Abstract
The upper layers (II–IV) are the most prominent distinguishing feature of mammalian neocortex compared with avian or reptilian dorsal cortex, and are vastly expanded in primates. Although the time-dependent embryonic generation of upper-layer cells is genetically instructed within their parental progenitors, mechanisms governing cell-intrinsic fate transitions remain obscure. POU-homeodomain transcription factors Pou3f3 and Pou3f2 (Brn1 and Brn2) are known to label postmitotic upper-layer cells, and are redundantly required for their production. We find that the onset of Pou3f3/2 expression actually occurs in ventricular zone (VZ) progenitors, and that Pou3f3/2 subsequently label neural progeny switching from deep-layer Ctip2+ identity to Satb2+ upper-layer fate as they migrate to proper superficial positions. By using an Engrailed dominant-negative repressor, we show that sustained neurogenesis after the deep- to upper-layer transition requires the proneual action of Pou3fs in VZ progenitors. Conversely, single-gene overexpression of any Pou3f in early neural progenitors is sufficient to specify the precocious birth of Satb2+ daughter neurons that extend axons to the contralateral hemisphere, as well as exhibit robust pia-directed migration that is characteristic of upper-layer cells. Finally, we demonstrate that Pou3fs influence multiple stages of neurogenesis by suppressing Notch effector Hes5, and promoting the expression of proneural transcription factors Tbr2 and Tbr1.
Keywords: development, differentiation, In utero electroporation, Neurogenesis, Upper layer
Introduction
The assembly of a 6-layered mammalian neocortex during development requires the production of neurons of the correct subtype in a tightly coordinated process, temporally and spatially (Caviness et al. 2009). It is well understood that in all mammals, including primates, deep-layer cells arise first and migrate first, whereas cells of the superficial layers are born and migrate later (Angevine and Sidman 1961; Rakic 1974). Furthermore, the production of distinct classes of pyramidal neurons follows a well-defined temporal sequence that is encoded in the molecular genetic [and perhaps epigenetic (Fasano et al. 2009; Okano and Temple 2009; Sanosaka et al. 2009)] makeup of their progenitor neural stem cells (Shen et al. 2006). However, the specific means that effect intrinsic fate transitions within the progenitor populations remain obscure. These progenitors reside along the ventricular surface, a long distance from the final positions of their daughter neurons, in the cortical plate (CP). As a result, migrating neurons, especially newborn pyramidal cells of the upper layers, are guided by radial glia (RG) fibers that span the entire cerebral wall (Rakic 1972).
The cortical upper layers, which are unique to Class Mammalia, probably underlie the enhanced cognitive abilities of mammals because they form connections between and within cortical areas (Marín-Padilla 1992; Hill and Walsh 2005). Compared with carnivorans and especially with rodents, the expansion of cortical surface in primates, including human, occurs concomitantly with expansion of the supragranular layers (II–III) (Hutsler et al. 2005). In humans, disturbances in the function of layers II and III have a broad spectrum that include cognitive disorders such as schizophrenia (Rajkowska et al. 1998). Additionally, failed neural migration and resulting heterotopias predominantly affect upper-layer cells (Ferland et al. 2009), and are a common etiology in epilepsy (Guerrini et al. 2008).
POU-homebox-domain-containing octamer-binding transcription factors Pou3f3/Brn1 and the very closely related Pou3f2/Brn2 have long been markers for upper-layer projection neurons (Hagino-Yamagishi et al. 1997) in cortex, but have been implicated in diverse functions such as cortical neural migration (McEvilly et al. 2002), upper-layer production (Sugitani et al. 2002), and neurogenesis (Castro et al. 2006). The first analysis of Brn1/2 knockout cortex demonstrated that Brn1 and Brn2 redundantly regulate key determinants for the radial migration of postmitotic neurons, and purported that their loss results in cortex laminar inversion (McEvilly et al. 2002). A later study showed that Brn1/2 was necessary for migration of Er81+ layer V cells, and for the actual production of layer II–IV cells (Sugitani et al. 2002); however, the mechanisms carrying out the latter process have remained elusive.
In this study, we have addressed this key issue by examining neurogenesis with respect to the spatiotemporal onset of Brn1/2 expression in mouse dorsal telencephalon. By using in utero electroporation, we have conducted precisely timed genetic manipulations of Brn1/2 in vivo to show when and how Pou3f factors are required for upper-layer neural specification and migration. Furthermore, we investigated the consequences of Brn1, Brn2, and Pou3f1/Oct6/Tst1/SCIP misexpression during development of the deep cortical layers to identify potential genetic effector pathways in their function as upper-layer neural determinants. In doing so, we concretely show that neural molecular identity, much of which is obtained postmitotically (Fishell and Hanashima 2008), has its roots in the function of molecules whose expression is governed spatio-temporally in the parental stem cells.
Materials and Methods
DNA Constructs
The coding sequences for Brn1, Brn2, Oct6, Hes1, Hes5, Hes6, NeuroD2, Ngn, Pax6, and Satb2 were amplified by PCR from E14.5 mouse embryo dorsal telencephalon cDNA with primers containg flanking NotI sites (or EcoRI for Hes5), and cloned into the NotI site (or EcoRI for Hes5) of pCAGEN (Matsuda and Cepko 2004). pUbC-NICD was generated by subcloning the mouse Notch1 intracellular domain with 6X myc tag (Kopan et al. 1994) into a customized vector based on pUB-EGFP (Matsuda and Cepko 2004). pIRES2-Tbr2 was generated by cloning the mouse Tbr2 coding sequence into pIRES2 (Clontech). pEGFP-dnMAML (Maillard et al. 2006) and pCAG-Brn2-DBD-EnR(“Brn2-EnR”) (Kim et al. 2008) have been described. pCAG-EnR(“EnR”) was made by digesting pCAG-Brn2-DBD-EnR at the EcoRI and AgeI sites, and ligating the pre-annealed primers AATTCTGAGCCGCCACCATG and CCGGCATGGTGGCGGCTCAG into the intervening region. pCAG-Brn1-DBD-EnR was constructed similarly to pCAG-EnR, except a fragment containing Brn1 DNA-binding domains analogous to those found in Brn2-DBD-EnR was ligated. pCAG-TagBFP_V5_NLS(“V5” or “Electroporated Nuclei”) contains the TagBFP coding sequence (Evrogen) with a C-terminal V5 epitope tag (“GKPIPNPLLGLDST”) followed by a 3× NLS (“DPKKKRKV”; dervied from the SV40 large T-antigen) cloned into pCAGEN. All electroporation experiments were carried out with DNA solution containing the following: 400 ng/μL pCAG-green fluorescent protein (Kwan et al. 2008), 800 ng/μL pEGFP-F (Clontech), 400 ng/μL pCAG-TagBFP_V5_NLS, and either 1.5 μg/μL (for Pou3f overexpression) or 1.3 μg/μL (for EnR) target DNA. Combination electroporations were performed with the EnR plasmid fixed at 1.3 μg/μL, with the other plasmid at the indicated ratio. Owing to an established ∼95% cotransfection yield with 2 electroporated plasmids (Ramos et al. 2006), a small fraction of V5+ or GFP+ electroporated cells will not exhibit exogenous gene-driven phenotypic changes, and vice versa.
Mice and Electroporation
Experiments and handling procedures were performed with permission and in compliance with the Institutional Animal Care and Use Committee (IACUC). Timed pregnant female CD-1 mice were purchased from Charles Rivers Laboratories and housed at the Yale animal care facility. In utero electroporation has been described previously (Gal et al. 2006); briefly, DNA solution was injected into embryonic telencephalic vesicles in utero via glass micropipetes, and 5 50 ms square fixed-potential pulses (ElectroSquarePorator ECM 830: BTX, Holliston, MA, United States of America) were administered by tweezertrodes (Fisher Scientific). The pulse potential was adjusted to maximize in utero electroporation yield, and varied from 23V(E11.5) to 42V(E13.5). Reproducibility was demonstrated in the phenotypic evaluation of at least 3 brains (at E14.5, E15.5, or P0) for every experimental condition, while quantifications utilized 2 or more brains with matching electroporations for each condition. Postnatal electroporation has been described previously (Boutin et al. 2008; Chesler et al. 2008); briefly, DNA solution was injected into the lateral ventricles of mixed sex anesthetized P0 mouse pups via glass micropipetes, and 135V electric pulses administered as per the above, except that tweezertrodes were coated with electrode gel (Parker Laboratories). For confirmation, we examined at least 6 postnatal brains for each condition that were electroporated in 2 or more distinct experimental trials. All anesthesia and postoperative care was administered in accordance with directives of the Yale IACUC.
Non-Human Primate Husbandry
Female rhesus monkeys were housed with a male for 3 days. Pregnancy was confirmed by ELISA testing of blood mCG levels at 12 and 18 days post-copulation. Rhesus macaque embryos (n = 3) were obtained by cesarean section. Rhesus macaque surgeries and post-operative care for mothers was coordinated with the Yale Veterinary clinical services. All procedures were approved by the Yale IACUC.
