position of the depolymerization front is shown by the white line, and the line used to generate the kymograph is shown by the black line along the fibers. See Supplemental Movie 6. (C) Kymograph of the region in B, showing the movement of the depolymerization front over time (dashed line, position vs. time).
Interestingly, within the aster we saw no evidence of stable microtubules on the side of the cut facing the spindle and retraction but little to no minus-end depolymerization on the side facing away. This lack of a stable subpopulation within a region of more uniformly oriented microtubules (the majority of plus ends pointing outward) and lack of minus-end depolymerization in the microtubules on the opposite side of the cut (predominantly minus ends generated from the same microtubules) support our hypotheses that the stable microtubule subpopulation within the spindle consisted of microtubule minus ends facing the cut and that microtubule minus ends in Xenopus egg extracts are stable to depolymerization.
DISCUSSION
Toward a Picture of Microtubule Life Histories inside the Metaphase Spindle
To fully characterize the forces and dynamics that govern spindle assembly, we need to describe the typical life history of a spindle microtubule, from nucleation through disappearance. Here, we have described the polymerization and depolymerization rates of microtubules in metaphase Xenopus egg extract spindles. This is the first quantitative analysis of polymerization rates for these spindles and depolymerization rates within any spindles. Our measurements did not detect appreciable variability of microtubule polymerization or depolymerization rates across the spindle longitudinally, nor did they reveal significant differences for polymerization and depolymerization rates inside vs. outside the spindle. Thus, microtubule plus-end polymerization and depolymerization rates are kept constant within the spindle. Our cutting experiments did clearly show that a subset of microtubules, which we believe to be minus end-exposed, was stable to cut-induced depolymerization. Ultimately, it will be useful to describe all the parameters of dynamic instability, the contribution of poleward flux, and the sites of new microtubule nucleation within the spindle. Reliable markers for microtubule minus ends and nucleation sites (which may only partially overlap, because of microtubule transport) as well as measurement of catastrophe, rescue, and pause frequencies, are needed to complete the picture.
Translational movements of microtubules toward the spindle poles also provide an important contribution to spindle organization. Coupled to minus-end depolymerization and plus-end polymerization, these movements produce poleward microtubule flux, shown by fluorescent markers to occur at a rate of 2-3 μm/min (Sawin and Mitchison, 1991; Waterman-Storer et al., 1998). Our measurements focused on microtubule plus-end dynamics rather than on these movements, but the polymerization rates we calculated may be falsely low (by ∼15%) and the depolymerization rates falsely high (by ∼7%) because of poleward translocation of the microtubule.
Our EB1 images revealed the selective accumulation of polymerizing microtubule plus ends in the middle of the Xenopus egg extract spindle, mostly oriented such that they are growing away from the chromatin. This was different from the relative uniformity of plus ends we saw in PtK cells (Tirnauer et al., 2002a). Two models, not mutually exclusive, may explain why polymerizing microtubule plus ends are densest in the middle of the spindle; these differ primarily in the position of microtubule minus ends. One predicts that the majority of microtubules emanate from the spindle poles and that their plus ends oscillate between polymerization and depolymerization phases of dynamic instability because of frequent rescues, keeping plus ends near the spindle equator. Alternatively, a considerable fraction of spindle microtubules could be nucleated within the equatorial spindle region and only grow to short lengths before undergoing catastrophe. These two models are not mutually exclusive, and components of both are likely to contribute to spindle assembly. Serial section EM alone will not suffice for differentiating between the two mechanisms (although it will be helpful) because it does not report directly on microtubule polarity. In lieu of directly visualizing microtubule minus ends, it may be possible to predict their positions by using a combination of FSM tracking to obtain a measure of spindle microtubule polarity and length, coupled with EB1 imaging to identify plus ends specifically.
