Abstract
Hsp90 functions in association with several cochaperones for folding of protein kinases and transcription factors, although the relative contribution of each to the overall reaction is unknown. We assayed the role of nine different cochaperones in the activation of Ste11, a Saccharomyces cerevisiae mitogen-activated protein kinase kinase kinase. Studies on signaling via this protein kinase pathway was measured by α-factor-stimulated induction of FIG1 or lacZ, and repression of HHF1. Several cochaperone mutants tested had reduced FIG1 induction or HHF1 repression, although to differing extents. The greatest defects were in cpr7Δ, sse1Δ, and ydj1Δ mutants. Assays of Ste11 kinase activity revealed a pattern of defects in the cochaperone mutant strains that were similar to the gene expression studies. Overexpression of CDC37, a chaperone required for protein kinase folding, suppressed defects the sti1Δ mutant back to wild-type levels. CDC37 overexpression also restored stable Hsp90 binding to the Ste11 protein kinase domain in the sti1Δ mutant strain. These data suggest that Cdc37 and Sti1 have functional overlap in stabilizing Hsp90:client complexes. Finally, we show that Cns1 functions in MAP kinase signaling in association with Cpr7.
INTRODUCTION
Among several major classes of molecular chaperone that have been characterized, Hsp90 appears to function primarily in folding of signal transducing proteins such as transcription factors and protein kinases (Caplan, 1999; Prodromou and Pearl, 2003). Hsp90 does not act alone in this capacity, but in association with several cochaperones that may also have chaperone function as defined by their ability to prevent polypeptide aggregation in vitro.
The current understanding of how Hsp90 cochaperones function derived from in vitro studies on progesterone receptors (PR) and glucocorticoid receptors (GR; Pratt and Toft, 1997, 2003). The results of these studies showed that Hsp70 and Hsp40 first interact with the receptor ligand-binding domain. Hsp90 is recruited into this complex by the cochaperone called Hop, which interacts with both Hsp90 and Hsp70. Subsequently, Hsp70 and Hop are displaced by other cochaperones, such as one of many immunophilins and p23. Folding occurs in association with ATP hydrolysis by Hsp90, which is stimulated by another cochaperone called Aha1 (and its paralog Hch1; Panaretou et al., 2002; Lotz et al., 2003). It is unclear whether folding occurs while the receptor is in association with Hsp90 or after its release. Whether the cochaperones are required for this reaction is not clear, because purified Hsp70 and Hsp90 can together facilitate folding of GR in vitro, whereas cochaperones such as Hop stimulate the reaction (Morishima et al., 2000; Rajapandi et al., 2000). Furthermore, deletion of yeast Hop (STI1), or p23 (SBA1) does not result in a slow growth phenotype except under stress conditions (Nicolet and Craig, 1989; Chang et al., 1997; Bohen, 1998; Fang et al., 1998).
Another question is whether the scheme described above for nuclear receptors reflects a set of general principles by which Hsp90 functions and whether it applies to other client types. Folding of protein kinases, for example, requires a cochaperone called Cdc37, that does not associate with GR or PR, although it does function in activation of androgen receptor and a viral reverse transcriptase (Fliss et al., 1997; Rao et al., 2001; Wang et al., 2002). Cdc37 interacts with many different protein kinases and is an essential gene (Hunter and Poon, 1997). It is required for cell cycle progression in yeast and mammals by regulating the activity of cyclin dependent kinases (Cdks; Reed, 1980a, 1980b; Stepanova et al., 1996). Cdc37 facilitates Cdk folding and may also be involved in assembly of Cdk:cyclin complexes (Lamphere et al., 1997). Cdc37 is also important for mitogen activated protein (MAP) kinase signaling. Studies using a Drosophila model system first demonstrated that mutation in Cdc37 affected signaling through a MAP kinase pathway (Cutforth and Rubin, 1994). Subsequent studies found that Cdc37 and Hsp90 were important for function of the MAP kinase kinase kinases (MAPKKKs) Raf1 and yeast Ste11 (Grammatikakis et al., 1999; Abbas-Terki et al., 2000). The mechanism of Cdc37 action is not clear; Cdc37 is a molecular chaperone because it can prevent polypeptide aggregation (Kimura et al., 1997) and this function is restricted to its N-terminal domain, which is the most conserved part of the protein (Grammatikakis et al., 1999; Lee et al., 2002). Cdc37 binds directly to Hsp90 via a region of ∼100 amino acids in the middle portion of the protein (Grammatikakis et al., 1999; Scholz et al., 2001) and partially inhibits Hsp90's ATPase (Siligardi et al., 2002). The C-terminal 118 amino acids (of 506) are completely dispensable for its function in yeast (Lee et al., 2002).
The relationship between Cdc37 and other Hsp90 cochaperones is slowly becoming characterized. Despite an early report showing competition between Hop and Cdc37 for binding to Hsp90, subsequent studies were more consistent with their coexistence on the same Hsp90 molecule (Silverstein et al., 1998; Hartson et al., 2000). Furthermore, yeast Cdc37 can interact with both Sti1/Hop and Cpr7 (an immunophilin) independently of Hsp90 (Abbas-Terki et al., 2002). Thus it is possible that Cdc37 enters the chaperone-dependent folding pathway via interaction with other cochaperones, although this remains to be tested.
In a previous report we showed that a defect in protein kinase folding in a sti1Δ yeast strain could be suppressed by CDC37 overexpression (Lee et al., 2002). In this report we further investigate the role of Hsp90 cochaperones in MAP kinase signaling and test the extent to which CDC37 overexpression can suppress defects arising from deletion of several different cochaperones. Our data support a model in which Sti1/Hop stabilizes Hsp90 and Cdc37 interactions with a client protein kinase.