Cell Cultures, Transfection, and Lysis
Mouse neuroblastoma 2A (N2a) cells were cultured in Dulbecco's D-MEM/F12 medium containing 9% fetal bovine serum (Invitrogen). Transfections were performed on 6-well plates at 80% confluency for immunoblotting (IB), or 12-well plates containing 50 000 cells for luciferase assays. 500 ng or 1 μg total DNA was incubated for 30 min (room temperature) in 40 or 80 μL Opti-MEM with 1 or 2 μL Dharmafect Duo reagent (Thermo Scientific) for luciferase or IB, respectively; following this incubation, the mixture was added to 300 or 900 μL serum-free medium in each well. Transfections for luciferase assays were with mixed with precise equimolar quantities of any DNA constructs used for side-by-side comparison. After 24 h, cells were washed in phosphate-buffered saline. For IB, cells were immediately scraped and lysed in radioimmunoprecipitation assay buffer (Cell Signaling) containing protease (Roche) and phosphatase (Sigma) inhibitor cocktails. Insoluble material was pelleted at 10k rpm for 10 min on a laboratory microcentrifuge, and total protein content of the supernatant was read using the Bradford assay.
Western and Luciferase Analysis
For immunoblot analysis, proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and then electrophoretically transferred to polyvinylidene fluoride membranes. Membranes were immunoblotted with anti-Myc (Cell Signaling 9B11, 1:1000), anti-GFP (abcam ab290, 1:2500), anti-glyceraldehyde 3-phosphate dehydrogenase (Millipore MAB374, 1:2000), anti-Satb2 (abcam ab51502, 1:200), anti-Tbr2 (abcam ab23345 1:1000), or anti-Brn2 (sc-6029, 1:400) overnight, and subsequently washed and incubated with goat anti-mouse (Biorad), goat anti-rabbit (Biorad), or donkey anti-goat (Jackon Immunoresearch) horseradish peroxidase conjugates. Exposure of enhanced chemiluminescence (General Electric Healthcare Life Sciences)-treated membranes allowed visualization of protein. Luciferase assays were performed using the Dual-Luciferase Reporter Assay System (Promega) and cotransfection of pRL-SV40 (Promega) with firefly luciferase reporters; PBS-washed cells were lysed and prepared according to manufacturer protocol. Samples were read on a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA, USA), with all conditions biologically quadruplicated for statistical analysis. All luciferase reporter plasmids were constructed according to depicted schemes by replacing the SV40 promoter in pGL3-Promoter (Promega) with the appropriate PCR-amplified mouse regulatory regions (with the exception that the Satb2-Enhancer reporters retain the minimal promoter). pGV-B-Hes1/5-Luciferase reporter plasmids have been described (Nishimura et al. 1998).
Tissue Preparation and Staining
P0 brains or whole embryo heads were immersion-fixed in 4% paraformaldehyde (PFA) for 4–8 h, cryoprotected to 30% sucrose via serial solutions, frozen, and sectioned on a cryostat (20 μm coronal sections; Leica Microsystems). To enhance the staining of nuclear markers, sections were incubated for 20 min in target retrieval solution at 95°C (DAKO Cytomation) prior to labeling. Postnatal brains were immersion-fixed overnight in 4% PFA and sectioned on a vibratome (80 μm coronal sections; Leica Microsystems). For immunolabeling, sections were incubated in primary antibodies in 5% normal donkey serum and 0.4% TX-100 overnight at 4°C (or room temperature for anti-Ngn2 or anti-CDP labeling). After 2 brief rinses in PBS, sections were incubated for 2h at room temperature in PBS with fluorophore-conjugated secondary antibodies raised in donkey hosts (Jackon Immunoresearch, 1:500). Following 2 brief rinses in PBS, sections were mounted in 2.5% polyvinyl alcohol with diazabicyclooctane (PVA-DABCO) (Evans et al. 2004). The following primary antibodies were used: Brn2 (Santa Cruz Biotechnology, goat, 1:250), Satb2 (abcam, mouse, 1:400), Ctip2 (abcam, rat, 1:500), Tbr2 (abcam, rabbit, 1:400), Tbr1 (Millipore, chicken, 1:2000), Tbr1 (abcam, rabbit, 1:1000), V5 (abcam, chicken, 1:200), GFP (abcam, chicken, 1:2000), Pax6 (Millipore, rabbit, 1:2000), Cux1 (“CDP,” Santra Cruz Biotechnology, rabbit, 1:50), Ngn2 (Santa Cruz Biotechnology, goat, 1:33), Sox2 (Santa Cruz Biotechnology, goat, 1:250).
Imaging and Quantification
Immunostained specimens were imaged with a Carl Zeiss AxioCam MRm coupled to Axioplan 2 or Axioimager Z2 epifluorescence microscopes (Carl Zeiss Microimaging, Thornwood, NY, United States of America) in Carl Zeiss ApoTome optical sectioning mode. We also utilized a Carl Zeiss LSM 510 Meta laser scanning confocal microscope, and a Coherent Chameleon Ultra laser (Coherent Laser Group, Santa Clara, CA, United States of America) coupled to an AxioObserver. Image processing was performed using Carl Zeiss Axiovision software and gimp 2.6 (http://www.gimp.org). Quantification of images was performed automatically by a custom-written, unbiased algorithm that measured the overlap of 2 color channels (i.e. red and blue) with a third channel (i.e. green). In brief, the program conducted a weak gaussian blur and gamma reduction on the third channel to eliminate image noise. The intensity of the third channel was then used as a weight for the amount of the other 2 colors on an individual pixel basis, which was then reported for each image in a spreadsheet. At least 5 images representing rostral, caudal, medial, and lateral regions were used to comprise each data point. Binning was performed with the same approach, except that red or blue colored boxes were drawn (with guidance from DAPI or laminar markers) in superposition of the GFP (green) channel such that per-pixel overlap with defined red–blue combinations reflected the position of the labeling. All statistical computations were performed with OpenOffice (http://openoffice.org) or R(http://www.r-project.org); 1- or 2-factor analysis of variance (ANOVA) was employed where appropriate (usually by § sign), but all P-values shown in figures reflect 2-tailed unpaired t-test results, except those provided with contingency tables, which reflect significance assessment by Fisher's exact test.
Results
Spatiotemporal Control of Neuronal Subtype Progression in Cortex is Identifiable by Brn1/2 Expression
It is well known that spatiotemporal gradients of neurogenesis govern the time-dependent formation of cortical layers in all studied mammals (Sidman and Rakic 1973; Sanderson and Weller 1990; Machon et al. 2007; Bayatti, Moss, et al. 2008; Bayatti, Sarma, et al. 2008), such that the onset of neurogenesis begins in a rostrolateral position and sweeps caudomedially; thus, the lateral cortex produces neurons earlier than medial cortex. Pax6 levels increase in the same rostrolateral-to-caudomedial gradient, exactly in lockstep with the appearance of Ngn2+ and Tbr2+ committed neural precursors (Machon et al. 2007). However, Pax6 eventually encompasses the entire cortical ventricular zone (VZ). Fate transitions that direct the production of progressively later-born neuronal subtypes within a progenitor lineage begin their first iteration in lateral cortex of the mouse around E11. Cortical upper-layer (II–IV) cells are eventually specified laterally around E12, but medial regions experience parallel transitions about 1 day or 2 thenceforth (Takahashi et al. 1999).
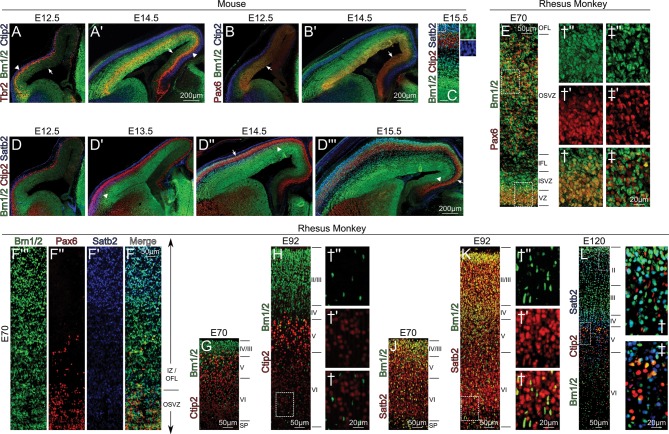
Using a Brn2 antibody that recognizes both Brn1 and Brn2 (Yamanaka et al. 2010), we examined the molecular distribution of Brn1/2 in the cortex from early through late corticogenesis (Fig. 1, Supplementary Figs 1 and 2). Fascinatingly, they are expressed in neural progenitors in a similar gradient as above, and their expression also spreads across the entire cortical lateral-to-medial axis during the course of neurogenesis (Fig. 1). The spatiotemporal onset of Brn1/2 expression lags behind that of Pax6 (Fig. 1B,B′), and therefore could correlate with mid-to-late neurogenic changes. Moreover, the Tbr2+ intermediate progenitor (INP) population is heterogeneous with respect to Brn1/2 expression; at E12.5, only the lateral-most INPs are Brn1/2+. However, by E14.5 the medial expansion of Brn1/2 also incorporates this INP population (Fig. 1A,A′).
Figure 1.