Microtubule Depolymerization Rates Are Rapid and Consistent between Spindle and Bundled Astral Fibers
A striking feature of our experiments was the rapid rate of spindle microtubule depolymerization. Pickett-Heaps observed UV-induced spindle fiber ARBs moving poleward at slow (∼1-4 μm/min) and rapid (∼20 μm/min) rates in metaphase and telophase, respectively (Leslie and Pickett-Heaps, 1984; Spurck et al., 1990). The second rate is similar to the depolymerization rate of pure microtubules in vitro (∼21 μm/min; Walker et al., 1989), astral microtubules in prometaphase newt lung epithelial cells (Hayden et al., 1990), and interphase microtubules in CHO (∼32 μm/min) and PtK (∼20 μm/min) cells (Shelden and Wadsworth, 1993). Our measured depolymerization rate of ∼30 μm/min for both spindle and astral microtubules in Xenopus egg extracts is similar to these previously reported, rapid rates, and slightly slower than reported for plus ends in clarified Xenopus egg extracts (Parsons and Salmon, 1997). This leads us to believe that this rate, twofold faster than the rate measured for fluorescent microtubules nucleated from centrosomes in Xenopus egg extracts (Belmont and Mitchison, 1996; Tournebize et al., 2000; Wilde et al., 2001; Tirnauer et al., 2002b) is a good estimate of an intrinsic microtubule depolymerization rate for microtubules in the spindle. At present we are unsure of the cause of the discrepancy in astral microtubule depolymerization rates measured by the methods here vs. on centrosome-nucleated microtubule asters. One possibility is that α-MCAK-induced microtubule bundling altered the depolymerization of astral microtubules in our system, causing them to acquire depolymerization properties of spindle microtubules. Another is that the methods used for single microtubule imaging underestimated depolymerization rates because of the need to add fluorescent tubulin as a probe or the need to image microtubules near the coverslip in very thin preparations. If it is true that microtubules in spindles and α-MCAK-induced asters depolymerize faster than single, isolated microtubules, there may exist in spindles depolymerization-enhancing factors with a significant role in regulating dynamics. XMAP215 can accelerate depolymerization (Vasquez et al., 1994) and is a candidate for such an activity.
We observed gradual reduction of contrast in the spindle depolymerization front as it approached the pole, which could have several causes. These include asynchronous rates of depolymerization, although we found the depolymerization front to be remarkably synchronous for the first few micrometers. Rescue events occurring on depolymerizing microtubules could reduce contrast as these microtubules resumed polymerization. Similarly, elongation of uncut or newly nucleated microtubules originating near the pole could replace those lost by depolymerization. Microtubule minus ends may also be distributed over a large area, as was recently suggested by high-resolution tracking of microtubule speckles in a Xenopus egg extract spindle (Valloton et al., 2003). Finally, translational movements of microtubules, due, for example, to microtubule release from centrosomes, could affect the degree of contrast in this region. We cannot formally distinguish among these possibilities, and we suspect that a combination is likely to be responsible.
A Subset of Spindle Microtubules Is Stable to Depolymerization
The current theory of dynamic instability predicts that when their internal GDP-tubulin lattice is exposed by severing, microtubules will depolymerize from their newly exposed plus ends. During our experiments we reproducibly observed that a subset of microtubules failed to depolymerize upon spindle cutting; we believe these to be microtubules of opposite polarity with minus ends exposed. Several types of experiments support the mixed polarity of microtubules in this system. Our own data from fluorescent EB1 imaging is consistent with the presence of antiparallel microtubules throughout the spindle. Additionally, electron microscopy with hook decoration as well as rigorous mathematical analysis of fluorescent speckle movements in Xenopus egg extract spindles both support the finding of antiparallel microtubule overlap in all spindle regions, greatest at the midspindle and extending to the spindle poles (Heald et al., 1997; Valloton et al., 2003). Extensive overlap of nonkinetochore microtubules was also observed by EM in PtK cells at metaphase (Mastronarde et al., 1993). Thus, most of our central cuts are predicted to expose approximately equal numbers of microtubule plus and minus ends. Cutting of kinetochore fibers in cells and pure microtubules in vitro resulted in rapidly depolymerizing plus ends and relatively stable minus ends (Walker et al., 1989; Spurck et al., 1997; Tran et al., 1997). In our asymmetric cuts, we observed preferential stability of microtubules in the larger fragment, consistent with the hypothesis that cut minus ends are differentially stable to depolymerization within the spindle. This does not rule out the possibility that microtubules are selectively stabilized by a factor in the midspindle that partitions with the larger spindle fragment; chromatin-associated proteins or proteins that recognize microtubule overlap could fulfill such a role.