MATERIALS AND METHODS
Materials, Yeast Strains, Growth Conditions, and Transformation
All strains are isogenic with BY4741 (MAT a) and were purchased from Research Genetics/Invitrogen (Carlsbad, CA). The genotype was verified by PCR analysis using strain-specific primers or by Western blot to check for the absence of the relevant protein. Strains were grown and transformed according to standard methods in either YPD or minimal media with amino acid supplements. Plasmids encoding galactose-inducible STE4 (pL19), STE11ΔN (pYGU-11ΔN), and STE12 (pGS3) were the kind gifts of Dr. Jeanne Hirsch.
RNA Preparation and Real-Time Reverse Transcriptase PCR
Total RNA was prepared from 50 ml yeast cultures grown to A600 = 0.3 in YPD. The pheromone α-factor (Sigma Chemical Co., St. Louis, MO), or water, was added to a final concentration of 5 μM and the cultures were incubated for 30 min at 30°C. Total RNA was prepared after cell lysis with glass beads (0.5 mm) using the Rneasy Mini kit (Qiagen, Chatsworth, CA). RNA, 1 μg, was used for reverse transcription using Omniscript reverse transcriptase (Qiagen) according to the manufacturer's instructions. Real-time PCR was performed with a Roche lightcycler with a Qiagen SYBR green kit. Primers, designed using the LC probe design software and chosen for having the same melting temperatures, were as follows: ARP5 forward: 5′ TGCTAAGGTGCCAGGA 3′; ARP5 reverse: 5′ CTAGAGCTGCCATACCC 3′; FIG1 forward: 5′ TGGGGTATTGGTGCGA 3′; FIG1 reverse: 5′ CCATTACTGCTGCCTTC 3′; HHF1 forward: 5′ CTGGTTTGATCTACGAAG 3′; and HHF1 reverse: 5′ GCATAAACAACATCCAAA 3′.
Amplification conditions, after an initial denaturation step for 15 min at 94°C were 94°C denaturing temperature (15 s), 55°C annealing temperature (25 s), and 72°C elongation temperature (10 s) for 45 cycles. Melting curve analysis was performed to check for a single amplicon that was verified by size using gel electrophoresis. Amplification was determined to be linear by analysis of a serial dilutions of cDNA. Lightcycler analysis software was used for determining crossing points. Data were analyzed by the 2-ΔΔCT method as described by Livak and Schmittgen (2001) and are presented as fold induction/repression of FIG1 and HHF1, respectively, normalized to ARP5 levels.
Pheromone-dependent Induction of lacZ
Yeast cells were transformed with pPRE-lacZL (LEU2), which contains the lacZ gene under control of Ste12 response elements (see below for plasmid construction). Yeast cells were grown to A600 = 0.2 in selective media and treated with or without α-factor (5 μM) for 3 h. Cells from 1.5 ml of culture were harvested by centrifugation and lysed by freeze thaw, and β-galactosidase activity was measured with a Galactostar kit (Tropix, Bedford, MA) according to the manufacturer's instructions. Fold induction was measured relative to β-galactosidase activity in uninduced cells.
Pheromone-independent β-galactoasidase activity was measured using the same procedure except that the cells were grown in raffinose-containing media and overexpressed STE4, STE11ΔN, and STE12 were induced by addition of 2% galactose for 6 h. Extracts were prepared and assayed for β-galactosidase as described above.
Plasmid Construction
The reporter plasmid pPRE-lacZL was prepared from pPRE-lacZ (gift of Dr. Kevin Morano), after digestion with SmaI and cotransformed into BY4741 with a 3.6-kb DNA fragment containing the LEU2 gene excised from pUL9 (gift of Dr. Jeanne Hirsch) with SmaI. The repaired plasmid was isolated after transformation of Escherichia coli with lysates from the transformed yeast.
Plasmids encoding His-tagged Ste7 and Fus3 were prepared by PCR amplification (30 cycles at 55°C) of the complete open reading frames followed by ligation into pYES2.1/V5-His-Topo (Invitrogen). The plasmid encoding Ste11ΔN was similarly prepared by amplification of the STE11 open reading frame corresponding to amino acids 341-717, adding a methionine codon to the 5′ primer. The same method was used to prepare a plasmid encoding full-length Cpr7. The plasmid encoding the TPR domain of Cpr7 was prepared by amplification of the CPR7 open reading frame corresponding to amino acids 201-393.
His-tagged protein kinases were induced by addition of galactose (2%) to the media and overnight incubation. The cells were lysed and the pull-down on Ni-NTA resin performed exactly as described previously except that the resin was incubated with the lysates for 1 h at 4°C. Western blots were performed as previously described.
Pull-down, Western Blot, and Kinase Assays
Wild-type and Hsp90 cochaperone mutants containing His6-V5-Ste11ΔN were grown in 200-ml 0.67% yeast nitrogen base plus 2% raffinose to A600 = 0.2. Galactose was then added to 2% to induce the expression of Ste11ΔN for 12 h. Cells were resuspended in extraction buffer (20 mM HEPES, pH 7.5, 100 mM KCl, 0.1 mM EDTA plus protease inhibitor cocktail tablets from Boehringer [Indianapolis, IN]). Extracts were prepared by glass bead lysis with 0.5-mm glass beads in mini-bead beater two times for 2-min bursts. His6-V5-Ste11ΔN was isolated after incubation of Ni-NTA resin with 0.5 ml of whole cell extracts at 3 mg/ml for 1 h at 4°C. The beads were washed three times with extraction buffer plus 10 mM imidazole. The proteins were eluted from the beads by the addition of 0.4 ml extraction buffer plus 150 mM imidazole. Eluted proteins were precipitated with 10% trichloroacetic acid and resuspended in SDS-PAGE sample buffer. Each sample was resolved by denaturing SDS-PAGE, and proteins were detected by Western blot using specific antisera.