The wild-type developing neocortex switches from producing deep-layer to upper-layer neurons in a lateral-to-medial graded fashion, which parallels the expression of Brn1/2. (A and A′) Tbr2+ (red) intermediate neural progenitors (INPs) are a heterogeneous population, in that they colabel with Brn1/2 (green) only in the lateral-most region of cortex at E12.5 (A), this boundary indicated by arrowhead. The medial spread of Brn1/2 (boundary indicated by arrows) in the VZ after E12.5 correlates with Brn1/2 persistence in INPs at E14.5 (A′). (B and B′) The lateral-to-medial induction of Brn1/2 (green) expression (arrows) parallels that of Pax6 (red) from E12.5 (B) to E14.5 (B′); however, Brn1/2 expression appears to lag slightly behind that of Pax6. (C) At E15.5, Brn1/2 (green) labels cells in all phases of differentiation; however, its persistence in postmitotic neurons correlates with Satb2 (blue) upper-layer cells and not Ctip2 (red) deep-layer cells. One caveat to this rule is that a population of prospective Ctip2+ layer V cells initiates weak Brn1/2 expression in postmigratory positions. Scale bar, 50 μm. (D and D′) Time-course for the onset of Brn1/2 (green) expression, and its correlation with Satb2+ (blue) migrating upper-layer neurons and not Ctip2+ (red) deep-layer residents. Brn1/2 sweeps across the entire lateral-to-mediate axis between E12.5 (D), E13.5 (D′), and E14.5 (D″), and is stably expressed throughout by E15.5 (D′″). Arrowheads in (D′ and D′″) indicate the medial boundary of the Brn1/2+/Satb2+ population in the IZ, which manifests itself in the CP with a lagging medial boundary (demarcated by arrows). The delay between these moving fronts probably correlates with the time required for the Brn1/2+/Satb2+ cells to migrate to subpial inside-out positions in the CP. (E) Proliferative compartment along the parietal ventricular surface in rhesus at E70. While the VZ proper(†) appears to be largely Pax6 (red)-Brn1/2 (green) double+, the OSVZ(‡) is heterogeneous with respect to the cellular expression of those markers. (F) Brn1/2 (green) cells presumably migrating out of the proliferative zone(Pax6, red) express Satb2 (blue) as they approach the parietal CP. (G and H) Monkey parietal CP at E70 (G) and E92 (H) exhibiting deep-layer Ctip2 (red) expression and upper-layer Brn1/2 (green) expression. (J and K) Monkey parietal CP at E70 (J) and E92 (K) exhibiting colabeling of the vast majority (see †) of Brn1/2 (green) cells with Satb2 (red), whereas many Brn1/2− cells throughout the CP express Satb2 at this time. (L) Brn1/2 (green), Ctip2 (red), and Satb2 (blue) labeling in parietal CP at E120, demonstrating upper-layer expression of Brn1/2. Similar to earlier time points, layer II cells near layer I form a dense band and are strongly Brn1/2 positive; these cells are likely the most recent pyramidal cells to have reached the CP, as their size and density reveal their relative immaturity.
At mid-corticogenesis in the mouse, Satb2 confers upper-layer identity, and is initially present in cells that project intracerebrally (Alcamo et al. 2008; Britanova et al. 2008; Chen et al. 2008). Conversely, Ctip2 labels all deep layer cells during embryogenesis, but is slowly restricted to a subpopulation of layer V corticospinal motor neurons (Kwan et al. 2008). In surveying Ctip2 and Satb2, we find that the developing mouse neocortex switches from the production of Ctip2+ deep-layer neurons to Satb2+ upper layer-neurons in a lateral-to-medial graded fashion (Fig. 1D,D′″). This process occurs in the mouse between E13 and E15, and parallels the lateral-to-medial graded onset of neurogenesis of 2 days prior. Interestingly, Brn1/2 protein only accumulates to sufficient levels to be inherited by neural progeny starting at mid-neurogenesis, subsequently labeling cells switching from Ctip2 expression to Satb2 in the intermediate zone (IZ) as they migrate to the upper layers of the CP (Fig. 1D,D′″). Brn1/2 is excluded from Ctip2+ deep-layer cells and their parent progenitors, although a small population of post-migratory layer V cells appears to express Brn1/2 after E15.5 (Supplementary Fig. 2). Thus, the onset and course of Brn1/2 expression is timed to be in precise concordance with the specification and differentiation of upper-layer cells.
To examine primate neurogenesis with a specific focus on upper-layer cell birth and differentiation, we investigated Brn1/2 localization in rhesus monkey embryos. At E70, which is around the time of mid-corticogenesis, Pax6+ cells of the VZ and OSVZ also expressed Brn1/2 (Fig. 1E)—similar to the VZ in mouse at E14.5. However, many Pax6+ cells, especially those in the OSVZ, remained Brn1/2 negative (Fig. 1E‡), and we are unsure about the nature of this OSVZ-specific heterogeneity at present. Nonetheless, presumptive Brn1/2+ cells migrating out of the proliferative zones and into the IZ subsequently express Satb2 (Fig. 1F), similar to migrating neurons at mid-corticogenesis in the mouse. In the rhesus CP at E70 and E92 (Fig. 1G,H), Brn1/2 prospective upper-layer cells are predominantly Ctip2-, and many are seen in the deep-layers with elongated nuclei characteristic of radially migrating cells (Fig. 1H† and Supplementary Fig. 3L). These migrating Brn1/2+ cells were also Satb2 positive (Fig. 1J,K) as expected.
Nevertheless, the primate CP exhibits Satb2 expression throughout layers II–VI from E70 to E92, unlike the rodent CP at equivalent stages. A comparative analysis (Supplementary Fig. 3) suggests that this may be due to post-migratory cellular maturation, which occurs within a time window that is exceeded by the long course of neurogenesis in primates (50 days+), but not by the rapid birth of cells belonging to the different layers in the mouse (∼5 days). Consistent with this notion, a subset of layer VI Tbr1+ cells in the mouse also expresses Satb2—by around P0 (Supplementary Fig. 3J), but not during embryogenesiss. Furthermore, Ctip2 labeling alone cannot distinguish the cells of layers V and VI at E14.5 in the mouse, whereas such a distinction is obvious by the size and intensity of Ctip2 nuclear staining of the primate cortex at E70, a semi-equivalent age (Supplementary Fig. 3C vs. D).
Early Overexpression of Pou3f Transcription Factors Promotes Aberrant Fate Switching to Upper-Layer Neuron Identities
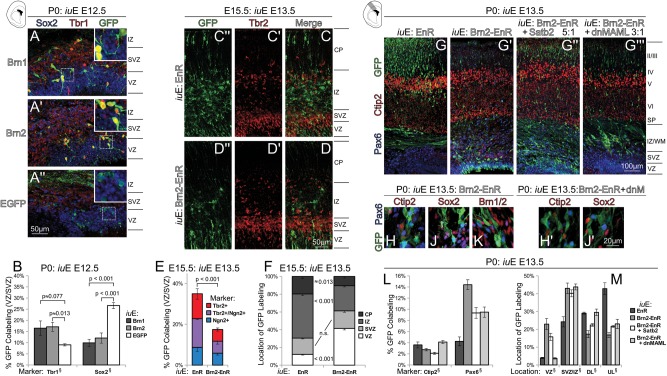
To determine whether Brn1/2 has a role in the switch to upper-layer neuronal identities, we overexpressed them by in utero electroporation in early developing cortex, when predominantly layer VI and V cells are specified. Overexpression of either Brn1 or Brn2 at both E11.5 and E12.5 effected substantial changes in the types of cells (Fig. 2A–D) produced. We examined Cux1 and Ctip2 expression at P0, which are normally associated with layers II–IV and layer V corticospinal tract neural identities, respectively. Compared with control electroporations, Brn1/2 caused a marked increase in the number of Cux1+ cells (Fig. 2C), and a concomitant decrease in Ctip2+ cells (Fig. 2D) was seen at P0.
Figure 2.

Overexpression of Brn1/2 in early cortex development is sufficient to specify the birth of Satb2+ neurons that are normally born later. (A and B″) Electroporations at E11.5 (A and A″) or E12.5 (B and B″) to force the expression of Brn1 (A′ and B′), Brn2 (A″ and B″), or EGFP alone (A and B) were compared at P0 and labeled for Cux1 (blue) and Ctip2 (red). Fewer V5+/Ctip2+ nuclei and far greater numbers of V5+/Cux1+ nuclei were seen at P0. (C and D) Quantifications of the overlap of V5+ nuclei and laminar markers Cux1 (C) and Ctip2 (D) at P0 in these electroporations reflect these substantial molecular genetic changes. Data from EGFP electroporations at E13.5 is used as a reference to depict the normal cell output during the time that Satb2+ or Cux1+ upper-layer cells are normally specified. § denotes 1-factor ANOVA P ≤ 0.02. Two-factor ANOVAs were also performed for Ctip2 and Cux1 labeling results, with age of electroporation and construct electroporated as independent variables; both factors demonstrated significant (P ≤ 0.03) contributions. (E and F″) Small unilateral medial electroporation of Brn2 at E12.5, fixed at E14.5. 10× magnification imaging of both cortical hemispheres at E14.5 (E″ and E) demonstrates that Satb2+(red) nuclei are normally absent in the medial CP (on the right, unelectroporated side) hemisphere, but sporadic Satb2+ cells are present on the left electroporated side. Higher-magnification optical sectioning (F″ and F) reveals a near complete overlap of these Satb2+ nuclei with V5+(green) nuclei of electroporated cells. Normally, Ctip2+(blue) cells are still being born at time in this position (right side). (G and H″) Intermediate positions and larger electroporations of Brn2 at E12.5 reveal the same at E14.5; normally, Ctip2+(blue) deep-layer cells are the first progeny of E12.5 electroporations (G″ and G); however, overexpression of Brn2 (H″ and H) causes a switching of these cells to Satb2+ upper-layer fate. (J and K′) Electroporation of EGFP in cortex at E11.5 strongly labels cells of deep laminar positions (arrows in J and J′) in coronal sections that project subcortically via the internal capsule (arrowheads in J and J′). However, overexpression of Brn2 (K and K′) by electroporation at E11.5 does not render this palisade of GFP in layer VI and the subplate. Furthermore, very few electroporated cells extend axons into the internal capsule (arrowheads, insets in K and K′), although numerous axons were seen traversing the corpus callosum and anterior commissure (asterisk in K′) toward the contralateral cortex.