Future Questions
If polymerization and depolymerization rates are not altered by the spindle interior environment, then how does the spindle form and maintain itself in these highly microtubule-destabilizing egg extracts? Our work suggests that the major parameters of dynamic instability altered within the spindle must be catastrophe, rescue, and/or new nucleation. Our results are consistent with the Ran hypothesis, as the major effect of constitutively active Ran on microtubule dynamics is to increase the frequency of rescues and reduce catastrophes, without dramatically affecting polymerization or depolymerization rates (Carazo-Salas et al., 2001; Wilde et al., 2001). The fact that we could see depolymerization extending at least 5 μm without significant blurring of the front leads us to suspect that rescues are rare for spindle microtubules near the poles. It will be interesting to see whether there is a gradient of increased microtubule rescues and/or reduced catastrophes around the chromatin that parallels the distribution of Ran activity. Another critical parameter that needs to be measured is the distribution of microtubule nucleation sites; these may be distributed throughout the spindle, with a predominance at poles and in the equatorial region. Quantitative modeling of these parameters will need to be combined with testing of molecular candidates for their regulation, including not only Ran but also major microtubule dynamics factors such as MCAK, XMAP215, and EB1.
Supplementary Material
Acknowledgments
We thank Mel Spiegel for advice on needles and use of the needle puller and Zach Perlman for advice on image analysis. We also thank members of the Woods Hole Cell Division Group (including Julie Canman, Lisa Cameron, Arshad Desai, Christine Field, Aaron Groen, Paul Maddox, and Karen Oegema), members of the Mitchison Lab, and Jennifer Waters-Shuler at the Nikon Imaging Center at Harvard Medical School. Mimi Shirasu-Hiza and Zach Perlman made insightful comments on the manuscript. This work was supported by a Universal Imaging Corporation Fellowship to the Cell Division Group at the Marine Biological Laboratories, Woods Hole, MA; and by National Institutes of Health Grants DK02578 and DK58766 (to J.S.T.), GM-24364 and GM-60678 (to E.D.S.), and GM-39565 (to T.J.M.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-11-0824. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-11-0824.
Abbreviations used: EM, electron microscopy; FSM, fluorescence speckle microscopy; MCAK, mitotic centromere associated kinesin (also called XKCM1 Xenopus kinesin central motor domain 1).
Online version of this article contains videos. Online version is available at www.molbiolcell.org.
References
- Belmont, L.D., and Mitchison, T.J. (1996). Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623-631. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R.E., Gruss, O.J., Mattaj, I.W., and Karsenti, E. (2001). Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3, 228-234. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Desai, A., Murray, A.W., Mitchison, T.J., and Walczak, C.E. (1999). The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385-412. [DOI] [PubMed] [Google Scholar]
- Forer, A., Kalnins, V.I., and Zimmerman, A.M. (1976). Spindle birefringence of isolated mitotic apparatus: further evidence for two birefringent spindle components. J. Cell Sci. 22, 115-131. [DOI] [PubMed] [Google Scholar]
- Harris, P.J., and Clason, E.L. (1992). Conditions for assembly of tubulin-based structures in unfertilized sea urchin eggs. Spirals, monoasters, and cytasters. J. Cell Sci. 102, 557-567. [DOI] [PubMed] [Google Scholar]
- Hayden, J.H., Bowser, S.S., and Rieder, C.L. (1990). Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J. Cell Biol. 111, 1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A., Drechsel, D., Kellogg, D., Salser, S., Sawin, K., Steffen, P., Wordeman, L., and Mitchison, T. (1991). Preparation of modified tubulins. Methods Enzymol. 196, 478-485. [DOI] [PubMed] [Google Scholar]
- Inoue, S., and Salmon, E.D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., Weis, K., and Heald, R. (2002). Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295, 2452-2456. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle: a self-made machine. Science 294, 543-547. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Arnal, I., Desai, A., Drechsel, D.N., and Hyman, A.A. (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340-1343. [DOI] [PubMed] [Google Scholar]