For kinase assays, the eluates from Ni-NTA resin were treated with 2 μg V5 antibody (Invitrogen) prebound to protein A Sepharose beads for immunoprecipitation in place of the trichloroacetic acid treatment described above. After 2 h of incubation, the beads were pelleted and washed three times with extraction buffer. Kinase reactions were started by incubating the beads in 20 μl kinase buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2), 100 mM ATP, and 10 μCi [γ-32P]ATP (Perkin Elmer-Cetus, Norwalk, CT) for 20 min at room temperature. The beads were washed three more times with extraction buffer, and the reactions were stopped by the addition of SDS sample buffer. The samples were then resolved by denaturing gel electrophoresis, and the activities were quantified on a phosphoimager.
RESULTS
Role of Hsp90 Cochaperones in MAP Kinase Signaling
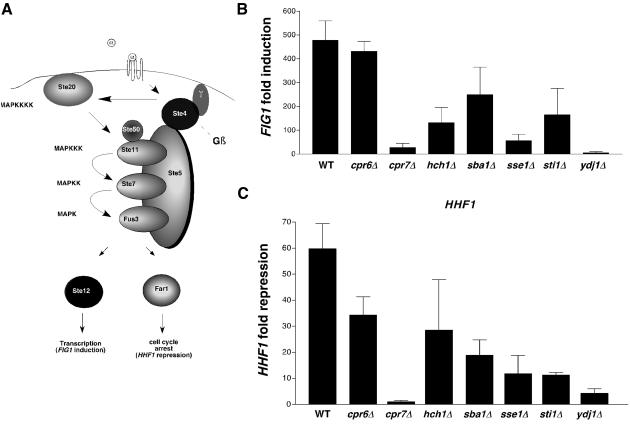
Previous studies established that the Saccharomyces cerevisiae MAPKKK Ste11 requires Hsp90 and Cdc37 for activity (Abbas-Terki et al., 2000). What is unknown is how many Hsp90 cochaperones besides Cdc37 are also required. To address this question, we assayed for MAP kinase signaling in several different mutant yeast strains, each deleted for a single cochaperone of Hsp90 but otherwise isogenic. Our procedure was to measure induction of pheromone responsive genes after a 30-min exposure to saturating amounts of α-factor. Two pheromone responsive genes were chosen for analysis: FIG1, a highly inducible gene that is a direct transcriptional target of Ste12, and HHF1, which is repressed as a consequence of G1 arrest. Levels of FIG1 and HHF1 mRNA were determined by quantitative real-time rtPCR relative to ARP5, whose mRNA levels do not change upon pheromone treatment (Roberts et al., 2000; our unpublished results). The cochaperones chosen for study have all been characterized previously as Hsp90 binding proteins, and include Cpr6, Cpr7, Hch1, Sba1, Sse1, and Sti1 (Duina et al., 1996; Chang et al., 1997; Bohen, 1998; Fang et al., 1998; Liu et al., 1999; Panaretou et al., 2002; Lotz et al., 2003). Ydj1, a cochaperone of Hsp70 was also included because it affects the activity of Hsp90 clients (Bohen et al., 1995). FIG1 was induced 478-fold in the wild-type strain but this induction was significantly reduced in all the mutant strains except for cpr6Δ (Figure 1B). Deletion of CPR7, SSE1, and YDJ1 resulted in the most severe decreases in FIG1 induction at 28-, 56-, and 6-fold, respectively, above background. HHF1 repression, a consequence of cell cycle arrest, followed a similar pattern in the mutants, although a modest defect was noted in the cpr6Δ mutant. Because cell cycle arrest reflects a more indirect function of MAP kinase signaling than FIG1 induction it is possible that additional chaperone dependent steps are involved, thus accounting for the role of Cpr6.
Figure 1.
Role of Hsp90 cochaperones in MAP kinase signaling. (A) Schematic of MAP kinase signaling. Yeast gene names are within objects and their generic designation as MAP kinases shown alongside. Adapted from Elion (2000). (B) Results of real-time rtPCR for induction of FIG1 30 min after α-factor treatment. Results are presented as fold induction measured against RNA levels in uninduced cells and normalized to levels of ARP5 mRNA. Results are the mean of n = 3 ± SE. (C) As in B (using same RNA preparations) except that levels of HHF1 were measured and represented as fold repression relative to samples from uninduced cells.
Similar experiments were carried out with a lacZ reporter gene under control of the Ste12 transcription factor. For these experiments, β-galactosidase levels were measured 3 h after pheromone treatment. The pattern of defects in the cochaperone mutant strains were similar to those measured by rtPCR, although the magnitude of the defect was reduced in each case. For comparison we also assayed a strain that had greatly reduced amounts of Hsp90 (hsc82Δ; Borkovich et al., 1989). β-galactosidase induction in this strain was reduced to the same extent as observed in the other mutant strains.