To visualize this effect in action, we conducted small unilateral medial electroporations of Brn1/2 at E12.5 and examined them at E14.5 (Fig. 2E,F″). At E14.5, the medial cortex has yet to undergo the switch to producing upper-layer cells, and is normally devoid of Satb2+ cells in the IZ and CP (Fig. 2E,F, right side). However, in Brn1/2 electroporations, Satb2+ nuclei were seen in electroporated cells (Fig. 2E,F). Dorsolateral electroporations demonstrated the same result (Fig. 2G,H) that Satb2 is positively regulated by Brn1/2. Interestingly, Brn1/2 electroporated cells did not express Satb2 until they had reached the IZ (Fig. 2H), indicating that there may be other determinants in Satb2 up-regulation. Forced co-expression with Sox11, a known transcriptional co-regulator with Brn2 (Tanaka et al. 2004) that is present only in postmitotic neurons of the IZ and CP (Ayoub et al. 2011), did not affect the kinetics of Satb2 induction by Brn2 (data not shown).
Neural identity also determines the type of axonal projections made. Upper-layer cells make Satb2-dependent commissural projections (Alcamo et al. 2008; Britanova et al. 2008), whereas deep-layer cells project subcortically via the internal capsule in a manner that is complexly regulated by Tbr1, FezF2, and Ctip2 (Chen et al. 2005, 2008; Molyneaux et al. 2005; Bedogni et al. 2010; Han et al. 2011; McKenna et al. 2011). Because control electroporations at E11.5 drive GFP expression in deep-layer neural progeny, prominent GFP-labeled axons are seen in the internal capsule at P0 (Fig. 2J,J′). Consistent with their role in promoting upper-layer neurogenesis, electroporations of Brn1 or Brn2 exhibit a near absence of GFP+ axons in the internal capsule at P0, yet GFP+ callosal projections persist (Fig. 2K,K′).
We also noticed that compared with E11.5 or E12.5 control electroporations, cells overexpressing Brn1 or Brn2 within the CP migrated into more superficial positions (Fig. 3B,B′). When we constructed laminar bins and quantified the position of GFP in either case, we found a significant increase in Brn1/2-transfected cells residing in layers II–IV with a coordinate decrease in cells located in layer VI and the SP (Fig. 3A). In electroporations conducted at E12.5, we observed many Brn1/2-transfected neurons obtaining very superficial positions just beneath or within layer I (Fig. 3C′) by P0; this was exceptionally unusual in control electroporations (Fig. 3C). Such findings are consistent with a model whereby cells endowed with an upper-layer genetic program can attain upper-layer positions despite being born prematurely (Frantz and McConnell 1996).
Figure 3.
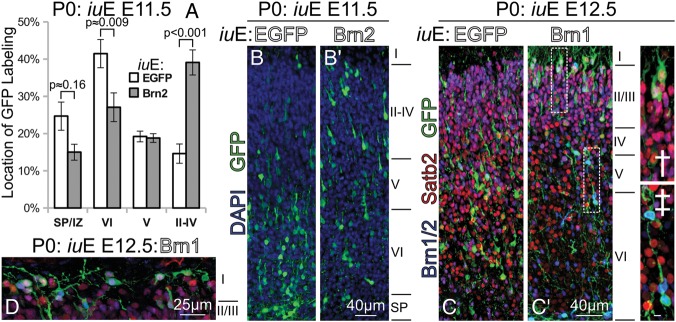
Brn1/2 overexpression confers cell-autonomous pial-directed neural migration to superficial positions. (A) Quantification of the position of GFP labeling at P0 in electroporations of Brn2 versus EGFP performed at E11.5. (B and B′) As shown in (A), Brn2-electroporated cells had a strong preference for superficial laminar positions (B′) compared with EGFP-only electroporated cells (B); this came at the expense of layer VI and subplate positioning. (C and C′) Similar electroporations, although performed at E12.5 with Brn1, result in a gross change in laminar position at P0. Most GFP+(green) cells in EGFP electroporations at E12.5 (C) assume positions predominantly within layer V and VI. Forced expression of Brn2 at E12.5 (C′) results in the layer II/III-targeted migration of GFP+(green)/Satb2+(red) cells at P0, many reaching positions just beneath layer I(C′†). Dashed boxes in (C′) indicate regions of interest enlarged in † and ‡. Some of the highest Brn1-expressing cells remained in the deep layers (C′‡); oddly, these cells labeled very strongly with Brn2 antibody (blue), and were Satb2-. Scale bar in (C′‡), 5 μm. (D) In the caudal cortex, Brn1/2-mediated superficially targeted migration was so robust that many Brn1-electroporated GFP+ (green) electroporated cells were seen in layer I at P0; those cells were immunolabeled, and were positive for Brn2 (blue) and Satb2 (red).
Still, a very small fraction Brn1/2-transfected neurons, especially the ones exhibiting the highest transgene expression, remained in deeper positions (Fig. 3C′‡) or failed to migrate altogether (Fig. 4A,A′). These cells were notable because they were overwhelmingly Satb2- (Fig. 3C′‡), whereas the much larger Satb2+ proportion of nonetheless more weakly transfected cells were chiefly seen in layers II–IV (Fig. 3C′†). On the whole, our data points to a substantial role for Brn1/2, and possibly Satb2, in directing an upper-layer migration process. Moreover, there were regional differences in the effects of Brn1/2 overexpression, such that caudal regions appeared more sensitive to the pro-migratory phenotype (Supplementary Fig. 4B′ vs. A′). We observed the migration of numerous transfected cells into layer I caudally at P0 (Fig. 3D).
Figure 4.
Brn1/2 are proneural genes that are required for neurogenesis in cortex at E13.5. (A and B) Besides prematurely instructing upper-layer identity and migration, Brn1/2 electroporations at E12.5 promoted neural differentiation at the expense of progenitor maintenance, seen at P0. Some of the strongest-expressing Brn1 (A) or Brn2 (A′)-electroporated cells remained in the VZ or SVZ at P0, presumably due to precocious differentiation. Almost no Sox2+(blue) progenitors remained in either Brn1 or Brn2 electroporation (insets), compared with electroporation of EGFP alone (A″). Tbr1 (red), which weakly labels a subpopulation of SVZ cells at P0, strongly labeled many of the neurons remaining in the VZ with Brn1/2 electroporation. A quantification of these findings in P0 brains electroporated at E12.5 is shown in (B) § denotes 1-factor ANOVA P < 0.01. (C and F) When compared with control EnR electroporations performed at E13.5 and examined at E15.5 (C″ and C), Brn2-EnR (D″ and D) electroporations exhibited fewer GFP+(green)/Tbr2+(red) intermediate neural precursors, and fewer GFP+ migrating neurons. This was apparently due to the blockade of early step in neural differentiation by Brn2-EnR; quantifications of electroporated cells demonstrated less Ngn2 and Tbr2 labeling (E), and an increase in the proportion of GFP remaining in the VZ(F). (G and M) Impaired neural differentiation by Brn2-EnR electroporation at E13.5 persisted to P0 (G′), whereas EnR electroporation alone (G) resulted in normal cortical development. Weak forced Satb2 expression was partially able to rescue defects associated with Brn2-EnR (G″), whereas weak forced dnMAML expression was nearly sufficient to overcome impairment of the initial step in neural differentiation by Brn2-EnR (G″′). Numerous Pax6+(blue) or Ctip2+(red) cells were noted in subcortical white-matter positions at P0 in Brn2-EnR electroporations at E13.5 (H); similar cells experienced a failure in migration and/or differentiation in dnMAML-rescued Brn2-EnR electroporations (H′); these cells were largely negative for Sox2 (J and J′) and Brn2 (K and K′) in either case. Quantifications in several sections from each electroporation condition are shown in (L and M: overlap of GFP(green) with Ctip2 (red) or Pax6 (blue)) in (L), while the proportional location of GFP labeling is shown in (M). § denotes 1-factor ANOVA P < 0.001. Pairwise t-test P-values for (L and M) are provided, along with replicate charts, in Supplementary Fig. 6C′–D′, respectively. UL = upper layer and DL = deep layer.
A third Pou3f, Oct6 (Pou3f1/Tst1/SCIP), is expressed during embryonic cortex development in a distribution very similar to that of Brn1/2 (Jones et al. 2009). To determine whether the above effects are specific to Brn1 and Brn2, we performed similar electroporations with Oct6. Although greater numbers of pCAG-Oct6-electroporated neurons failed to migrate out of the progenitor zones such as the highest-expressing pCAG-Brn1/2 cells (Supplementary Fig. 5G–J), those that did reach the CP by P0 were comparably skewed toward upper-layer molecular identities (Supplementary Fig. 5A–D) and positions (Supplementary Fig. 5E′,F). Interestingly, Oct6 exhibits much stronger transcriptional activity than Brn1 and Brn2 across all reporters studied (Supplementary Fig. 5Q,T′), confirming that this precocious differentiation within the VZ could be gene-dosage–related. Correspondingly, prior studies have demonstrated that while Brn1, Brn2, and Oct6 exhibit congruent DNA-binding specificities, Oct6 has a more robust transactivation domain than Brn1 (Schreiber et al. 1997).