- Leslie, R.J., and Pickett-Heaps, J.D. (1984). Spindle microtubule dynamics following ultraviolet-microbeam irradiations of mitotic diatoms. Cell 36, 717-727. [DOI] [PubMed] [Google Scholar]
- Maddox, P., Aaron, S., Coughlin, P., Mitchison, T.J., and Salmon, E.D. (2003). Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N., McDonald, K.L., Ding, R., and McIntosh, J.R. (1993). Interpolar spindle microtubules in PtK cells. J. Cell Biol. 123, 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., and Kirschner, M.W. (1985). Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J. Cell Biol. 101, 766-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec, F., Surrey, T., and Karsenti, E. (2003). Self-organization and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 15, 118-124. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B., Lee, G.M., Rieder, C.L., and Rupp, G. (1989). Mechanically cut mitotic spindles: clean cuts and stable microtubules. J. Cell Sci. 94, 415-423. [DOI] [PubMed] [Google Scholar]
- Parsons, S.F., and Salmon, E.D. (1997). Microtubule assembly in clarified Xenopus egg extracts. Cell Motil. Cytoskel. 36, 1-11. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and Khodjakov, A. (2003). Mitosis through the microscope: advances in seeing inside live dividing cells. Science 300, 91-96. [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., and Mitchison, T.J. (1991). Poleward microtubule flux in mitotic spindles assembled in vitro. J. Cell Biol. 112, 941-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden, E., and Wadsworth, P. (1993). Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J. Cell Biol. 120, 935-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurck, T.P., Forer, A., and Pickett-Heaps, J.D. (1997). Ultraviolet microbeam irradiations of epithelial and spermatocyte spindles suggest that forces act on the kinetochore fibre and are not generated by its disassembly. Cell Motil. Cytoskel. 36, 136-148. [DOI] [PubMed] [Google Scholar]
- Spurck, T.P., Stonington, O.G., Snyder, J.A., Pickett-Heaps, J.D., Bajer, A., and Mole-Bajer, J. (1990). UV microbeam irradiations of the mitotic spindle. II. Spindle fiber dynamics and force production. J. Cell Biol. 111, 1505-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., Canman, J.C., Salmon, E.D., and Mitchison, T.J. (2002a). EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell 13, 4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., Grego, S., Salmon, E.D., and Mitchison, T.J. (2002b). EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell 13, 3614-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize, R. et al. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13-19. [DOI] [PubMed] [Google Scholar]
- Tran, P.T., Walker, R.A., and Salmon, E.D. (1997). A metastable intermediate state of microtubule dynamic instability that differs significantly between plus and minus ends. J. Cell Biol. 138, 105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieselmann, N., and Wilde, A. (2002). Ran localizes around the microtubule spindle in vivo during mitosis in Drosophila embryos. Curr. Biol. 12, 1124-1129. [DOI] [PubMed] [Google Scholar]
- Valloton, P., Ponti, A., Waterman-Storer, C.M., Salmon, E.D., and Danuser, G. (2003). Recovery, visualization, and analysis of actin and tubulin polymer flow in live cells: a fluorescent speckle microscopy study. Biophys. J. 85, 1289-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez, R.J., Gard, D.L., and Cassimeris, L. (1994). XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 127, 985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.A., Inoue, S., and Salmon, E.D. (1989). Asymmetric behavior of severed microtubule ends after ultraviolet-microbeam irradiation of individual microtubules in vitro. J. Cell Biol. 108, 931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., Desai, A., Bulinski, J.C., and Salmon, E.D. (1998). Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8, 1227-1230. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., and Salmon, E.D. (1997). Pathways of spindle assembly. Curr. Opin. Cell Biol. 9, 37-43. [DOI] [PubMed] [Google Scholar]
- Wilde, A., Lizarraga, S.B., Zhang, L., Wiese, C., Gliksman, N.R., Walczak, C.E., and Zheng, Y. (2001). Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3, 221-227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.