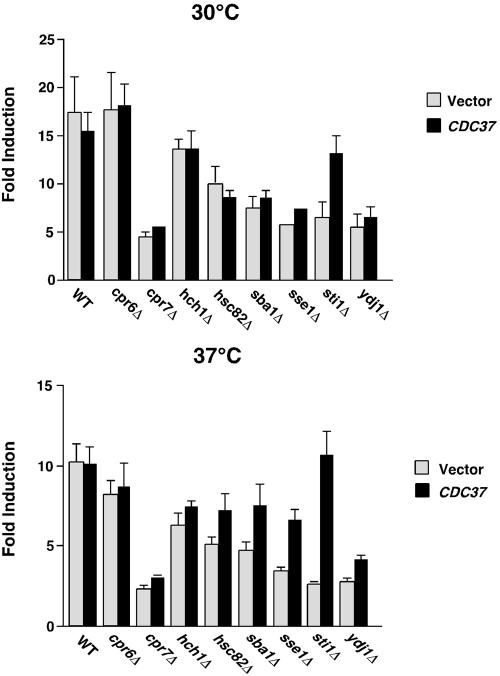
Our previous studies showed that CDC37 overexpression suppressed the defect for v-Src folding in a sti1Δ mutant. Sti1 is thought to function in the transfer of a client from Hsp70 to Hsp90 by bridging these two chaperones, thus bringing them into close proximity. Our hypothesis was that Cdc37 bypassed Hsp90 recruitment to the client, perhaps by functioning in an Hsp90-independent manner (Lee et al., 2002). To gain more insight into Cdc37 function, we tested whether overexpressed CDC37 could suppress the defects in MAP kinase signaling in other cochaperone mutant strains. At 30°C, however, only the defect in sti1Δ was suppressed (Figure 2), even though similar amounts of overexpressed Cdc37 protein were observed in all the strains tested (our unpublished observations). When assayed at 37°C, the defect in the sti1Δ mutant was more acute, yet CDC37 overexpression still suppressed the defect to wild-type levels. Partial suppression of the MAP kinase defect at 37°C was noted in several other mutants, with the greatest effect after sti1Δ being in the sse1Δ mutant (approximately twofold suppression). Notably, there was no suppression of the cpr7Δ mutant phenotype, indicating that Cdc37 cannot bypass the function of Cpr7 for MAP kinase signaling. Collectively, these data indicate that Cdc37 and Sti1 share a functional relationship that is distinct from the relationship that Cdc37 has with other cochaperones.
Figure 2.
Suppression of MAP kinase signaling defects by CDC37 overexpression. Wild-type and mutant yeast cells (as indicated) were treated with pheromone for 3 h before assaying induction of a β-galactosidase reporter gene. Cells were transformed by plasmid overexpressing CDC37 (black bars) or by the vector alone (gray bars). Data are presented as fold induction of β-galactosidase relative to uninduced cells (treated without pheromone). The experiments were done with cells incubated at 30°C (top panel) or 37°C (bottom panel). N = 3 ± SE.
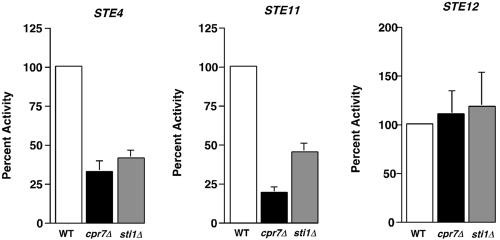
Further studies were performed to determine whether deletion of cochaperones affected processes other than signaling. This was accomplished by stimulating the pheromone responsive signaling pathway at different stages in a α-factor independent manner. The pathway was induced by overexpression of the G-protein β-subunit, STE4, a dominantly active form of STE11 (STE11ΔN) and overexpression of the transcription factor STE12. In each case, the lacZ reporter gene was induced in a pheromone-independent manner in wild-type cells (Figure 3). To analyze the effect of chaperones, we analyzed two mutants, cpr7Δ and sti1Δ, each of which were defective for MAP kinase signaling. In both strains, however, the induction was reduced compared with the wild-type for STE4 and STE11ΔN inducers, but not for STE12. These data indicate that deletion of CPR7 or STI1 affects signaling at the level of, or downstream of, Ste4 and Ste11 but upstream of the transcription factor Ste12. Chaperone action therefore appears to be restricted to the signaling pathway.
Figure 3.
Pheromone-independent activation of MAP kinase signaling pathway in wild-type and mutant cells. Wild-type (WT), cpr7Δ or sti1Δ cells overexpressing STE4, STE11ΔN (STE11), or STE12 were grown to midlog phase and assayed for β-galactosidase activity. Data are presented as percentage of β-galactosidase compared with the wild-type control. N = 3 ± SE.
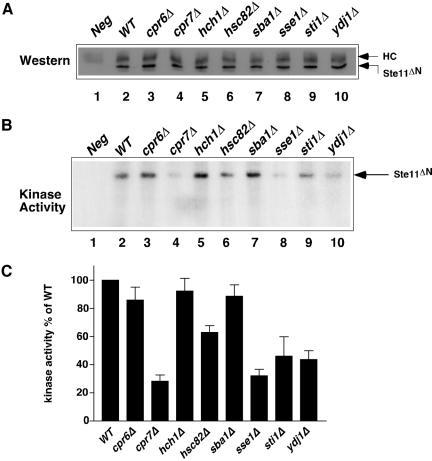
The studies to this point reflect how chaperone loss of function affected signaling via the MAP kinase pathway as a whole. Our next approach was to test directly whether the pattern of defects observed above was reflected in loss of protein kinase activity. To this end we expressed a double-tagged version of Ste11ΔN that contains the kinase domain of the MAPKKK. The kinase was isolated from cell extracts by a two-step procedure involving Ni-NTA agarose and subsequent immunopurification with anti-V5. We then assayed the levels of Ste11ΔN self-phosphorylation by incorporation of 32P-phosphate (Neiman and Herskowitz, 1994). As shown in Figure 4A, similar amounts of Ste11ΔN were isolated from wild-type and all the cochaperone deletion strains, yet their capacity for self-phosphorylation was markedly different (Figure 4, B and C). Significantly, the capacity for self-phosphorylation was reduced the most in those strains that also had the least ability for MAP kinase signaling such as cpr7Δ, and sse1Δ. There was little defect observed in the hch1Δ and sba1Δ mutants, even though there was reduced pheromone responsiveness in these strains. We attribute this to the decreased sensitivity of the kinase assay compared with the real-time PCR measurements of pheromone responsiveness.