But if Oct6 is more transcriptionally powerful than Brn1/2, why are Brn1/2 double-knockouts sufficient to abrogate upper-layer neurogenesis? Quantitative measures of different mRNA species derived from RNA-seq data from E14.5 embryonic cortex (Ayoub et al. 2011) revealed that Oct6 is the most weakly expressed of the 3 (Supplementary Fig. 5P). Combining the luciferase reporter and in vivo expression data, we were able to estimate the relative transcriptional contributions of each Pou3f gene to neurogenesis at E14.5; Oct6 makes up less than 20% of this weighted total (Supplementary Fig. 5U′).
Transcriptional Impairment of Brn1/2 Inhibits Neurogenesis and Migration Within a Specific Temporal Window
As stated above, the VZs of Brn1/2-electroporated brains demonstrate a handful of very highly-expressing transfected cells that appear to differentiate into neurons in situ, without migrating at all (Fig. 4A,A′). Because such a finding is characteristic of precocious differentiation, possibly incited by a loss in Notch signaling (Hashimoto-Torii et al. 2008), we sought to determine if any progenitors remained in these electroporations by P0. With control electroporations at E12.5, asymmetric division results in progenitor maintenance (Fig. 4A″,B) side-by-side with neuron production. Interestingly, we could barely find any Brn1/2- or Oct6-electroporated cells that were Sox2+ (Fig. 4A,B, Supplementary Fig. 5J), or that possessed the characteristic shape of RG or other VZ-resident progenitors. Instead, the neuron-like cells in the VZs of Brn1/2- or Oct6-electroporated brains expressed high levels of Tbr1 (Fig. 4A,B, Supplementary Fig. 5K), a characteristic marker for differentiating neurons (Englund et al. 2005) that is expressed in all pyramidal cells in the cortex at P0 (though layer VI cells exhibit the highest expression). In summary, these data suggest that the Pou3fs are genuine proneural transcription factors (Castro et al. 2006).
Because Brn1/2 knockout cortices exhibit little, if any, production of upper layer cells (Sugitani et al. 2002), we next asked whether their proneural activity was a key determinant in upper-layer neurogenesis. Brn1 and Brn2 proteins bind similar DNA sequences (Friedrich et al. 2005); the amino-acid sequence of their DNA-binding domains is identical except for a N → S substitution in the homeobox and a T → S substitution in the linker region. Thus, we used a construct containing the DNA-binding domains of Brn2 fused to an Engrailed repressor domain (EnR) (Kim et al. 2008) to ablate Brn1/2-mediated transcriptional activation.
We initially electroporated Brn2-EnR at E13.5, a time when upper-layer cells are specified in dorsolateral cortex. Within 48-h after electroporation, major differences were seen between Brn2-EnR and EnR alone (Fig. 4C–F). Specifically, Brn2-EnR prevented the departure of cells from the VZ, with very few reaching the CP (Fig. 4D vs. C,F). Instead, the GFP+ cells in Brn2-EnR electroporations exhibited columnar, bipolar, and RG shapes characteristic of progenitors, with few expressing Tbr2 (Fig. 4D). Quantifications demonstrated that Brn2-EnR impaired neurogenesis at an early step (Fig. 4E), as it resulted in the tandem reduction of Ngn2 and Tbr2. We used a cleaved Caspase-3 antibody to investigate the possibility that apoptosis may also be involved in this reduction of neurogenesis; cell death events were sporadic, but did not appear to be more heavily associated with Brn2-EnR (data not shown).
By P0, consequences of the neurogenic blockade by Brn2-EnR starting at E13.5 were very apparent; instead of producing GFP+ neurons as normal (Fig. 4G), Brn2-EnR expressing cells remained in the VZ as Pax6+ progenitors (Fig. 4G′,L,M). The effect was only partially mitigated by low-concentration pCAG-Satb2 co-expression with Brn2-EnR (Fig. 4G″ and L,M). On the other hand, co-electroporation with low-concentration pEGFP-dnMAML (which ablates Notch-directed transcription) led to a substantial, but incomplete, rescue of neurogenesis, as evidenced by fewer GFP+ cells in the VZ (Fig. 4G′″ and L,M), and fewer Pax6+ cells, compared with EnR alone.
Many of the neurons generated were arrested in the IZ/white matter (WM) at P0 in both Brn2-EnR and dnMAML rescue conditions; some expressed high levels of Pax6 (Fig. 4H,H′), but were otherwise Sox2− (Fig. 4J,J′) and Brn2− (Fig. 4K). Others expressed Ctip2 (Fig. 4H,H′). The IZ/WM-arrested cells that expressed Pax6 but not Sox2 could be undergoing a failed sequence of neural differentiation, because Pax6 is known to be negatively regulated by the induction of neurogenesis (Kawaguchi et al. 2004; Bel-Vialar et al. 2007). Although dnMAML restored the initial step of neurogenesis in Brn2-EnR electroporations, cells were impaired at subsequent stages of differentiation, as evidenced by excessive numbers of Tbr2+/Ngn2+ cells near the SVZ at P0, and of Tbr1+/NeuroD1+ cells within the IZ/WM (Supplementary Fig. 7).
Overall, there were slight regional biases in these effects; we found that Brn2-EnR had its strongest phenotype in more rostral regions of cortex, whereas caudal regions seemed to be less susceptible to its anti-migratory effects (data not shown). Spatiotemporal differences in neurogenesis, as a consequence of spatiotemporal molecular disparities, likely underlie these findings.
Thus, we wanted to know if impairment of Brn1/2-directed transcription could prevent neurogenesis in dorsal telencephalon at other times. We electroporated Brn2-EnR at E12.5, a time when predominantly layer V and IV cells are specified, and examined brains at P0. Similar to electroporations at E13.5, Brn2-EnR prevented neurogenesis and migration (Fig. 5A,B), increasing the number of Pax6+/VZ-resident cells; however, blockade of the initial step of neurogenesis was far less severe. When we plotted differences in GFP location at P0 between Brn2-EnR and EnR control electroporations performed at E13.5 versus E12.5, we found that VZ retention, vis-à-vis neurogenesis impairment, was nearly 3-fold (Fig. 5C) more severe at E13.5 than at E12.5, whereas migration of cells to the upper layers of CP was equally hindered.
Figure 5.
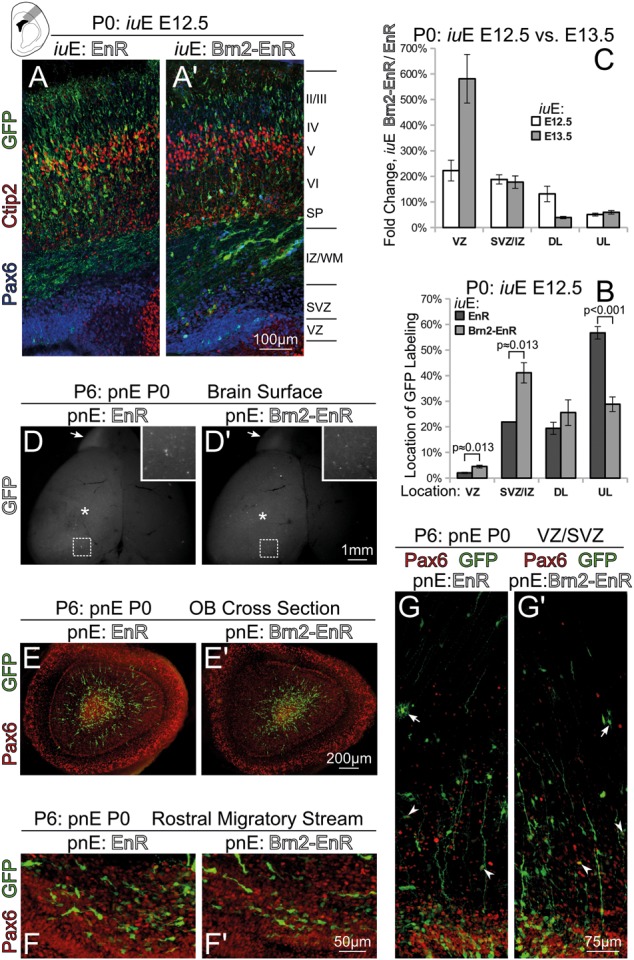
Brn1/2 is required for neurogenesis specifically at the time of the births of upper-layer projection neurons. (A and B) P0 sections electroporated at E12.5 with either EnR (A) or Brn2-EnR (A′), stained with Ctip2 (red), Pax6 (blue), and GFP (green). Brn2-EnR has an inhibitory effect on neurogenesis and migration; however, the impairment of neurogenesis at E12.5 is not as strong as Brn2-EnR overexpression performed at E13.5. The proportional location of GFP labeling in E12.5 EnR and Brn2-EnR electroporations is shown (B). (C) Comparison in E12.5 and E13.5 electroporations (examined at P0) of the Brn2-EnR over EnR quotient (i.e. fold change) of GFP proportional labeling for any compartment. This chart demonstrates a much stronger impairment in the VZ departure of Brn2-EnR electroporated cells at E13.5 than at E12.5. (D and G′) Brn2-EnR or EnR was electroporated at P0 and examined at P6; postnatal genesis of VZ/SVZ olfactory bulb interneurons is unimpaired by Brn2-EnR. Although Brn2-EnR electroporations appeared weaker than EnR electroporations (D and D′), GFP+ radial glia endfeet were observed on the cortical surface (* and insets in D and D′) in both cases, and GFP+ cells were seen in OB (arrows in D and D′). Coronal cross-sections of either OB or rostral cortex demonstrated the presence of GFP+ prospective neurons in OB (E and E′) and in the rostral migratory stream (F and F′) in either case. Furthermore, the VZ/SVZ did not appear different in Brn2-EnR versus EnR electroporations; GFP(green) labeled both Pax6†(arrowheads in G and G′) and Pax6- (arrows in G and G′) astrocyte-like cells. UL = upper layer and DL = deep layer.
Additionally, postnatal genesis of olfactory bulb (OB) interneurons was not impaired by Brn2-EnR (Fig. 5D–F′). We conducted electroporations of Brn2-EnR or EnR in P0 pups, finding GFP+ neurons both in OB (Fig. 5D,D′) and the rostral migratory stream (RMS, Fig. 5F,F′) at P6. Although the Brn2-EnR electroporations appeared weaker in GFP fluorescence intensity than controls, we could not detect any gross differences in the appearance of GFP+ cells in the postnatal SVZ (Fig. 5G,G′). In summary, these findings suggest that Brn1/2 has a concrete role in neurogenesis, but primarily during the time that upper-layer projection neurons are specified and born.
Finally, we devised a Brn1-EnR construct similar to Brn2-EnR to determine whether any functionality could be attributable to differences between the protein DNA-binding domains. E13.5 electroporation of Brn1-EnR generated a similar, albeit slightly weaker phenotype to that of Brn2-EnR, when brains were examined at P0 (Supplementary Fig. 5L′,L): It resulted in a parallel failure of transfected cells to downregulate Pax6 expression, although did not appear to impact radial migration as severely (Supplementary Fig. 5M,N). More importantly, however, the effect of Brn1-EnR was substantially more pronounced when electroporations were performed at E13.5 than at E12.5 (data not shown). Thus, although some subtle disparities in in vivo DNA binding may exist between Brn1 and Brn2, they seem to share an overlapping function in directing upper-layer neurogenesis.
Pou3fs Promote Neurogenesis by Upregulating Other Proneural Genes, as Well as by Diminishing Notch-Directed Transcription of Hes5
All data gathered henceforth have suggested that the Pou3f factors are true proneural genes capable of biasing progenitor cells toward a neural fate at the expense of progenitor maintenance. To uncover potential pathways involved in this process, we examined their ability to influence both the program of neural differentiation and Notch signaling. In progenitors, if Notch signaling is low, Ngn1/2 becomes highly active, setting off a well-programmed cascade of differentiation (Kawaguchi et al. 2004; Englund et al. 2005; Sessa et al. 2008; Sansom et al. 2009). The core of this cascade (Ngn2 → Tbr2 → NeuroD → Tbr1) is conserved in the differentiation of all glutamatergic cortical neurons.
To this end, we designed luciferase reporters for Ngn2 and Tbr2 following published findings (Fig. 6A,B). We observed that Brn2 overexpression in N2a cells elicited the endogenous expression of Tbr2 (Fig. 6C,D,H), but not Ngn2 (Fig. 6E–G). On the other hand, Pax6 overexpression in N2a cells caused endogenous Ngn2, but not Tbr2, expression (Fig. 6E,F,H). Nonetheless, Brn1/2 drove positive transcriptional activity from both Ngn2 and Tbr2 reporter plasmids that was increased by cotransfection of combinations of factors (Fig. 6I,J). We also found considerable in vitro and in vivo evidence of Pou3f regulation of Tbr1, a downstream target in the cortical neurogenic program (Fig. 7). Furthermore, we identified regulatory sequences at the Satb2 locus, and have determined that octamer-binding transcription factors (such as Pou3f factors) likely contribute to the regulation of Satb2 (Supplementary Results and Supplementary Fig. 8)—all Pou3f factors tested induce Satb2 in vivo (Supplementary Figs. 8E–H and 5F) and in vitro (Supplementary Fig. 5Q and 8A–C). Taken together, these results imply that Pou3fs are important facilitators of differentiation at multiple distinct steps in the neurogenic cascade.
Figure 6.
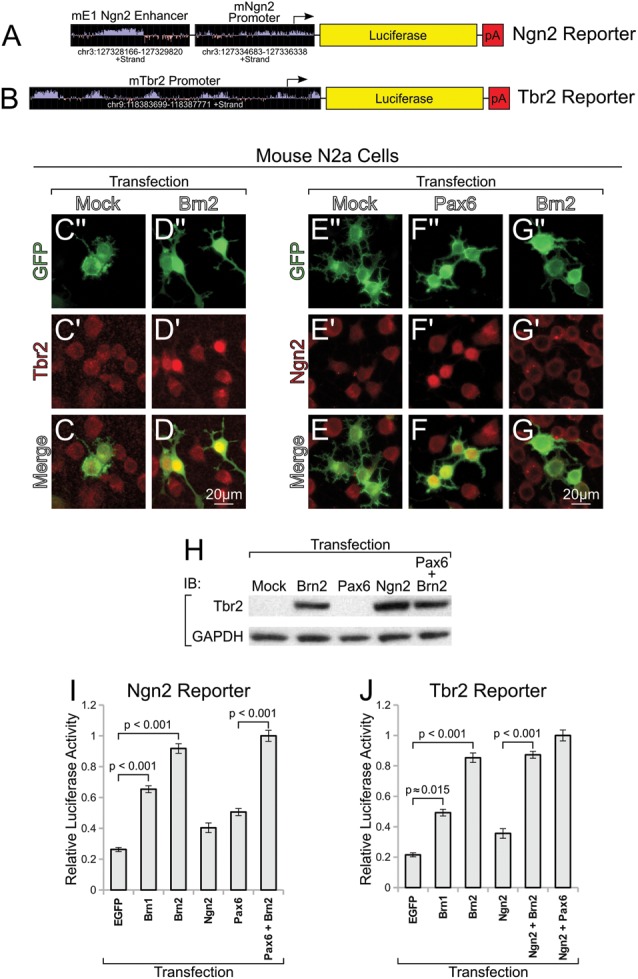
Brn1/2 regulate pro-neural genes Ngn2 and Tbr2 in vitro. (A and B) The design of Ngn2 and Tbr2 reporters is similar to those described previously (Shimojo et al. 2008; Ochiai et al. 2009). (C″ and G) N2a cells at 24-h post-transfection and serum starvation labeled for Tbr2 (C″ and D), Ngn2 (E″ and G), or immunoblotted (H) for Tbr2. Mock transfected cells do not express either Tbr2 (C″ and C) or Ngn2 (E″ and E) endogenously. Sporadic Brn2-transfected cells (but not all) express detectable levels of Tbr2 (D″ and D), and sporadic Pax6-transfected cells (but not all) express detectable levels of Ngn2 (F″ and F); Brn2 does not stimulate endogenous Ngn2 expression under these conditions (G″ and G). Immunoblot analysis for Tbr2 demonstrates similar findings (H). (I and J) Ngn2 and Tbr2 reporter activity showns strong activation by Brn1/2 alone and in combination with known upstream proneural regulators. All luciferase values were normalized to the transfection condition generating the maximal activity.
Figure 7.
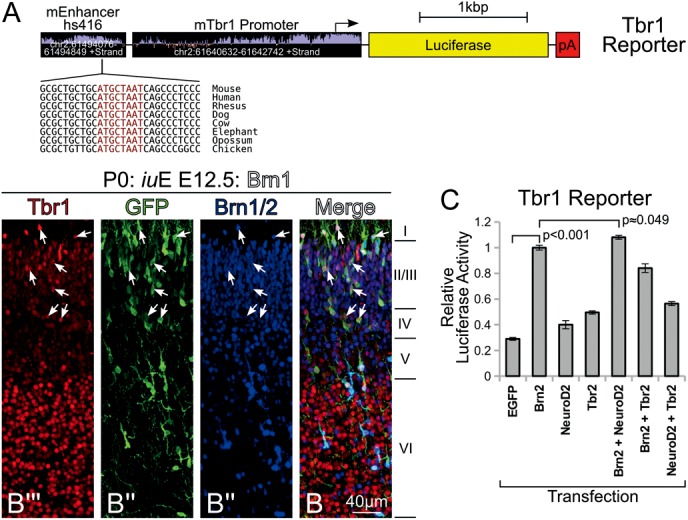
Brn1/2 regulate Tbr1 in vitro and in vivo. (A) The design of the Tbr1 reporter used in this study is entirely novel, and incorporates approximately 2 kb of upstream promoter sequence, as well as an enhancer located about 150 kb upstream; this enhancer has known activity that corresponds to the precise location of Tbr1 expression in the mouse embryo at E11.5 (Visel et al. 2007). Also shown is a conserved octamer sequence found in the enhancer. (B″′ and B) P0 cortical plate showing electroporation of Brn1 at E12.5 labeled for Tbr1 (red), GFP (green), and Brn1/2 (blue). Of note, electroporated cells (arrows), even those migrating into subpial layer II, exhibit higher levels of Tbr1 expression than their neighbors. (C) Luciferase assay of Tbr1 reporter demonstrates induction by Brn2 in N2a cells at 24-h post-transfection and serum starvation; activity shown is normalized to Brn2 transfection alone.