Figure 4.
Kinase activity of Ste11ΔN. (A) Ste11ΔN was isolated from extracts derived from wild-type and mutant strains by Ni-NTA resin and immunoprecipitated with anti-V5. The relative amount of Ste11ΔN was determined by Western blot. Heavy chain, HC. (B) Kinase activity of immunoprecipitated Ste11ΔN in same strains as A, determined by autophosphorylation using 32PATP followed by SDS-PAGE and exposure of the dried gel to x-ray film. (C) Quantitation of Ste11ΔN kinase acitivity after phosphoimaging. Data are from three independent experiments, ±SEM.
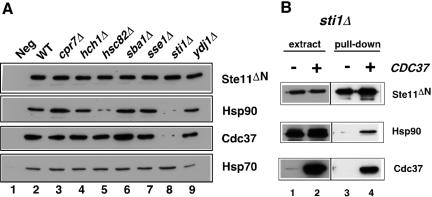
We further investigated the contribution of individual cochaperones to the ability of Hsp90 to stably complex with Ste11ΔN. These studies involved isolation of Ste11ΔN using Ni-NTA resin from wild-type and cochaperone deletion strains as shown in Figure 5. Similar amounts of Ste11ΔN protein were recovered from each strain. The affinity isolated Ste11ΔN was resolved by SDS-PAGE and probed for the presence of copurifiying Hsp90, Hsp70, and Cdc37 by Western blot. The results of these experiments showed that Hsp90 and Cdc37 binding to Ste11ΔN were strongly affected by loss of Sti1 function. Deletion of HSC82 also affected the binding of Hsp90, but had relatively little effect on the levels of Cdc37 that were bound to Ste11ΔN. There was no significant change in Hsp90 or Cdc37 binding in any of the other mutants, suggesting that their functions are independent of Hsp90 and Cdc37 assembly with the protein kinase. Hsp70 binding to Ste11ΔN was generally unaffected by deletion of the cochaperones except in the ydj1Δ mutant, where a reduction was observed. We further investigated whether CDC37 overexpression affected Hsp90 binding to Ste11ΔN in the sti1Δ mutant using the same approach. As shown in Figure 5B, Ste11ΔN was able to stably complex with Hsp90 in sti1Δ cells that overexpressed CDC37. These data suggest that Sti1 normally stabilizes Hsp90 and Cdc37 with protein kinase clients. The ability of CDC37 overexpression to suppress loss of Sti1 function is therefore correlated with the ability of Cdc37 protein to also stabilize client:Hsp90 interactions. On the other hand, it is also clear that stabilization of such client:Hsp90 interactions is insufficient for proper folding. The most significant defects in Ste11ΔN kinase activity occurred in mutants such as cpr7Δ and sse1Δ, where normal levels of Hsp90 and Cdc37 were found to complex with the client (compare Figure 4 with Figure 5).
Figure 5.
Hsp90 and Cdc37 binding to Ste11. (A) Ni-NTA resin pull-down experiment of His-tagged Ste11ΔN from the strains indicated, followed by Western blot analysis for Ste11ΔN (with anti-His6), Hsp90, Cdc37, and Hsp70 as indicated in the figure. Extracts from cells not expressing Ste11ΔN were used in lane 1. (B) Pull-down experiment of Ste11ΔN from sti1Δ cell extracts with or without overrexpressed CDC37. Western blots of extracts from these strains (lanes 1 and 2) are compared with Western blots of the eluates (lanes 3 and 4) from the pull-down, which were probed with anti-His6 (for Ste11ΔN), anti-Hsp90, and anti-Cdc37.
Suppression of cpr7Δ by CNS1 for MAP Kinase Signaling
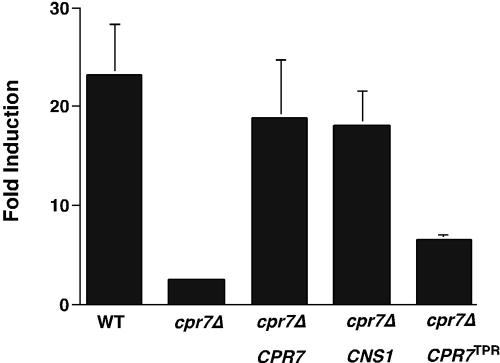
The results above in Figures 1, 2, and 4 showed that deletion of CPR7 strongly affected MAP kinase signaling but in a manner than cannot be suppressed by CDC37 overexpression. This indicates that Cpr7 has a separate function from Cdc37, although both chaperones are clearly part of the same Hsp90 machinery and indeed have been shown to interact with each other (Abbas-Terki et al., 2002). Previous studies established that the growth phenotype of cpr7Δ could be suppressed by CNS1 overexpression, as could a defect in GR activation (Dolinski et al., 1998; Marsh et al., 1998). On the other hand, it was recently found that temperature-sensitive cns1 mutants that also displayed a defect in GR signaling were wild-type for MAP kinase signaling (Tesic et al., 2003). On this basis we investigated whether the defect in MAP kinase signaling in the cpr7Δ mutant could be suppressed by CNS1 overexpression. As shown in Figure 6, CNS1 completely suppressed the phenotype of the cpr7Δ mutant, consistent with these two proteins functioning together in the same Hsp90 subcomplex. Furthermore, the isolated TPR domain of Cpr7 also displayed partial suppression of the cpr7Δ phenotype, although this was not more than twofold above the levels found in the cpr7Δ strain by itself. Together these data show that Cns1 and Cpr7 share a functional relationship that is distinct from Cdc37 and Sti1.