At the start of neurogenesis, Notch and RBPJ/κ signaling control self-renewal via induction of the widely studied hairy/enhancer-of-split (Hes) genes (Ohtsuka et al. 1999). Hes1/5 negatively regulates transcription of neurogenesis-promoting genes Neurogenins (Ngn) 1 and 2, thereby effecting progenitor maintenance and inhibiting neurogenesis (Imayoshi et al. 2008; Shimojo et al. 2008; Ochiai et al. 2009). We studied the effects of Brn2 expression on Hes1 and Hes5 reporters, finding that its presence strongly inhibited Hes5 (Fig. 8E), but had minimal effect on Hes1 (Fig. 8D). Cotransfection of the proneural bHLH factor Hes6, but not Ngn2, followed a similar trend (Fig. 8D,E); we thus examined Hes6 more closely. The Pou3fs drove positive Hes6 reporter activity, although forced expression of Ngn2 or a Pou3f in combination with Ngn2 showed higher reporter induction (Fig. 8F). Lastly, we found that Brn2-EnR transfection substantially increased the ability of NICD to activate either the Hes1 or Hes5 reporter (Fig. 8G), corroborating our earlier in vivo findings (Fig. 4 and Supplementary Fig. 7). Because the Brn2-EnR construct contains only the Brn2 DNA-binding domains and a repressor domain, this positive transcriptional influence on Hes1 and Hes5 is probably due to the suppression of a Notch transcriptional repressor such as Hes6.
Figure 8.
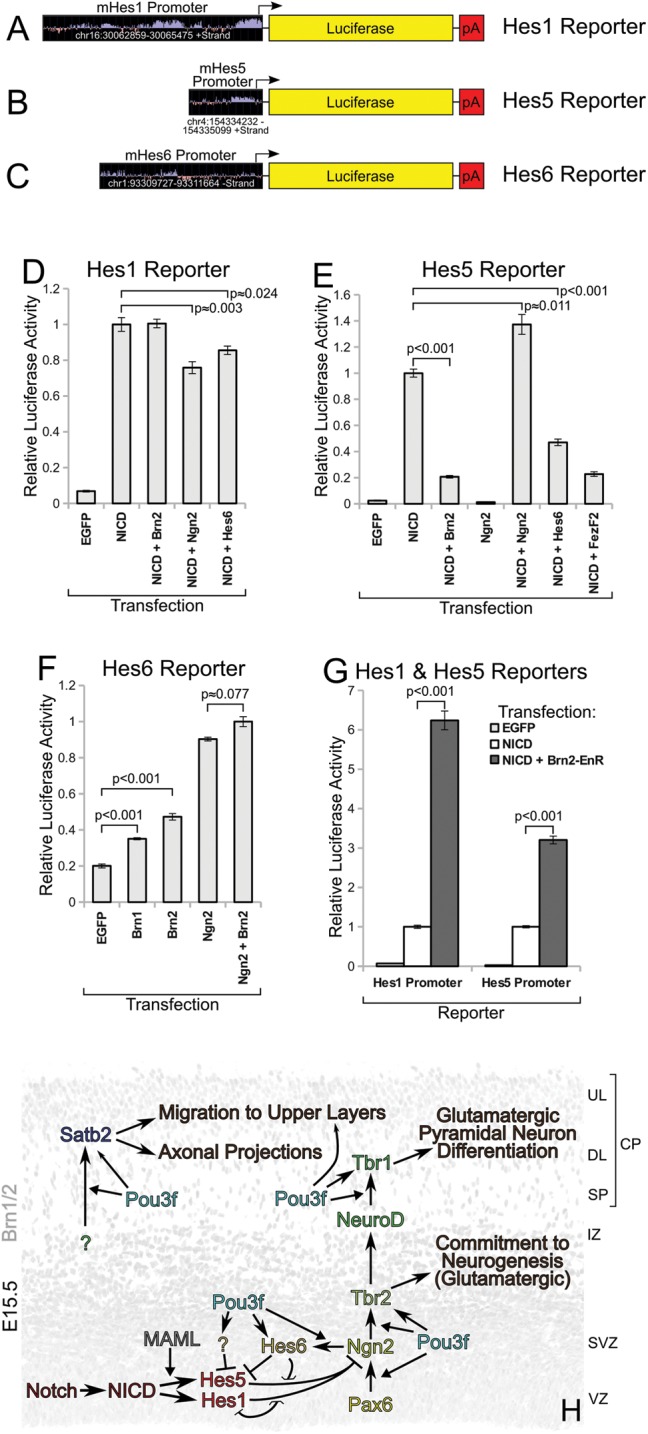
Brn1/2 interfere with Notch activity by suppressing Hes5 and activating Hes6 expression. Luciferase assays were performed in N2a cells at 24-h post-transfection and serum starvation. (A through C) Luciferase promoter reporters used in this study. (D) Hes1 reporter activities, relative to NICD transfection, demonstrating weak inhibitory effects of cotransfection with Ngn2 or Hes6, but not Brn2. (E) Reporter activities for Hes5, relative to NICD transfection, demonstrating strong inhibition by Brn2, Hes6, and FezF2. (F) Hes6 reporter activities, relative to Ngn2 + Brn2 cotransfection condition, demonstrating induction of Hes6 by proneural factors. (G) Hes1 and Hes5 reporter data, individually normalized to NICD transfection condition, showing a substantial increase in activity when cotransfected with Brn2-EnR. (H) Model depicting the mid-corticogenesis roles of the Pou3fs in Notch-directed transcription, and cascade of neural differentiation; this model incorporates previously published findings and summarizes all data from this study. Chevron-shaped arrows show direct transcriptional pathways, whereas triangular arrows show facilitation of pathways. Smaller arrowheads represent relatively weaker pathways.
Discussion
We show, using a plethora of in vivo and in vitro molecular tools, that Brn1/2 (and to a lesser extent, Oct6) is a crucial regulator of the transition from early- to mid-neurogenesis in developing mouse cortex. The beginning of their expression correlates with the progression of neurogenic laminar fates in progenitors, and they persist in a manner that is coincidental with a change in the identity of the progeny neurons.
Our studies reveal that in the absence of Brn1/2 transcription, the production of upper-layer cells is strongly reduced; this correlates well with knockout animals, which exhibit a near absence of these cells altogether (Sugitani et al. 2002). The necessity for Brn1/2 in neurogenesis increases 3-fold between E12.5 and E13.5, coincident with the increase in Brn1/2’s own expression in dorsal telencephalic VZ—progenitors express Brn1/2 just in time to meet the requirement for it in subsequent neuron production. Starting at E13.5, the presence or absence of other factor(s) could have a role in prompting the requirement for the Pou3f TFs in neurogenesis, and/or in inducing their expression. Indeed, the upstream regulation of Pou3f genes might be very revealing; future work can identify whether such factors as Pax6, Tlx (Schuurmans et al. 2004), Hes1/5 (Imayoshi et al. 2008), or others contribute to these functions. Intriguingly, many cells rescued from Brn2-EnR expression at E13.5 with dnMAML are largely unable to complete neural differentiation and migrate fully into the CP. Thus, Brn1/2 is not only necessary for the initiation of neural commitment after this time via, but also for subsequent steps in neurogenic differentiation.
Our results firmly agree with prior Brn1/2 knockout analysis in demonstrating that Brn1/2 is essential for producing cortical neurons of the upper layers. However, our results reach beyond current knowledge in several critical ways. We have harnessed the power of in utero electroporation to conduct knockout studies in only a subset of cells in a temporally controllable manner. When Brn1/2-directed transcription is lost, neural differentiation and migration is imperiled at E12.5, but neurogenesis is lost after E13.5—coincidental with the births of upper-layer neurons. Furthermore, we show that Brn1, Brn2, and Oct6 are bona fide proneural transcription factors that promote neurogenesis at the expense of progenitor renewal. Misexpression of any of these Pou3fs in early cortex development is sufficient alone to drive the production of neurons, a large proportion adopting the migratory and molecular characteristics of later-born upper-layer cells. Thus, these Pou3fs are necessary for late-born neurogenesis, but their presence also confers that upper-layer specification in molecular identity (Fig. 2A–G), axonal projection (Fig. 2J,K), and migratory behavior (Fig. 3).
Of substantial interest is the Pou3f-directed cell-autonomous migration to superficial laminar positions in the CP. Classical transplantation studies of mid-neurogenic progenitors into earlier brains have shown that the sequence of neuronal subtype birth is cell-intrinsic, and that older donor progenitors produce neurons migrating to the upper layers although they are born concurrently with the deep layers of the host (Frantz and McConnell 1996). Our data show that the propensity for upper-layer neurons to migrate into superficial laminar positions is genetically instructed by Pou3fs, and is not based solely on the fact that they are normally born after deeper resident neurons. Knowing that Satb2 is crucial for radial migration (Britanova et al. 2008), and that it can partially rescue the phenotype associated with Brn2-EnR, we define a Pou3f → Satb2 pathway that instructs cell-intrinsic upper-layer specification.
In conclusion, it is thought that the basic Pax6 → Ngn2 → Tbr2 (Englund et al. 2005) sequence is obeyed as the first step of differentiation in all cortical pyramidal neurons. We show that during the birth of prospective upper-layer neurons, the initial step of this sequence cannot proceed in the absence of Pou3f-directed transcription. Our work provides a glimpse of the molecular cellular changes encoding the intrinsic fate transitions in progenitor cell lineages. Indeed, such transitions underlie the proper birth sequence of pyramidal neurons that serve to generate a vast neural diversity in cortex (Shen et al. 2006).
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by the Yale Medical Science Training Program, the Kavli Institute for Neuroscience at Yale, the Patterson Trust Fellowship in Brain Circuitry (A.E.A.), as well as by grants R01-DA023999 (P.R.) and R01-NS038296 (P.R.) from the NIH.
Supplementary Material
Notes
We thank Mary Pappy, Marianne Horn, and Steve R. Wilson for their ongoing technical assistance and veterinary expertise. Conflict of Interest: None declared.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. doi:10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. doi:10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc Natl Acad Sci USA. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayatti N, Moss JA, Sun L, Ambrose P, Ward JFH, Lindsay S, Clowry GJ. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cereb Cortex. 2008;18:1536–1548. doi: 10.1093/cercor/bhm184. doi:10.1093/cercor/bhm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayatti N, Sarma S, Shaw C, Eyre JA, Vouyiouklis DA, Lindsay S, Clowry GJ. Progressive loss of PAX6, TBR2, NEUROD and TBR1 mRNA gradients correlates with translocation of EMX2 to the cortical plate during human cortical development. Eur J Neurosci. 2008;28:1449–1456. doi: 10.1111/j.1460-9568.2008.06475.x. doi:10.1111/j.1460-9568.2008.06475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RAM, Beyer RP, Bammler TK, Rubenstein JLR, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. doi:10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S, Medevielle F, Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. doi:10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Boutin C, Diestel S, Desoeuvre A, Tiveron M-C, Cremer H. Efficient in vivo electroporation of the postnatal rodent forebrain. PLoS ONE. 2008;3:e1883. doi: 10.1371/journal.pone.0001883. doi:10.1371/journal.pone.0001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. doi:10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Göttgens B. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. doi:10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Nowakowski RS, Bhide PG. Neocortical neurogenesis: morphogenetic gradients and beyond. Trends Neurosci. 2009;32:443–450. doi: 10.1016/j.tins.2009.05.003. doi:10.1016/j.tins.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. doi:10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. doi:10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler AT, Le Pichon CE, Brann JH, Araneda RC, Zou D-J, Firestein S. Selective gene expression by postnatal electroporation during olfactory interneuron nurogenesis. PLoS ONE. 2008;3:e1517. doi: 10.1371/journal.pone.0001517. doi:10.1371/journal.pone.0001517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RAM, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are Expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. doi:10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Janson AM, Nyengaard JR. Quantitative Methods in Neuroscience: A Neuroanatomical Approach. Oxford: Oxford University Press; 2004. [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23:561–574. doi: 10.1101/gad.1743709. doi:10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Batiz LF, Neal J, Lian G, Bundock E, Lu J, Hsiao Y-C, Diamond R, Mei D, Banham AH. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum Mol Genet. 2009;18:497–516. doi: 10.1093/hmg/ddn377. doi:10.1093/hmg/ddn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Hanashima C. Pyramidal neurons grow up and change their mind. Neuron. 2008;57:333–338. doi: 10.1016/j.neuron.2008.01.018. doi:10.1016/j.neuron.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Frantz GD, McConnell SK. Restriction of late cerebral cortical progenitors to an upper-layer fate. Neuron. 1996;17:55–61. doi: 10.1016/s0896-6273(00)80280-9. doi:10.1016/S0896-6273(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Friedrich RP, Schlierf B, Tamm ER, Bösl MR, Wegner M. The class III POU domain protein Brn-1 can fully replace the related Oct-6 during schwann cell development and myelination. Mol Cell Biol. 2005;25:1821–1829. doi: 10.1128/MCB.25.5.1821-1829.2005. doi:10.1128/MCB.25.5.1821-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. doi:10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini R, Dobyns WB, Barkovich AJ. Abnormal development of the human cerebral cortex: genetics, functional consequences and treatment options. Trends Neurosci. 2008;31:154–162. doi: 10.1016/j.tins.2007.12.004. doi:10.1016/j.tins.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Hagino-Yamagishi K, Saijoh Y, Ikeda M, Ichikawa M, Minamikawa-Tachino R, Hamada H. Predominant expression of Brn-2 in the postmitotic neurons of the developing mouse neocortex. Brain Res. 1997;752:261–268. doi: 10.1016/s0006-8993(96)01472-2. doi:10.1016/S0006-8993(96)01472-2. [DOI] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MMS, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. doi:10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Torii K, Torii M, Sarkisian MR, Bartley CM, Shen J, Radtke F, Gridley T, Sestan N, Rakic P. Interaction between Reelin and Notch signaling regulates neuronal migration in the cerebral cortex. Neuron. 2008;60:273–284. doi: 10.1016/j.neuron.2008.09.026. doi:10.1016/j.neuron.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. doi:10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Lee D-G, Porter KK. Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 2005;1052:71–81. doi: 10.1016/j.brainres.2005.06.015. doi:10.1016/j.brainres.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. doi: 10.1242/dev.021535. doi:10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. doi:10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Ogawa M, Saito K, Matsuzaki F, Okano H, Miyata T. Differential expression of Pax6 and Ngn2 between pair-generated cortical neurons. J Neurosci Res. 2004;78:784–795. doi: 10.1002/jnr.20347. doi:10.1002/jnr.20347. [DOI] [PubMed] [Google Scholar]
- Kim DS, Matsuda T, Cepko CL. A core paired-type and POU homeodomain-containing transcription factor program drives retinal bipolar cell gene expression. J Neurosci. 2008;28:7748–7764. doi: 10.1523/JNEUROSCI.0397-08.2008. doi:10.1523/JNEUROSCI.0397-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. doi:10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O, Backman M, Machonova O, Kozmik Z, Vacik T, Andersen L, Krauss S. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev Biol. 2007;311:223–237. doi: 10.1016/j.ydbio.2007.08.038. doi:10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. doi:10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Padilla M. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J Comp Neurol. 1992;321:223–240. doi: 10.1002/cne.903210205. doi:10.1002/cne.903210205. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. doi:10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. doi:10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JLR, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. doi:10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. doi:10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Isaka F, Ishibashi M, Tomita K, Tsuda H, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and enhancer of split. Genomics. 1998;49:69–75. doi: 10.1006/geno.1998.5213. doi:10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, Kainuma M, Masaoka M, Minobe S, Namihira M, Nakashima K, Sakakibara A, Ogawa M. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. doi:10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. doi:10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. doi:10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Selemon LD, Goldman-Rakic PS. Neuronal and glial somal size in the prefrontal cortex: a postmortem morphometric study of schizophrenia and Huntington disease. Arch Gen Psychiatr. 1998;55:215–224. doi: 10.1001/archpsyc.55.3.215. doi:10.1001/archpsyc.55.3.215. [DOI] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. doi:10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. doi:10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Ramos RL, Bai J, LoTurco JJ. Heterotopia formation in rat but not mouse neocortex after RNA interference knockdown of DCX. Cereb Cortex. 2006;16:1323–1331. doi: 10.1093/cercor/bhj074. doi:10.1093/cercor/bhj074. [DOI] [PubMed] [Google Scholar]
- Sanderson KJ, Weller WL. Gradients of neurogenesis in possum neocortex. Dev Brain Res. 1990;55:269–274. doi: 10.1016/0165-3806(90)90208-g. doi:10.1016/0165-3806(90)90208-G. [DOI] [PubMed] [Google Scholar]
- Sanosaka T, Namihira M, Nakashima K. Epigenetic mechanisms in sequential differentiation of neural stem cells. Epigenetics. 2009;4:89–92. doi: 10.4161/epi.4.2.8233. doi:10.4161/epi.4.2.8233. [DOI] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan D-J, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. doi:10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J, Enderich J, Sock E, Schmidt C, Richter-Landsberg C, Wegner M. Redundancy of class III POU proteins in the oligodendrocyte lineage. J Biol Chem. 1997;272:32286–32293. doi: 10.1074/jbc.272.51.32286. doi:10.1074/jbc.272.51.32286. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin L-M, Seibt J, Tang H. Sequential phases of cortical specification involve neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. doi:10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao C-A, Hadjantonakis A-K, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. doi:10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. doi:10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in Notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. doi:10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Rakic P. Neuronal migration, with special reference to developing human brain: a review. Brain Res. 1973;62:1–35. doi: 10.1016/0006-8993(73)90617-3. doi:10.1016/0006-8993(73)90617-3. [DOI] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K-I, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. doi:10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS. Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834–8846. doi: 10.1128/MCB.24.20.8834-8846.2004. doi:10.1128/MCB.24.20.8834-8846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. doi:10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Tosaki A, Miyazaki H, Kurosawa M, Furukawa Y, Yamada M, Nukina N. Mutant huntingtin fragment selectively suppresses Brn-2 POU domain transcription factor to mediate hypothalamic cell dysfunction. Hum Mol Genet. 2010;19:2099–2112. doi: 10.1093/hmg/ddq087. doi:10.1093/hmg/ddq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.