Figure 6.
MAP kinase signaling in cpr7Δ mutants. cpr7Δ cells expressing CPR7, CNS1, or a CPR7 truncation with just the TPR domains (CPR7TPR) were induced with α-factor and β-galactosidase levels measured 3 h later. Data are presented as fold-induction vs. uninduced controls. N = 3 ± SE.
DISCUSSION
Hsp90 functions with many different cochaperones, and the results shown here represent a comparative analysis of their overall contribution to the folding of a protein kinase. We chose to use deletion mutants so that the measurements reflected complete loss of function, at least for genes that have no paralogs, such as STI1 and SBA1. This was not always possible, however, because several cochaperones exist as gene pairs or part of a larger gene family. SSE1 and HCH1, for example, are two genes that have paralogs in the form of SSE2 and AHA1 (Mukai et al., 1993; Panaretou et al., 2002; Lotz et al., 2003). The same is true of YDJ1, which has functional overlap with SIS1, and both genes belong to the larger hsp40 family (Caplan and Douglas, 1991; Cheetham and Caplan, 1998). On the other hand, the expectation that paralogs are homologous is not always predictable. CPR6 and CPR7 have significant sequence similarity with each other but completely different functions (Figure 1). Also, defects associated with deletion of CPR7 cannot be compensated for by overexpressing CPR6 (Dolinski et al., 1998; Marsh et al., 1998). Two essential cochaperones that have no paralogs, Cdc37 and Cns1, were studied by their ability to suppress defects associated with sti1Δ and cpr7Δ strains, respectively, and will be discussed in more detail below.
What is clear from the comparative analysis is that some cochaperones are more important than others for MAP kinase signaling. Deletion of CPR7, SSE1, and YDJ1 resulted in stronger defects in MAP kinase signaling than deletion of CPR6, STI1, or SBA1. Furthermore, comparison of the results presented here with those previously reported using heterologous clients reveals some common trends. Deletion of CPR6 or SBA1, for example, results in a only a slight defect in v-Src or nuclear receptor signaling, whereas deletion of CPR7 or YDJ1 resulted in strong signaling defects (Kimura et al., 1995; Dey et al., 1996; Duina et al., 1996; Warth et al., 1997; Fang et al., 1998). By correlation, cochaperones that are essential for viability are also likely to play essential roles in the folding pathway; examples here are Cdc37 and Cns1.
Our results demonstrate a very close correlation between loss of Ste11 protein kinase activity and signaling via the MAP kinase pathway in terms of FIG1 and HHF1 gene expression. This suggests that loss of Ste11 activity as a result of cochaperone gene deletion represents the main defect in the signaling pathway in the different mutant strains tested. Furthermore, we failed to detect interaction between Hsp90 and two other protein kinases in the pathway, Ste7 and Fus3 (our unpublished results). However, we cannot completely rule out a role for Hsp90 in folding Ste7 or Fus3 on this basis alone, because the levels of Ste7 were found to be reduced in an hsp82 mutant strain (Louvion et al., 1998). On the other hand, not all kinases require Hsp90 for activity, and in animal cells some kinases downstream of Raf1 are insensitive to the action of geldanamycin, the Hsp90 inhibitor (Miyata et al., 2001; Boudeau et al., 2003).
The results shown above demonstrate the use of a genetic approach to identify functional relationships among Hsp90 cochaperones. Specifically, Cdc37 and Sti1 appear to have some functional overlap because overexpression of CDC37 suppressed sti1Δ for defects in Ste11 and v-Src activity (Lee et al., 2002). The mechanism of this suppression was revealed by analysis of Hsp90 binding to Ste11ΔN in sti1Δ strains that overexpressed CDC37. In the absence of STI1, there was little stable binding of Hsp90 to the kinase, but in the same strain overexpressing CDC37, Hsp90 binding was recovered (Figure 5B). These data support the hypothesis that client loading onto Hsp90 depends on cochaperones such as Sti1 and Cdc37, which also inhibit Hsp90's ATPase (Prodromou et al., 1999; Siligardi et al., 2002; Prodromou and Pearl, 2003). It is interesting to note that yeast Cdc37 is a much less potent inhibitor of Hsp90's ATPase than is Sti1, at least in vitro (Prodromou et al., 1999; Siligardi et al., 2002). If this proves so in vivo, then Sti1 will provide the primary function of preparing Hsp90 for client loading. This is supported by our findings that deletion of STI1 prevents stable complex formation between the client and Hsp90. Because Cdc37 can also interact with the client, Hsp90 and Sti1 (Abbas-Terki et al., 2002), it is likely that multiple interactions lead to stable formation of the subsequent chaperone:client complexes. On the other hand, it still remains unclear how such stability contributes to the overall efficiency of the folding reaction. In previous studies we noted that CDC37 overexpression suppressed v-Src loss of function in the sti1Δ mutant. Similar to the results found here, we also observed that Hsp90 binding to v-Src was severely decreased in the sti1Δ mutant. In this case, however, CDC37 overexpression failed to restore stable Hsp90 binding to v-Src in the sti1Δ mutant (Lee et al., 2002). Bypass suppression of v-Src activity by CDC37 overexpression, therefore, did not correlate with formation of stable Hsp90:client complexes and may reflect Hsp90-independent activity of Cdc37 (Lee et al., 2002). On the other hand, our results do show that stable binding of Cdc37 or Hsp90 with a client kinase is insufficient for proper folding when other factors are missing. Deletion of genes that caused the greatest defect in Ste11ΔN activity (CPR7, SSE1, and YDJ1) did not appreciably affect Cdc37 or Hsp90 binding to the client.
The results of this study also show that Cpr7 functions in association with Cns1, confirming previous analyses with these two proteins (Tesic et al., 2003). In addition, deletion of CPR7 does not affect the ability of Hsp90 or Cdc37 to interact with Ste11ΔN. Although it is difficult to speculate on the mechanism underlying Cpr7/Cns1 function, it appears to be independent of Cpr7's peptidyl prolyl isomerase activity (Figure 6). Previous studies have also established that Cpr7 has chaperone activity (Mayr et al., 2000), so it is possible that Cpr7 and Cns1 function as an additional and important chaperone in the folding of Ste11. Our studies of Ydj1, which also has chaperone activity (Cyr, 1995), similarly show that it is required in a function that is beyond its ability to facilitate complex assembly between the client and Hsp90/Cdc37. If this were so we might have expected as little Hsp90 binding to Ste11ΔN as was found in the sti1Δ mutant strain, which was clearly not the case (Figure 5A). It therefore seems likely that protein kinase folding requires several chaperone activities in addition to Hsp90 and Cdc37. Further studies will be needed to determine how the action of these chaperones is coordinated with each other during the folding process.
Acknowledgments
We thank Drs. J. Hirsch and K. Morano for plasmids and J. Hirsch for critical comments on the manuscript.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0480. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0480.
References
- Abbas-Terki, T., Briand, P.A., Donze, O., and Picard, D. (2002). The Hsp90 co-chaperones Cdc37 and Sti1 interact physically and genetically. Biol. Chem. 383, 1335-1342. [DOI] [PubMed] [Google Scholar]
- Abbas-Terki, T., Donze, O., and Picard, D. (2000). The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS Lett. 467, 111-116. [DOI] [PubMed] [Google Scholar]
- Bohen, S.P. (1998). Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol. Cell. Biol. 18, 3330-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen, S.P., Kralli, A., and Yamamoto, K.R. (1995). Hold 'em and fold 'em: chaperones and signal transduction [comment]. Science 268, 1303-1304. [DOI] [PubMed] [Google Scholar]
- Borkovich, K.A., Farrelly, F.W., Finkelstein, D.B., Taulien, J., and Lindquist, S. (1989). hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9, 3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau, J., Deak, M., Lawlor, M.A., Morrice, N.A., and Alessi, D.R. (2003). Heat-shock protein 90 and Cdc37 interact with LKB1 and regulate its stability. Biochem. J. 370, 849-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, A.J. (1999). Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 9, 262-268. [DOI] [PubMed] [Google Scholar]
- Caplan, A.J., and Douglas, M.G. (1991). Characterization of YDJ 1, a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114, 609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.C., Nathan, D.F., and Lindquist, S. (1997). In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17, 318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham, M.E., and Caplan, A.J. (1998). Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3, 28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutforth, T., and Rubin, G.M. (1994). Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell 77, 1027-1036. [DOI] [PubMed] [Google Scholar]
- Cyr, D.M. (1995). Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 359, 129-132. [DOI] [PubMed] [Google Scholar]
- Dey, B., Caplan, A.J., and Boschelli, F. (1996). The Ydj1 molecular chaperone facilitates formation of active p60 v-src in yeast. Mol. Biol. Cell 7, 91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski, K.J., Cardenas, M.E., and Heitman, J. (1998). CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol. Cell. Biol. 18, 7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina, A.A., Chang, H.C., Marsh, J.A., Lindquist, S., and Gaber, R.F. (1996). A cyclophilin function in Hsp90-dependent signal transduction [see comments]. Science 274, 1713-1715. [DOI] [PubMed] [Google Scholar]
- Elion, E.A. (2000). Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3, 573-581. [DOI] [PubMed] [Google Scholar]
- Fang, Y., Fliss, A.E., Rao, J., and Caplan, A.J. (1998). SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18, 3727-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss, A.E., Fang, Y., Boschelli, F., and Caplan, A.J. (1997). Differential in vivo regulation of steroid hormone receptor activation by Cdc37p. Mol. Biol. Cell 8, 2501-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis, N., Lin, J.H., Grammatikakis, A., Tsichlis, P.N., and Cochran, B.H. (1999). p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell. Biol. 19, 1661-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartson, S.D., Irwin, A.D., Shao, J., Scroggins, B.T., Volk, L., Huang, W., and Matts, R.L. (2000). p50 Cdc 37 is a nonexclusive hsp90 cohort which participates intimately in Hsp90-mediated folding of immature kinase molecules. Biochemistry 39, 7631-7644. [DOI] [PubMed] [Google Scholar]
- Hunter, T., and Poon, R.Y.C. (1997). Cdc 37, a protein kinase chaperone? Trends Cell Biol. 7, 157-161. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., Rutherford, S.L., Miyata, Y., Yahara, I., Freeman, B.C., Yue, L., Morimoto, R.I., and Lindquist, S. (1997). Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 11, 1775-1785. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., Yahara, I., and Lindquist, S. (1995). Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways [see comments]. Science 268, 1362-1365. [DOI] [PubMed] [Google Scholar]
- Lamphere, L., Fiore, F., Xu, X., Brizuela, L., Keezer, S., Sardet, C., Draetta, G.F., and Gyuris, J. (1997). Interaction between Cdc37 and Cdk4 in human cells. Oncogene 14, 1999-2004. [DOI] [PubMed] [Google Scholar]
- Lee, P., Rao, J., Fliss, A., Yang, E., Garrett, S., and Caplan, A.J. (2002). The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 159, 1051-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.D., Morano, K.A., and Thiele, D.J. (1999). The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274, 26654-26660. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402-408. [DOI] [PubMed] [Google Scholar]
- Lotz, G.P., Lin, H., Harst, A., and Obermann, W.M. (2003). Aha1 binds to the middle domain of hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 278, 17228-17235. [DOI] [PubMed] [Google Scholar]
- Louvion, J.F., Abbas-Terki, T., and Picard, D. (1998). Hsp90 is required for pheromone signaling in yeast. Mol. Biol. Cell 9, 3071-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, J.A., Kalton, H.M., and Gaber, R.F. (1998). Cns1 is an essential protein associated with the hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Delta cells. Mol. Cell. Biol. 18, 7353-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, C., Richter, K., Lilie, H., and Buchner, J. (2000). Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J. Biol. Chem. 275, 34140-34146. [DOI] [PubMed] [Google Scholar]
- Miyata, Y., Ikawa, Y., Shibuya, M., and Nishida, E. (2001). Specific association of a set of molecular chaperones including HSP90 and Cdc37 with MOK, a member of the mitogen-activated protein kinase superfamily. J. Biol. Chem. 276, 21841-21848. [DOI] [PubMed] [Google Scholar]
- Morishima, Y., Kanelakis, K.C., Silverstein, A.M., Dittmar, K.D., Estrada, L., and Pratt, W.B. (2000). The Hsp organizer protein hop enhances the rate of but is not essential for glucocorticoid receptor folding by the multiprotein Hsp90-based chaperone system. J. Biol. Chem. 275, 6894-6900. [DOI] [PubMed] [Google Scholar]
- Mukai, H., Kuno, T., Tanaka, H., Hirata, D., Miyakawa, T., and Tanaka, C. (1993). Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132, 57-66. [DOI] [PubMed] [Google Scholar]
- Neiman, A.M., and Herskowitz, I. (1994). Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA 91, 3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet, C.M., and Craig, E.A. (1989). Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou, B. et al. (2002). Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell 10, 1307-1318. [DOI] [PubMed] [Google Scholar]
- Pratt, W.B., and Toft, D.O. (1997). Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18, 306-360. [DOI] [PubMed] [Google Scholar]
- Pratt, W.B., and Toft, D.O. (2003). Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228, 111-133. [DOI] [PubMed] [Google Scholar]
- Prodromou, C., and Pearl, L.H. (2003). Structure and functional relationships of Hsp90. Curr. Cancer Drug Targets 3, 301-323. [DOI] [PubMed] [Google Scholar]
- Prodromou, C., Siligardi, G., O'Brien, R., Woolfson, D.N., Regan, L., Panaretou, B., Ladbury, J.E., Piper, P.W., and Pearl, L.H. (1999). Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18, 754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapandi, T., Greene, L., and Eisenberg, E. (2000). The molecular chaperones Hsp90 and Hsc70 are both necessary and sufficient to activate hormone binding by glucocorticoid receptor. J. Biol. Chem. 275, 22597-22604. [DOI] [PubMed] [Google Scholar]
- Rao, J., Lee, P., Benzeno, S., Cardozo, C., Albertus, J., Robins, D.M., and Caplan, A.J. (2001). Functional interaction of human Cdc37 with the androgen receptor but not with the glucocorticoid receptor. J. Biol. Chem. 276, 5814-5820. [DOI] [PubMed] [Google Scholar]
- Reed, S.I. (1980a). The selection of amber mutations in genes required for completion of start, the controlling event of the cell division cycle of S. cerevisiae. Genetics 95, 579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, S.I. (1980b). The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95, 561-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J. et al. (2000). Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873-880. [DOI] [PubMed] [Google Scholar]
- Scholz, G.M., Cartledge, K., and Hall, N.E. (2001). Identification and characterization of Harc, a novel Hsp90-associating relative of Cdc37. J. Biol. Chem. 276, 30971-30979. [DOI] [PubMed] [Google Scholar]
- Siligardi, G., Panaretou, B., Meyer, P., Singh, S., Woolfson, D.N., Piper, P.W., Pearl, L.H., and Prodromou, C. (2002). Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277, 20151-20159. [DOI] [PubMed] [Google Scholar]
- Silverstein, A.M., Grammatikakis, N., Cochran, B.H., Chinkers, M., and Pratt, W.B. (1998). p50(cdc37) binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J. Biol. Chem. 273, 20090-20095. [DOI] [PubMed] [Google Scholar]
- Stepanova, L., Leng, X., Parker, S.B., and Harper, J.W. (1996). Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 10, 1491-1502. [DOI] [PubMed] [Google Scholar]
- Tesic, M., Marsh, J.A., Cullinan, S.B., and Gaber, R.F. (2003). Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 278, 32692-32701. [DOI] [PubMed] [Google Scholar]
- Wang, X., Grammatikakis, N., and Hu, J. (2002). Role of P50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 277, 24361-24367. [DOI] [PubMed] [Google Scholar]
- Warth, R., Briand, P.A., and Picard, D. (1997). Functional analysis of the yeast 40 kDa cyclophilin Cyp40 and its role for viability and steroid receptor regulation. Biol. Chem. 378, 381-391. [DOI] [PubMed] [Google Scholar]